Circadian mRNA Abundance Changes in Microdissected Proximal Convoluted and Straight Tubules from Mouse Kidney
Method: Proximal convoluted tubules (PCTs [S1]) and proximal straight tubules (PSTs [S2]) were microdissected from kidneys of mice (male C57BL/6, 4-6 weeks) every 4 hours. Each time point was represented by 3 samples, each from a different mouse. RNA-Seq used the NEBNext(TM) Low Input RNA library preparation kit and a NovaSeq6000 DNA sequencer. The data were analyzed using the JTK_CYCLE algorithm to identify transcripts that vary with time of day with a 20 or 24 hour period (Hughes et al. PMID: 20876817). To see only those transcripts that significantly vary in a circadian pattern, select a sort option in the drop-down menu below.
This website was created by Molly Bingham, Chin-Rang Yang, Hyun Jun Jung, Joe Chou, Viswanathan Raghuram, Mark Knepper and Margo Dona (July 2021). Download data. Contact us with questions or comments: knep@helix.nih.gov
Sort transcripts by:
"Transcripts that significantly vary" was defined as having P<0.05 and a period of either 20 or 24 hours and are highlighted in gray.
To search, use browser's Find command (Ctrl-F)
| Ensembl ID | Gene Symbol | Annotation | PCT | |||
| P-value | Period, hr | Phase, hr | Amplitude (Log2[TPM]) | |||
| ENSMUSG00000021414 | Fam217a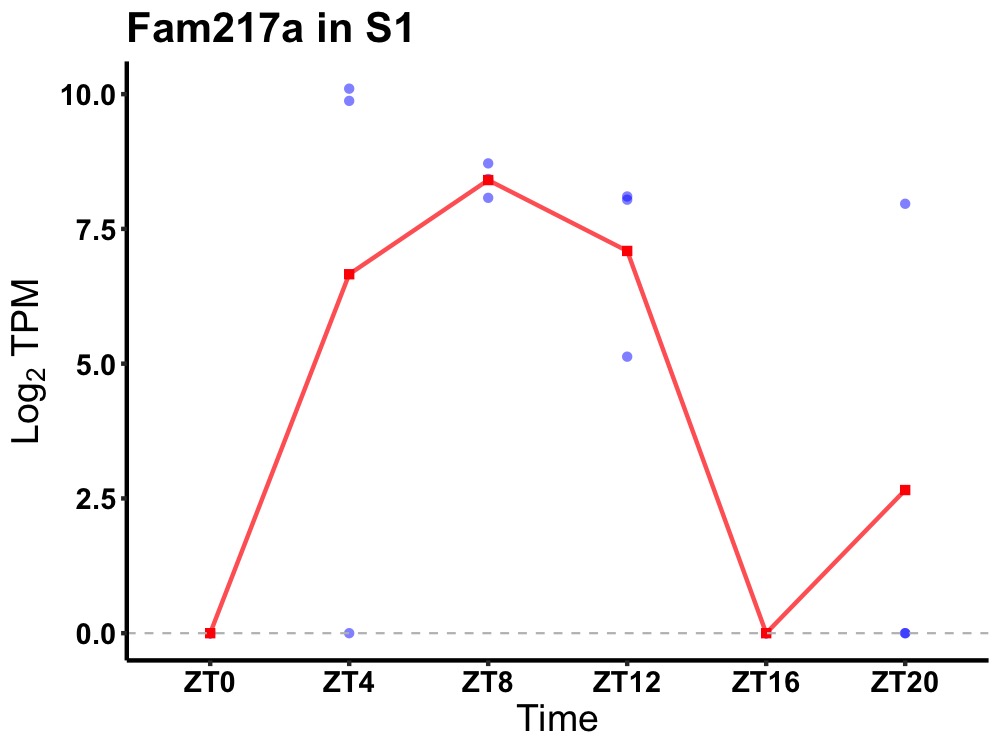 |
protein FAM217A | 0.023 | 20 | 8 | 5.96 |
| ENSMUSG00000034601 | 2700049A03Rik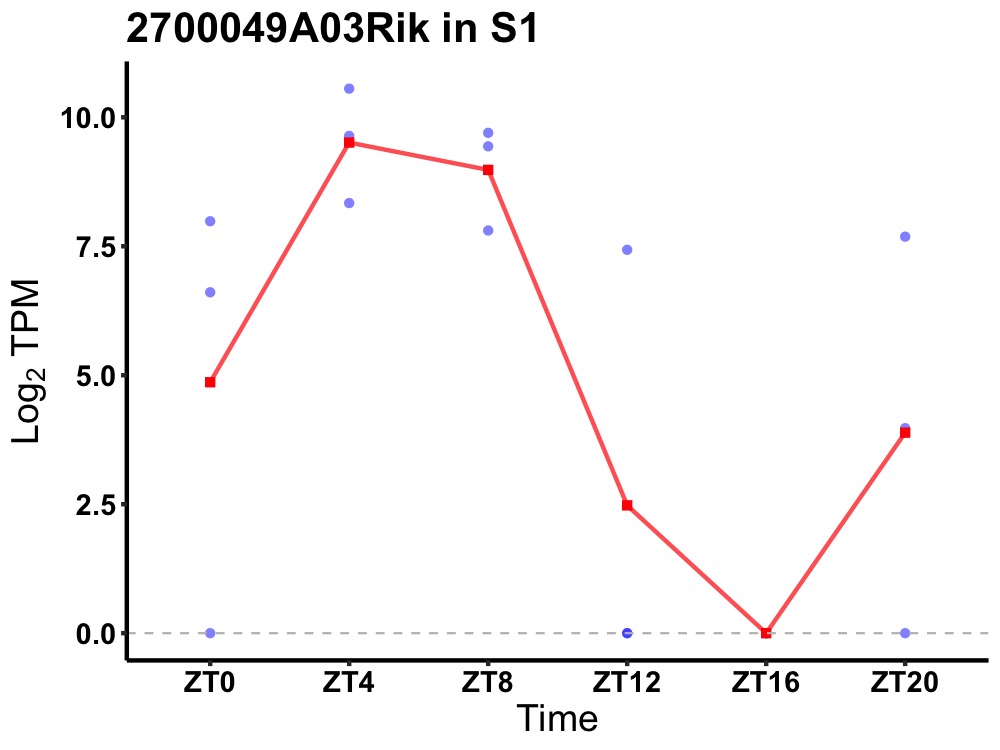 |
protein TALPID3 | 0.004 | 20 | 6 | 5.90 |
| ENSMUSG00000024298 | Xpo6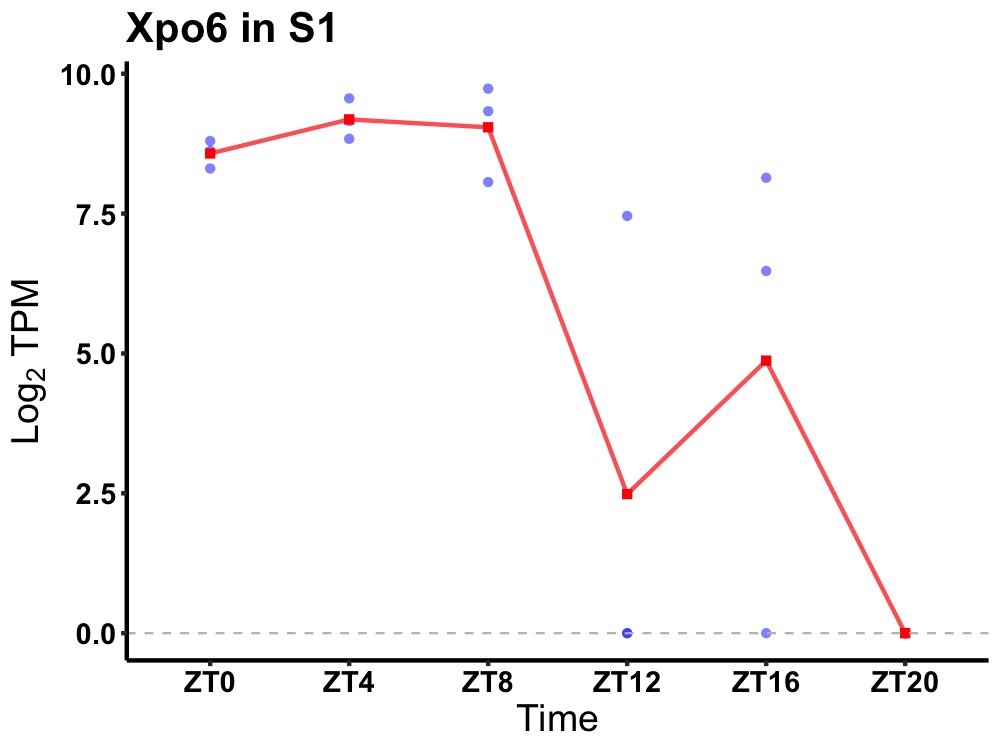 |
exportin-6 | 0.016 | 24 | 6 | 5.87 |
| ENSMUSG00000020889 | Nr1d1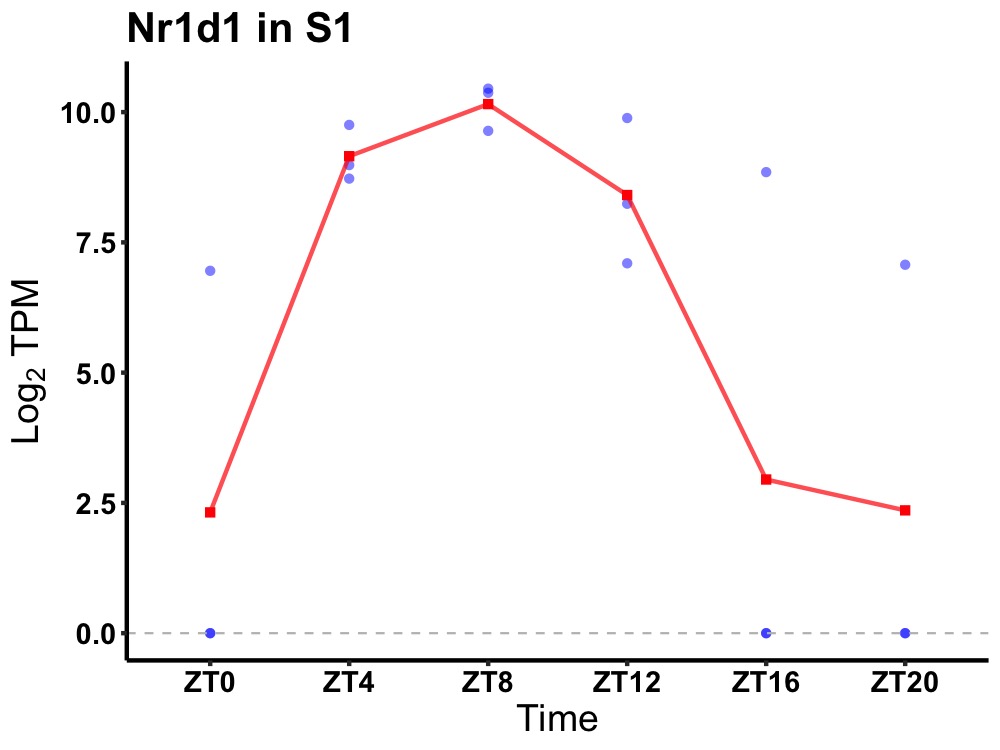 |
nuclear receptor subfamily 1 group D member 1 | 0.005 | 24 | 8 | 5.84 |
| ENSMUSG00000025316 | Banp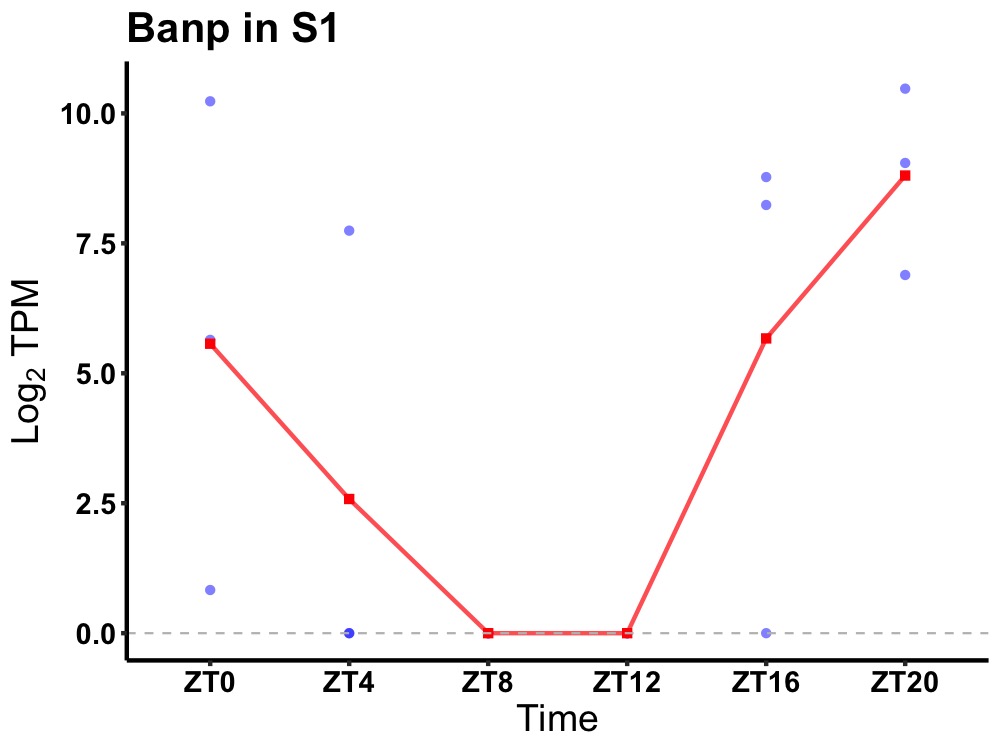 |
protein BANP | 0.023 | 24 | 22 | 5.83 |
| ENSMUSG00000011427 | Zfp790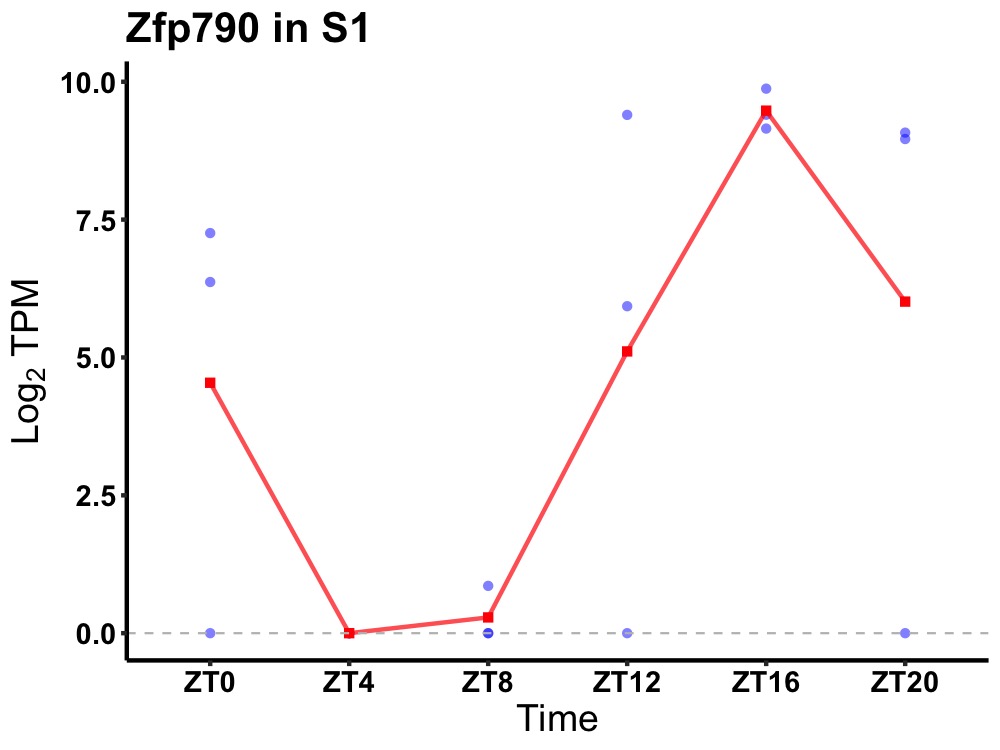 |
zinc finger protein 790 | 0.019 | 24 | 18 | 5.81 |
| ENSMUSG00000067942 | Zfp160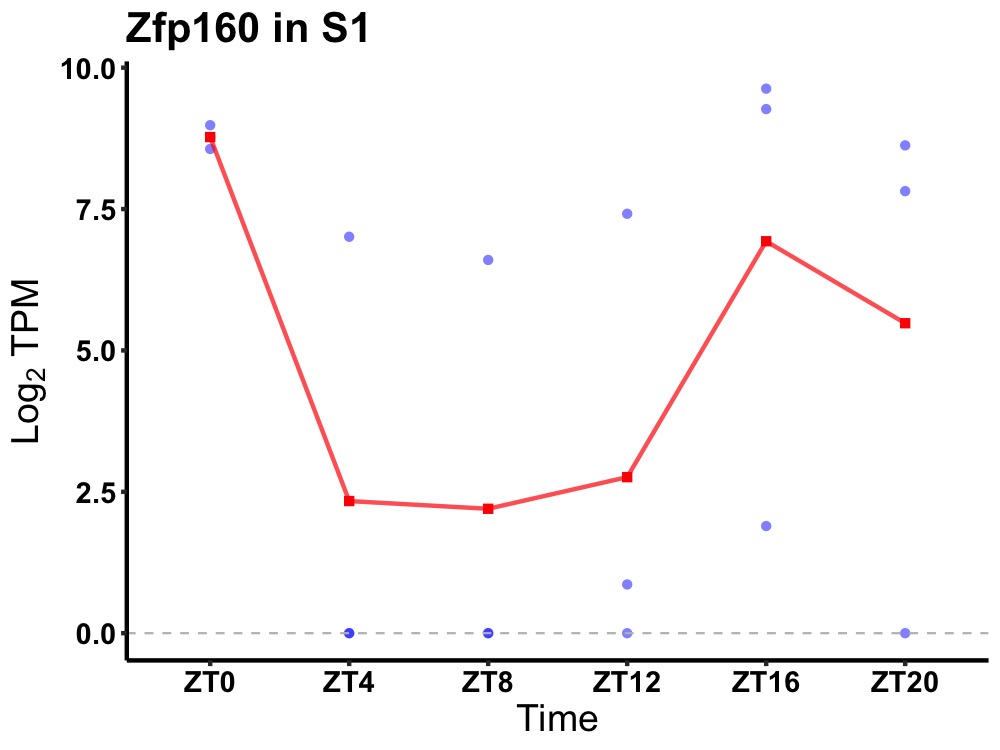 |
zinc finger protein 160 | 0.016 | 20 | 18 | 5.74 |
| ENSMUSG00000036986 | Pml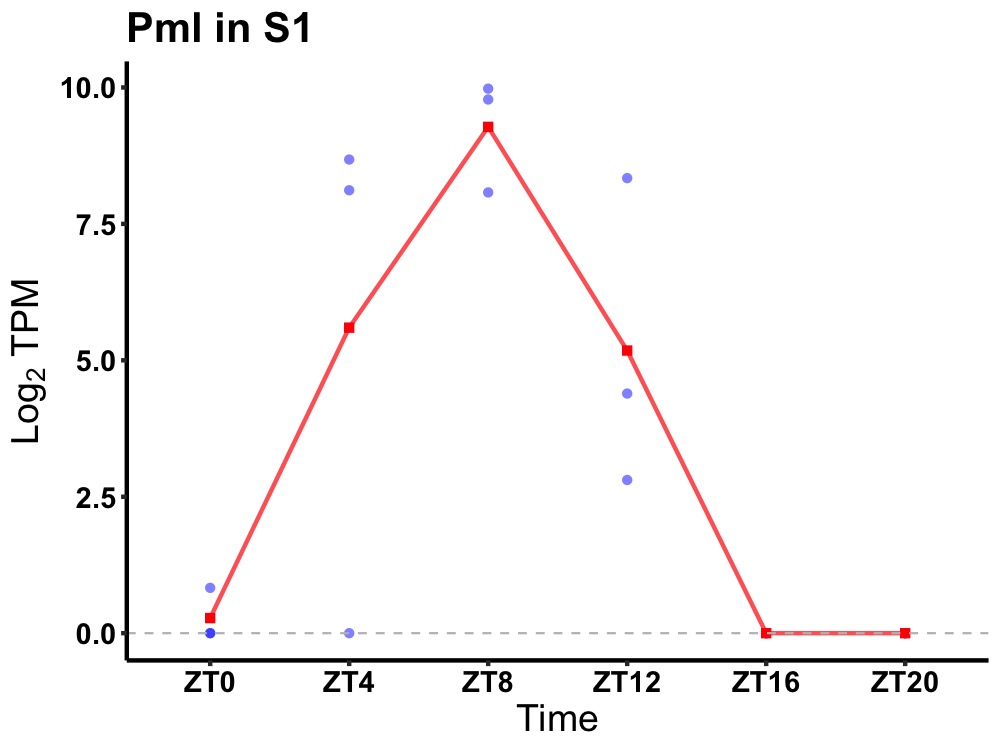 |
protein PML | 0.006 | 24 | 8 | 5.71 |
| ENSMUSG00000021733 | Slc4a7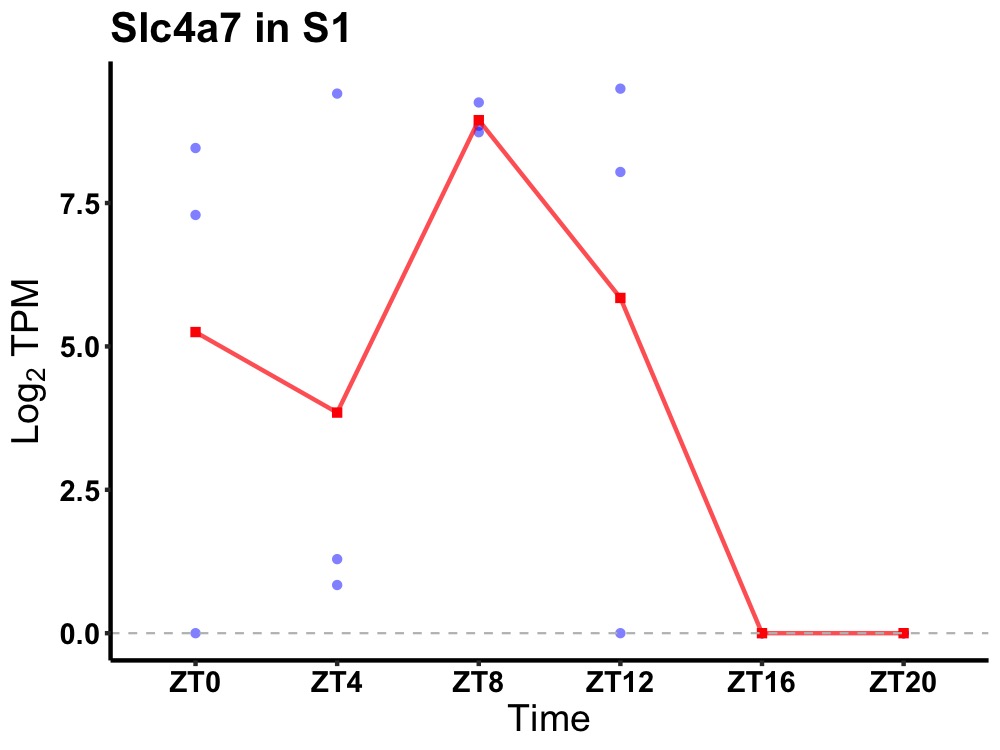 |
sodium bicarbonate cotransporter 3 | 0.049 | 24 | 8 | 5.69 |
| ENSMUSG00000020429 | Igfbp1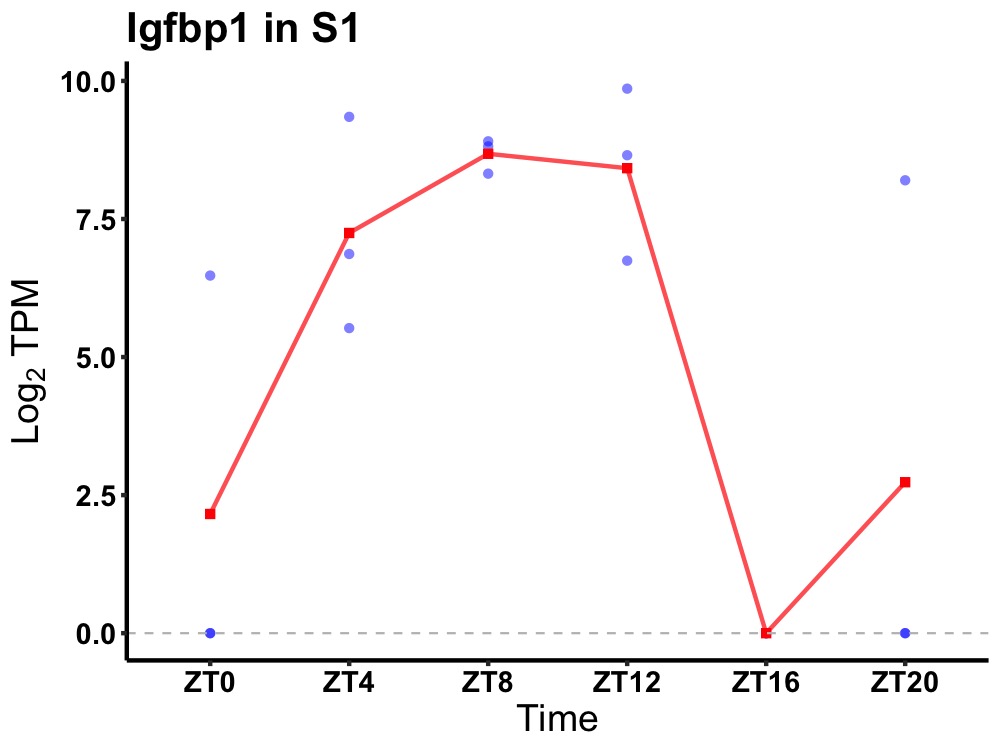 |
insulin-like growth factor-binding protein 1 | 0.027 | 20 | 8 | 5.59 |
| ENSMUSG00000020744 | Vps37c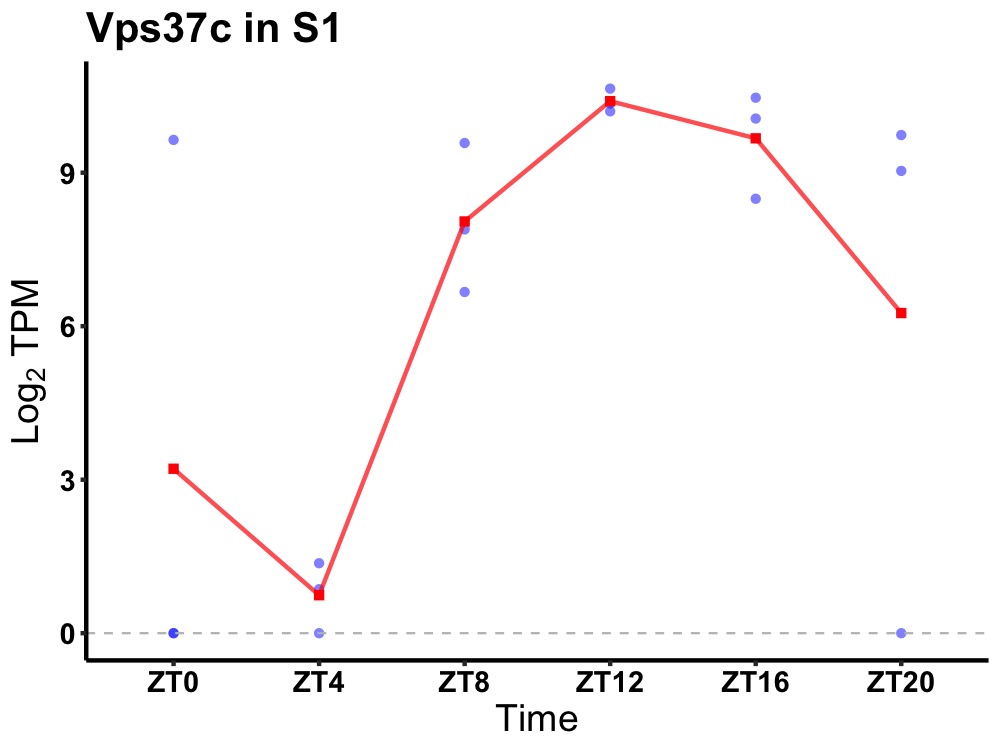 |
vacuolar protein sorting-associated protein 37C | 0.008 | 24 | 14 | 5.58 |
| ENSMUSG00000039328 | Rnf122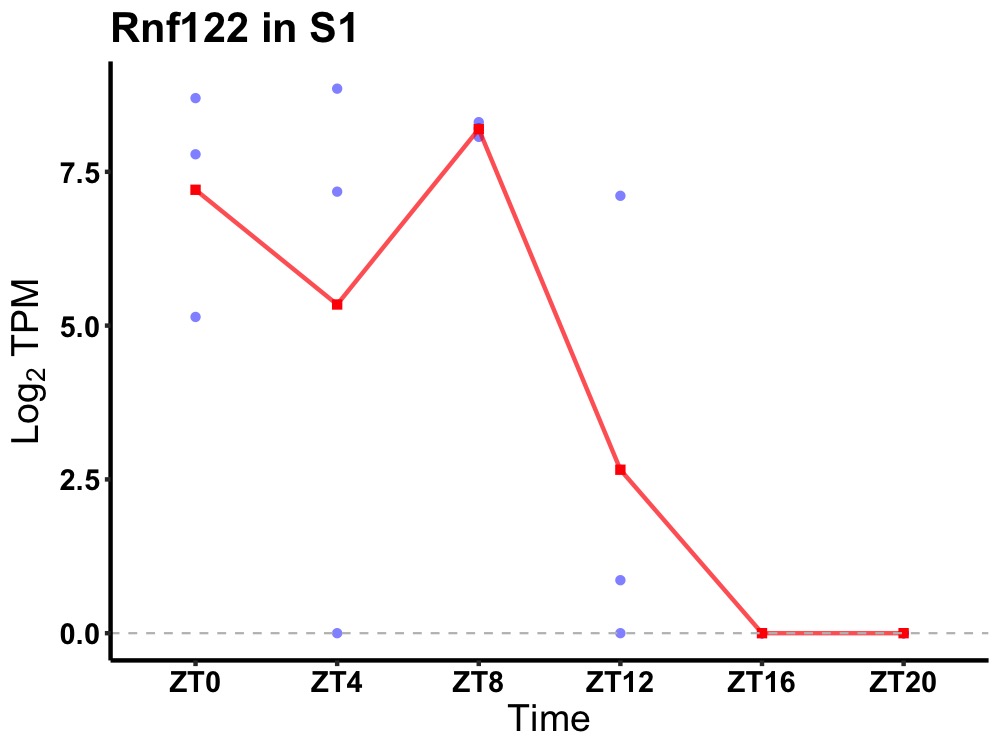 |
RING finger protein 122 | 0.036 | 24 | 6 | 5.52 |
| ENSMUSG00000028886 | Eya3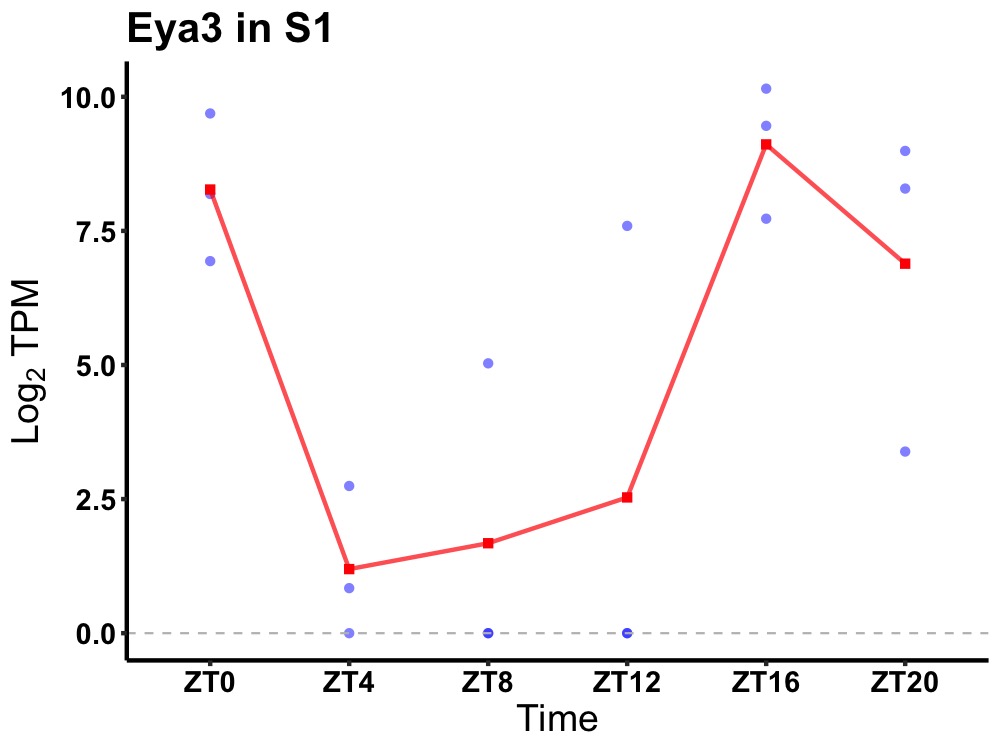 |
eyes absent homolog 3 | 0.014 | 20 | 18 | 5.46 |
| ENSMUSG00000049411 | Tmem241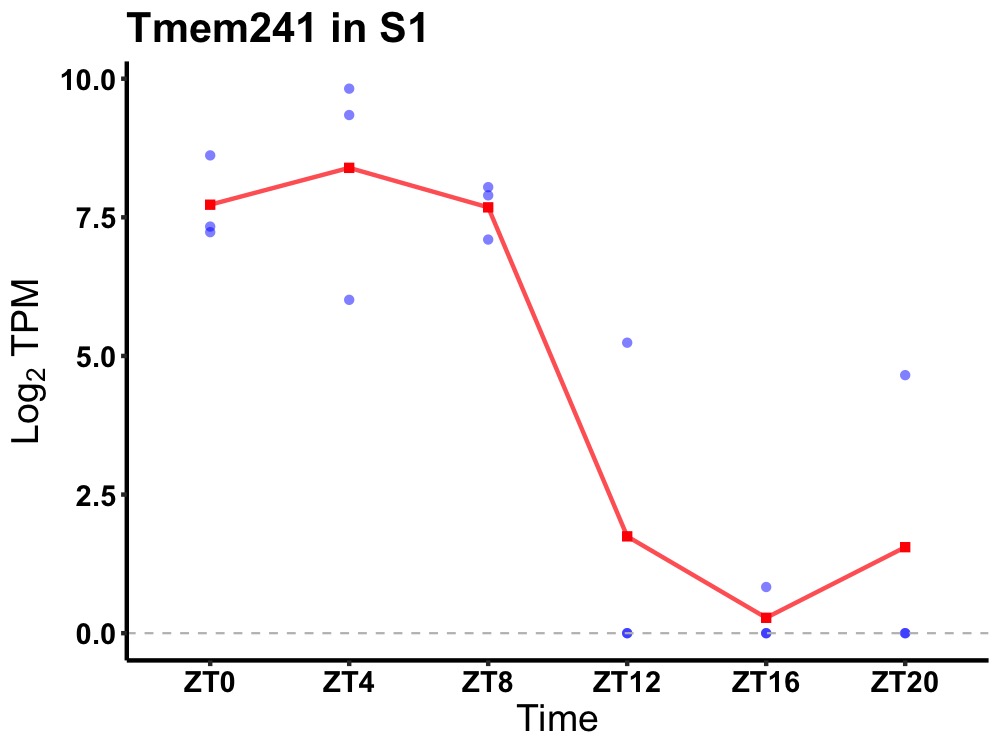 |
transmembrane protein 241 | 0.007 | 24 | 6 | 5.19 |
| ENSMUSG00000054920 | Klhl5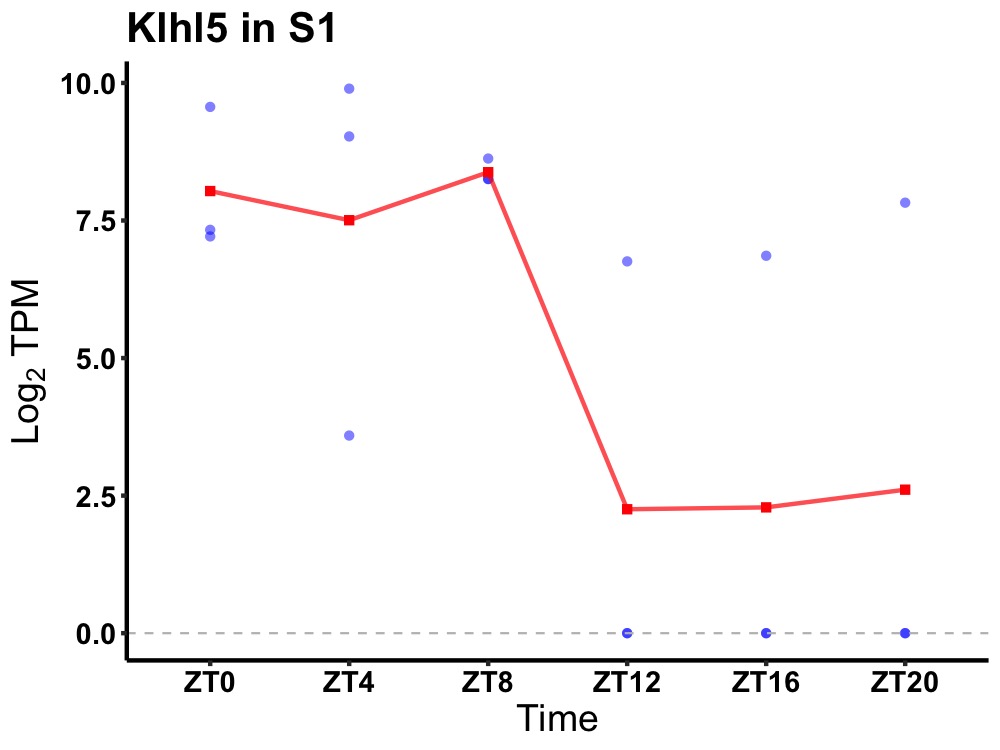 |
kelch-like protein 5 | 0.049 | 24 | 6 | 5.10 |
| ENSMUSG00000008035 | Mid1ip1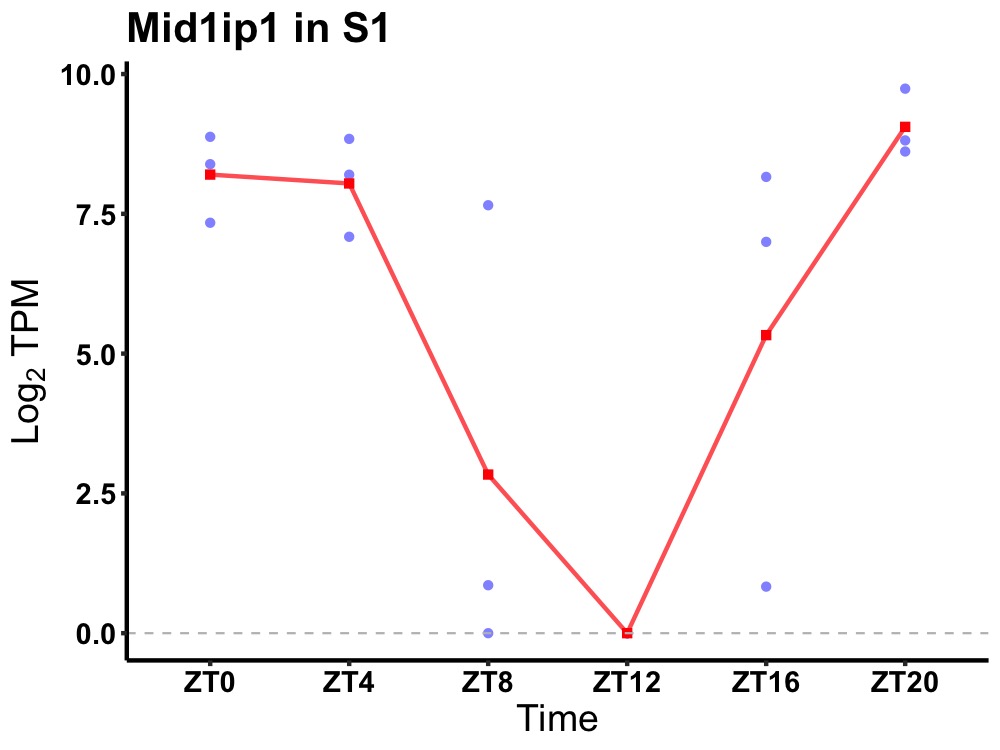 |
mid1-interacting protein 1 | 0.000 | 20 | 2 | 5.05 |
| ENSMUSG00000008200 | Fnbp4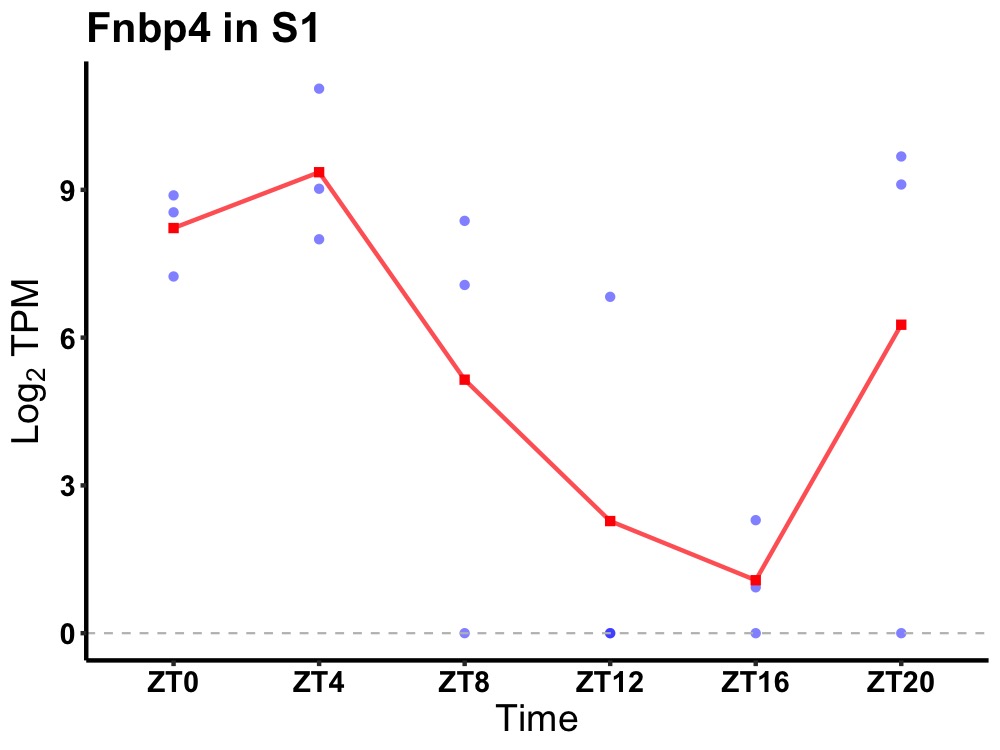 |
formin-binding protein 4 | 0.023 | 20 | 4 | 5.04 |
| ENSMUSG00000031652 | N4bp1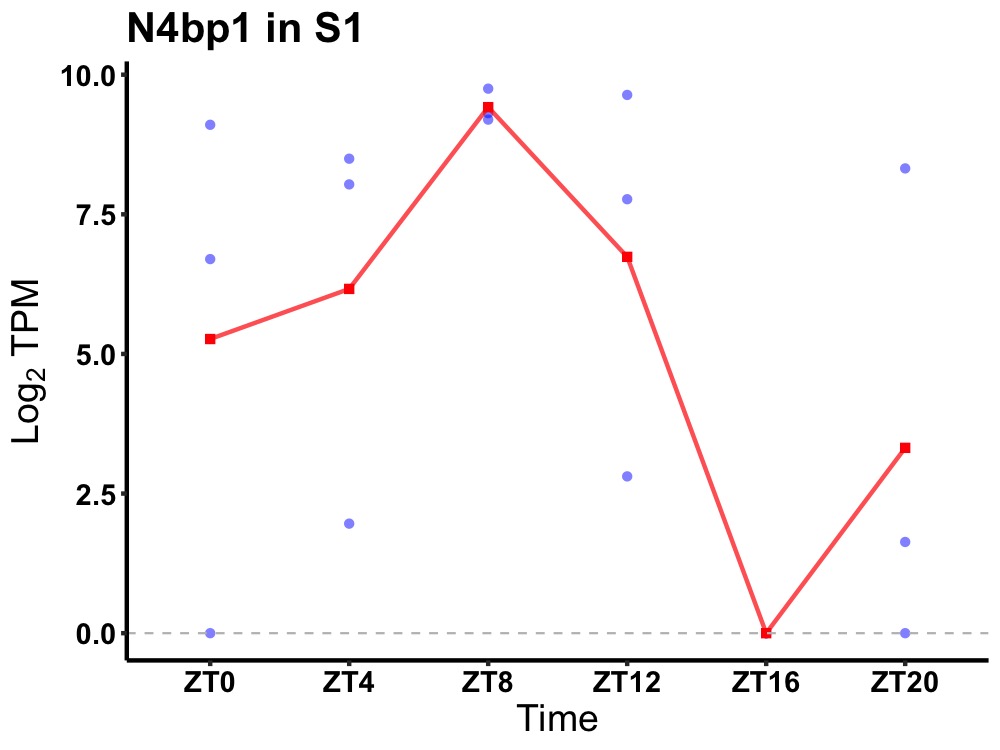 |
NEDD4-binding protein 1 | 0.023 | 20 | 8 | 5.02 |
| ENSMUSG00000004994 | Ccdc130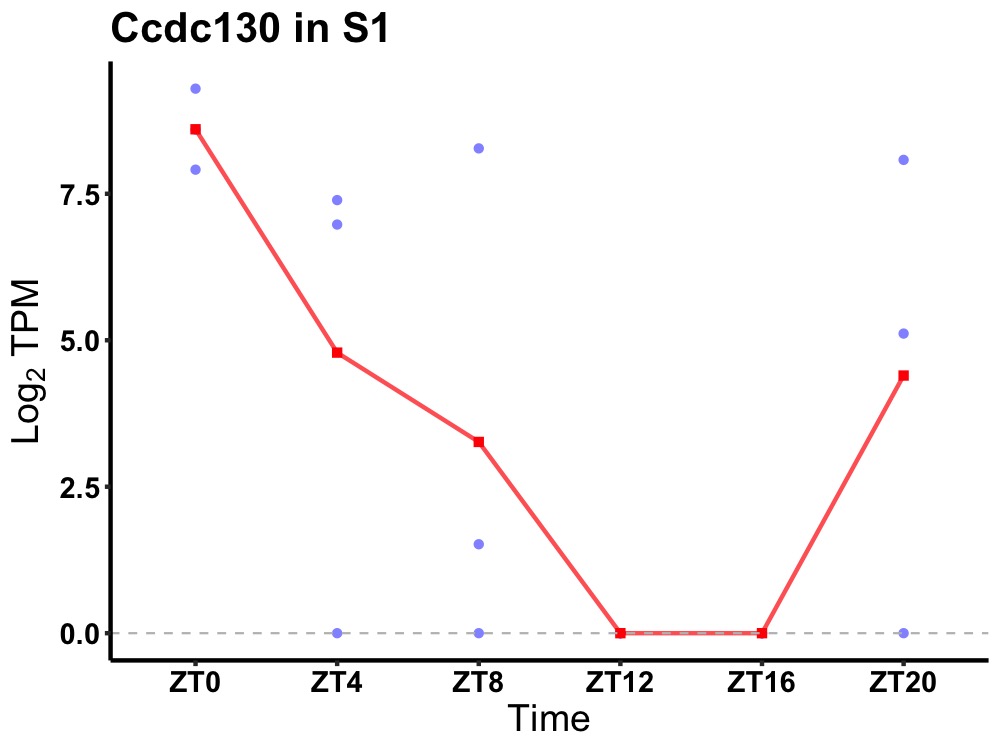 |
probable splicing factor YJU2B | 0.049 | 24 | 2 | 4.93 |
| ENSMUSG00000006456 | Rbm14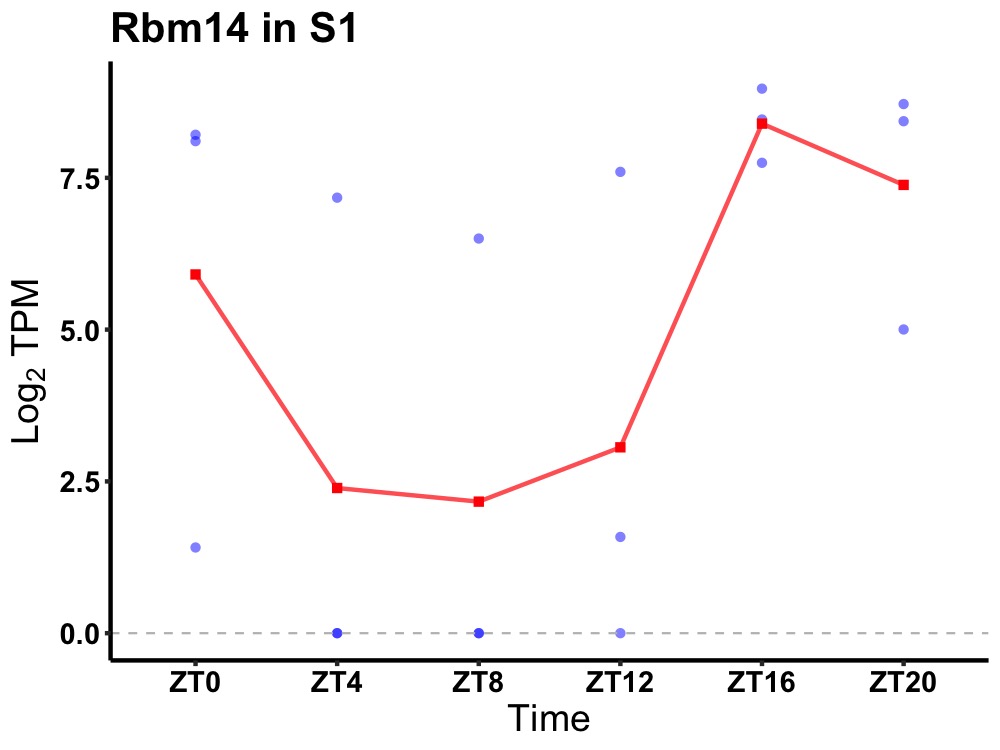 |
RNA-binding protein 14 | 0.027 | 24 | 20 | 4.84 |
| ENSMUSG00000051786 | Tubgcp6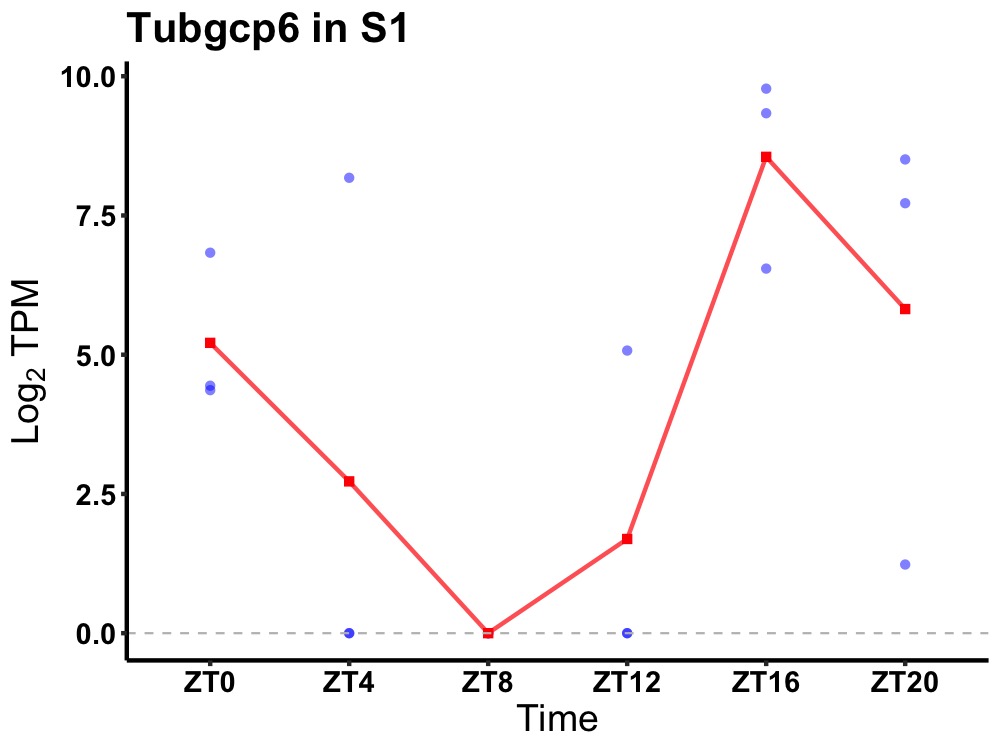 |
gamma-tubulin complex component 6 | 0.049 | 20 | 18 | 4.83 |
| ENSMUSG00000000127 | Fer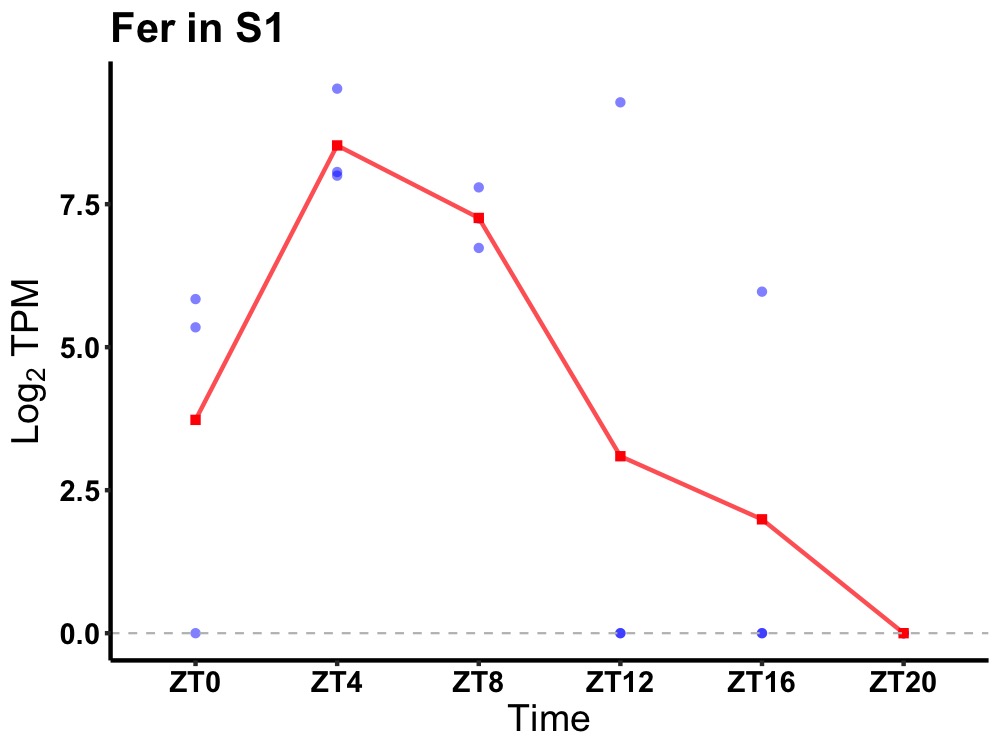 |
tyrosine-protein kinase Fer | 0.031 | 24 | 6 | 4.76 |
| ENSMUSG00000031371 | Haus7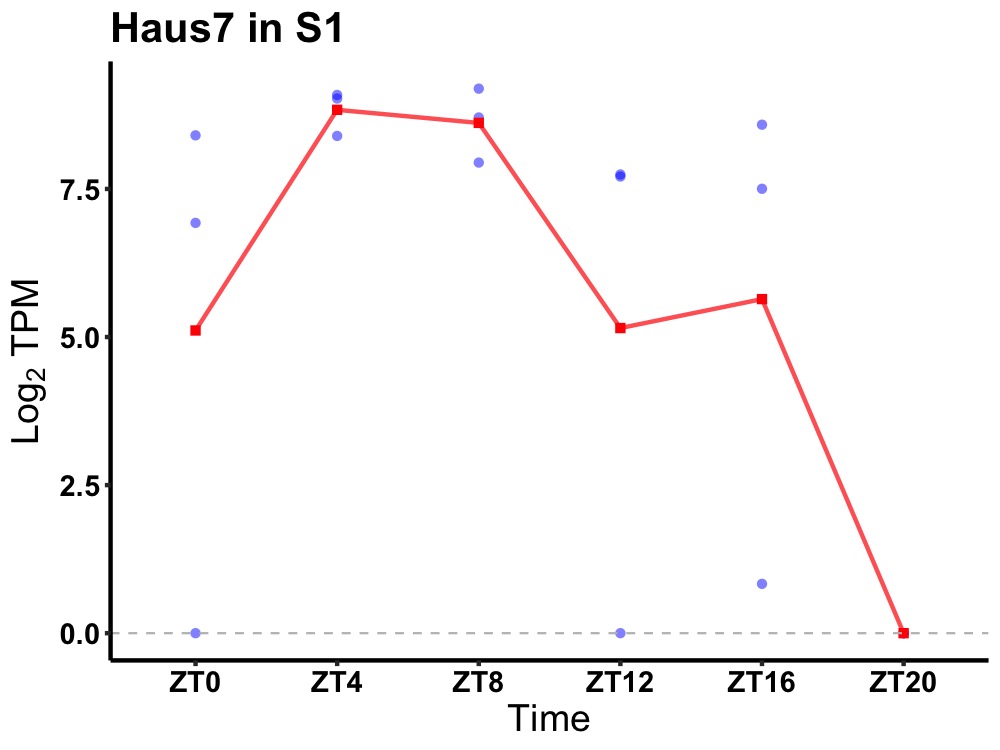 |
HAUS augmin-like complex subunit 7 | 0.031 | 24 | 8 | 4.74 |
| ENSMUSG00000041303 | Gtf3c3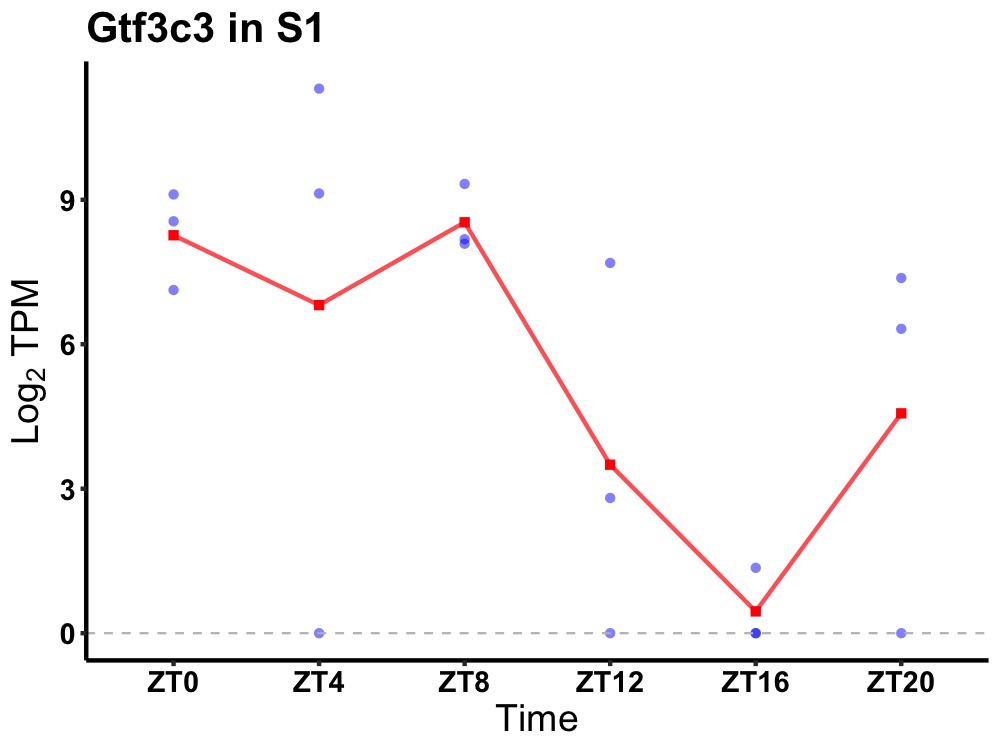 |
general transcription factor 3C polypeptide 3 | 0.043 | 24 | 6 | 4.63 |
| ENSMUSG00000038550 | Ciart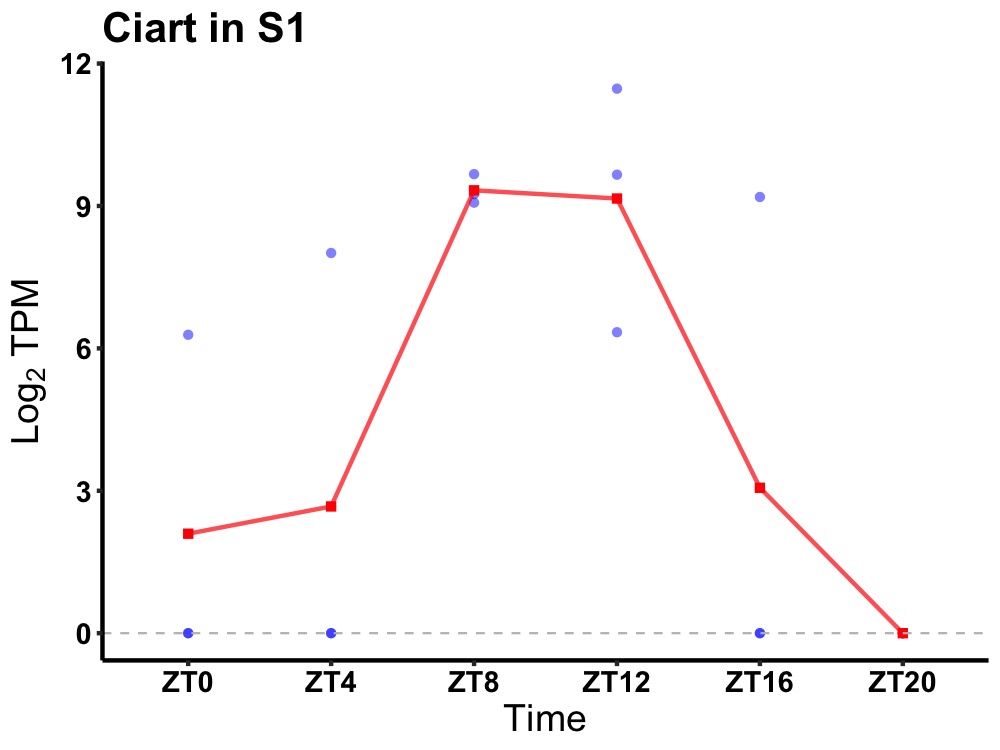 |
circadian-associated transcriptional repressor | 0.036 | 20 | 12 | 4.48 |
| ENSMUSG00000016946 | Kctd5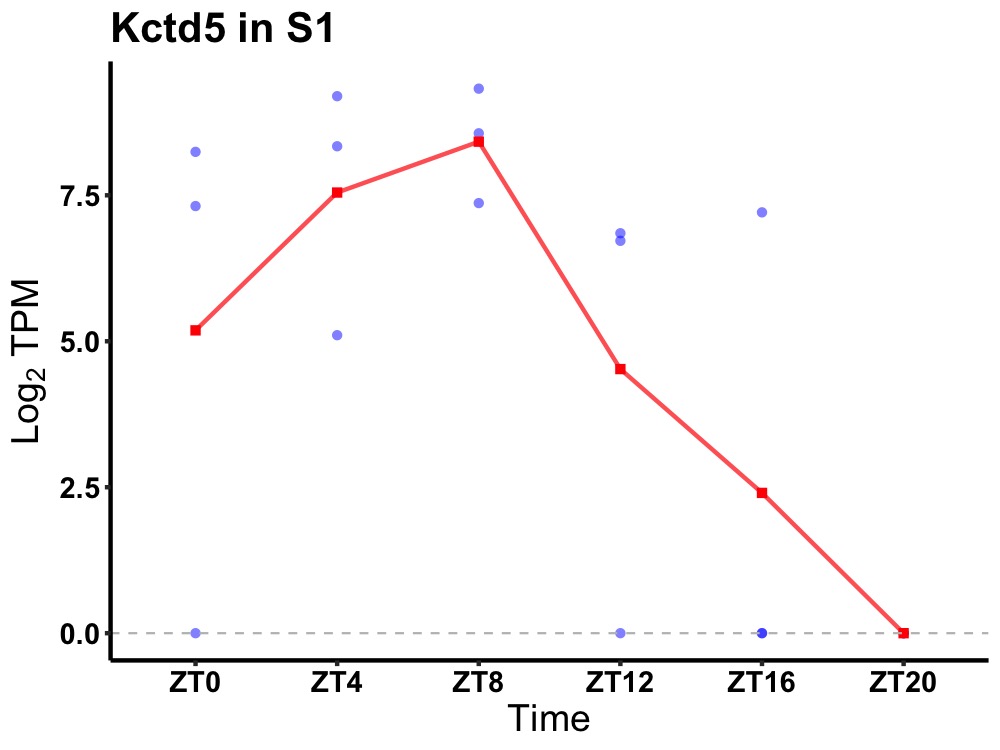 |
BTB/POZ domain-containing protein KCTD5 | 0.031 | 24 | 8 | 4.36 |
| ENSMUSG00000021697 | Depdc1b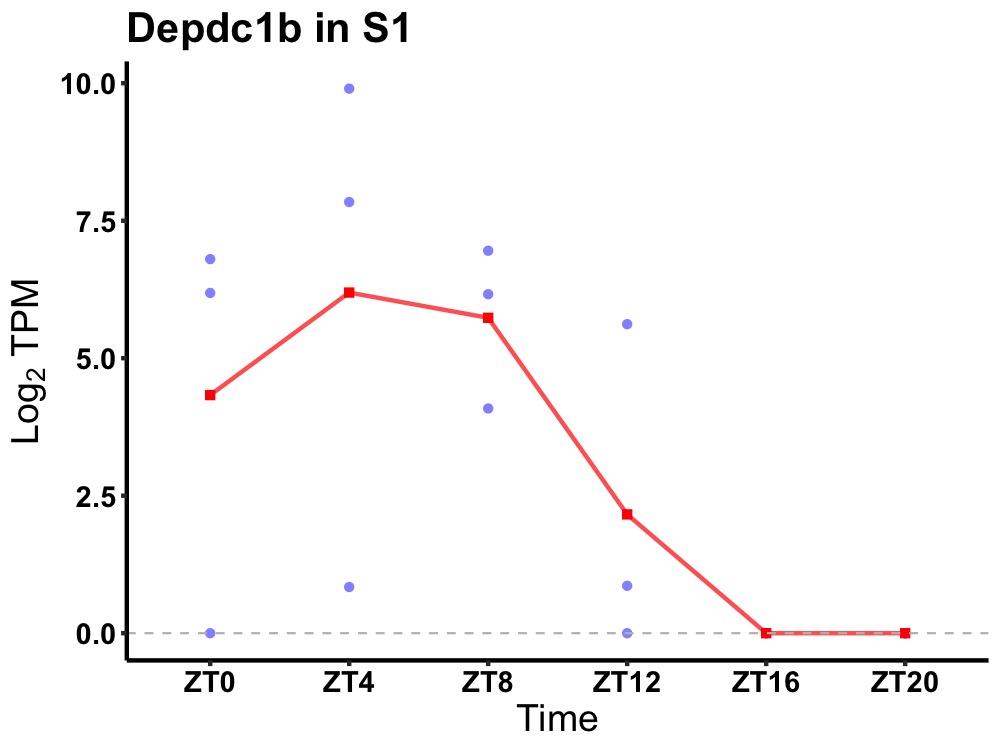 |
DEP domain-containing protein 1B | 0.014 | 24 | 6 | 4.36 |
| ENSMUSG00000028793 | Rnf19b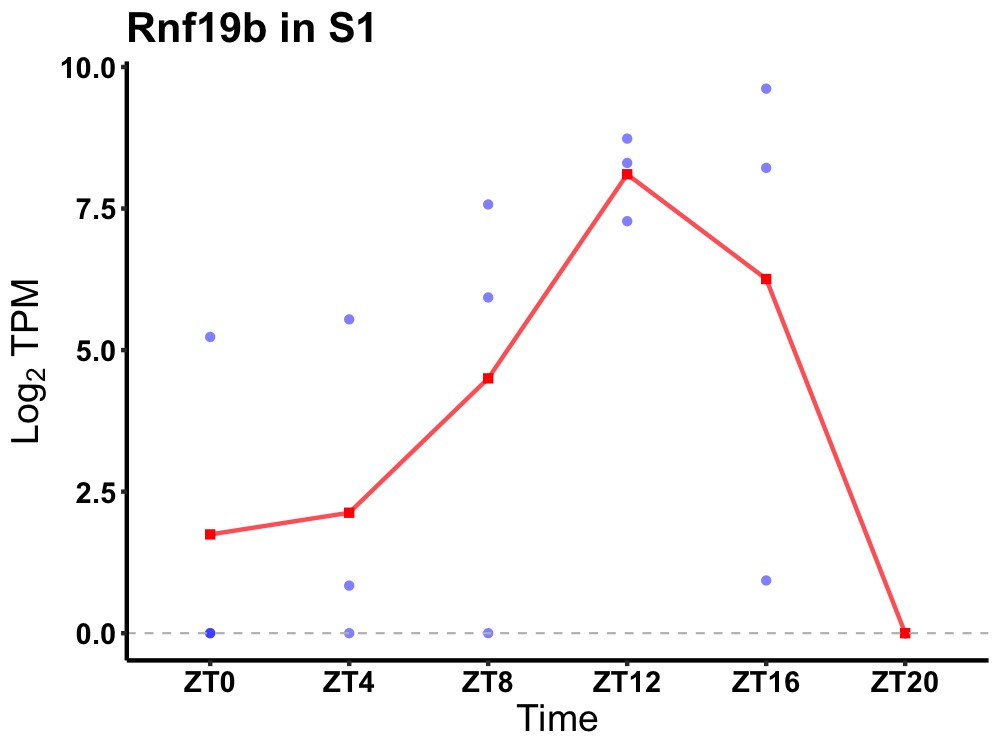 |
E3 ubiquitin-protein ligase RNF19B | 0.031 | 20 | 12 | 4.00 |
| ENSMUSG00000038976 | Ppp1r9b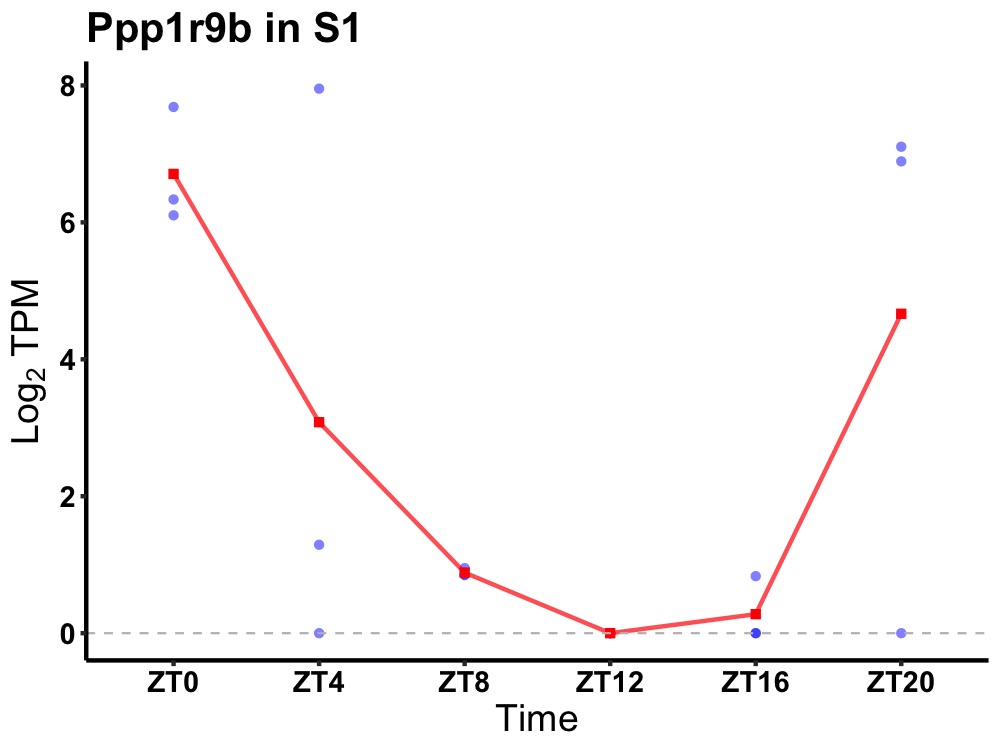 |
neurabin-2 | 0.031 | 24 | 2 | 3.89 |
| ENSMUSG00000043300 | B3galnt1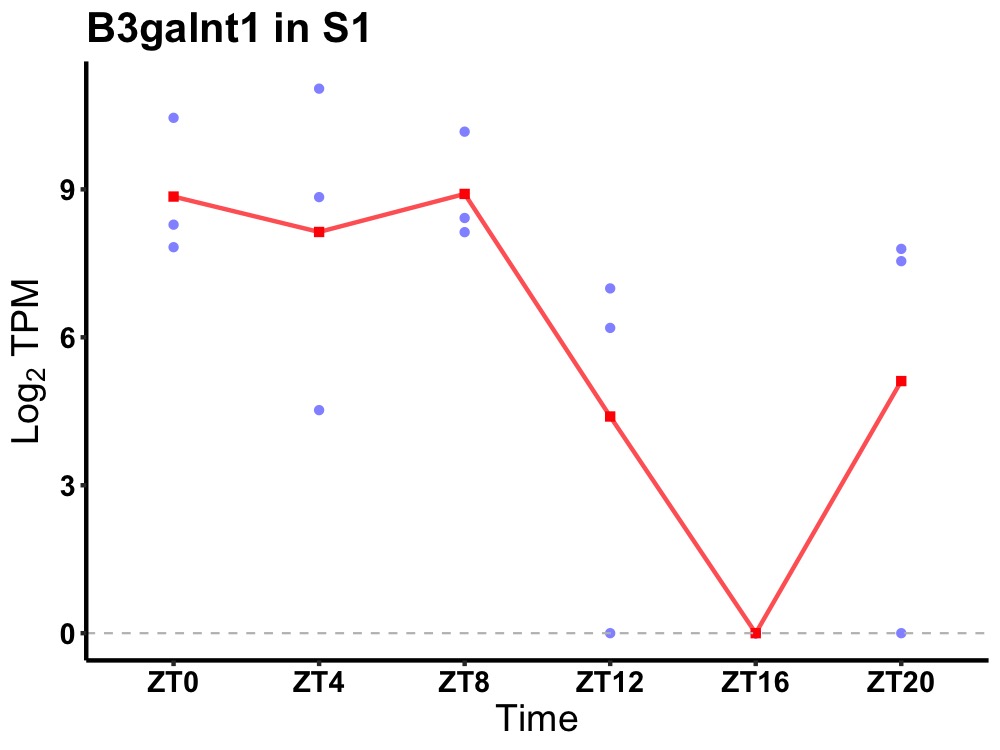 |
UDP-GalNAc:beta-1,3-N-acetylgalactosaminyltransferase 1 | 0.006 | 24 | 4 | 3.79 |
| ENSMUSG00000072662 | Mansc4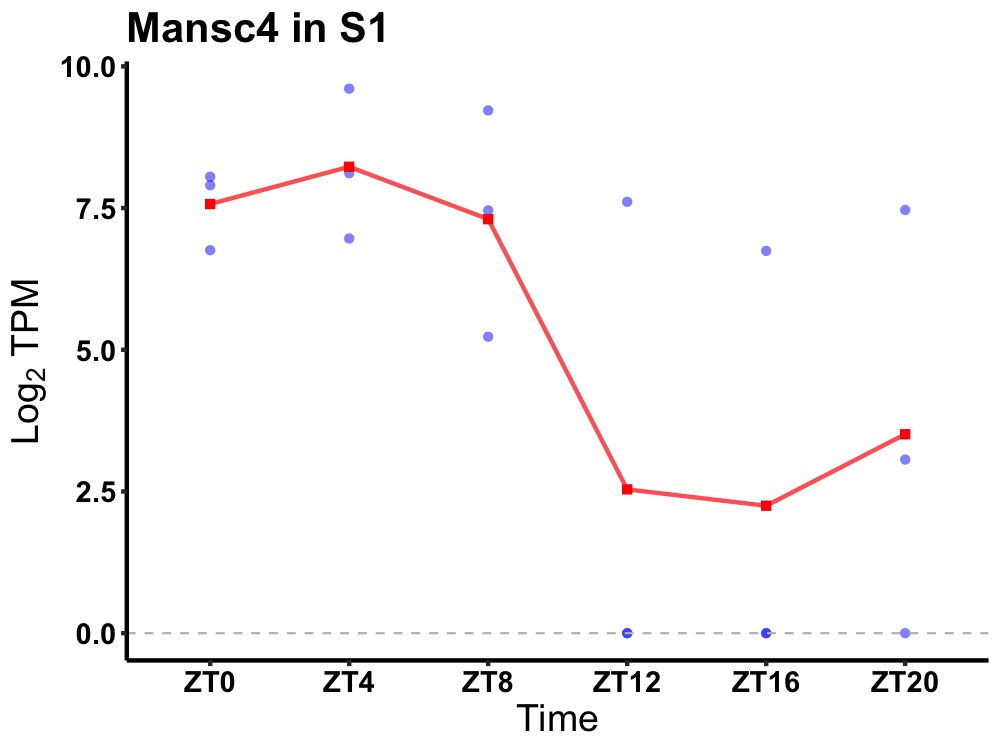 |
MANSC domain-containing protein 4 | 0.049 | 24 | 4 | 3.70 |
| ENSMUSG00000049321 | Zfp2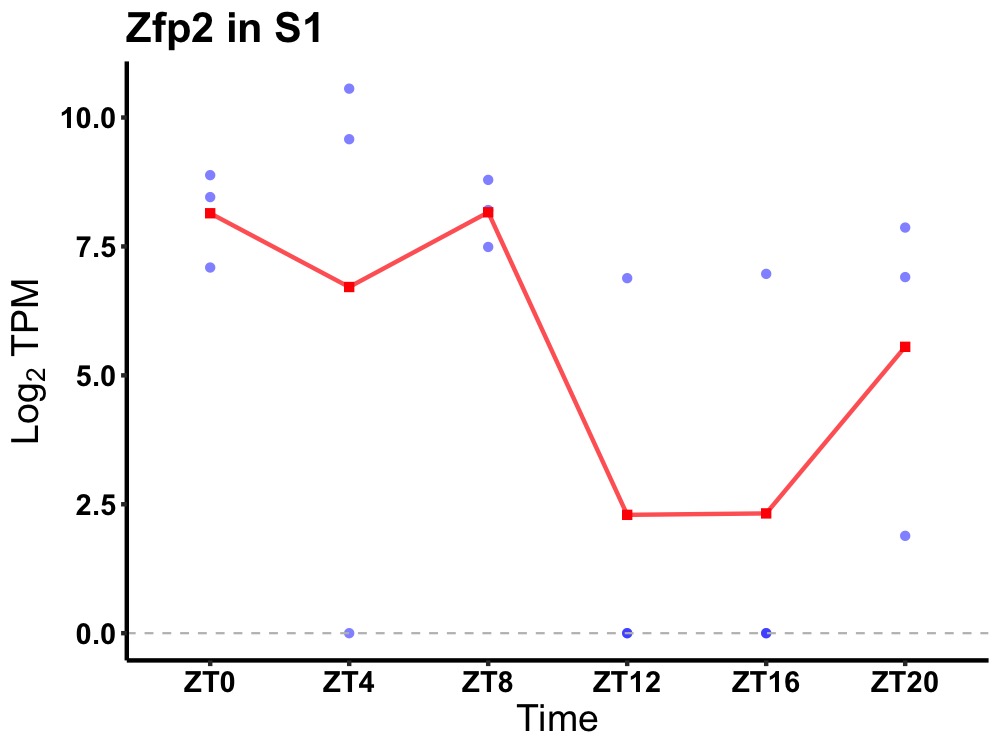 |
zinc finger protein ZFP2 | 0.027 | 24 | 4 | 3.63 |
| ENSMUSG00000029569 | Vars2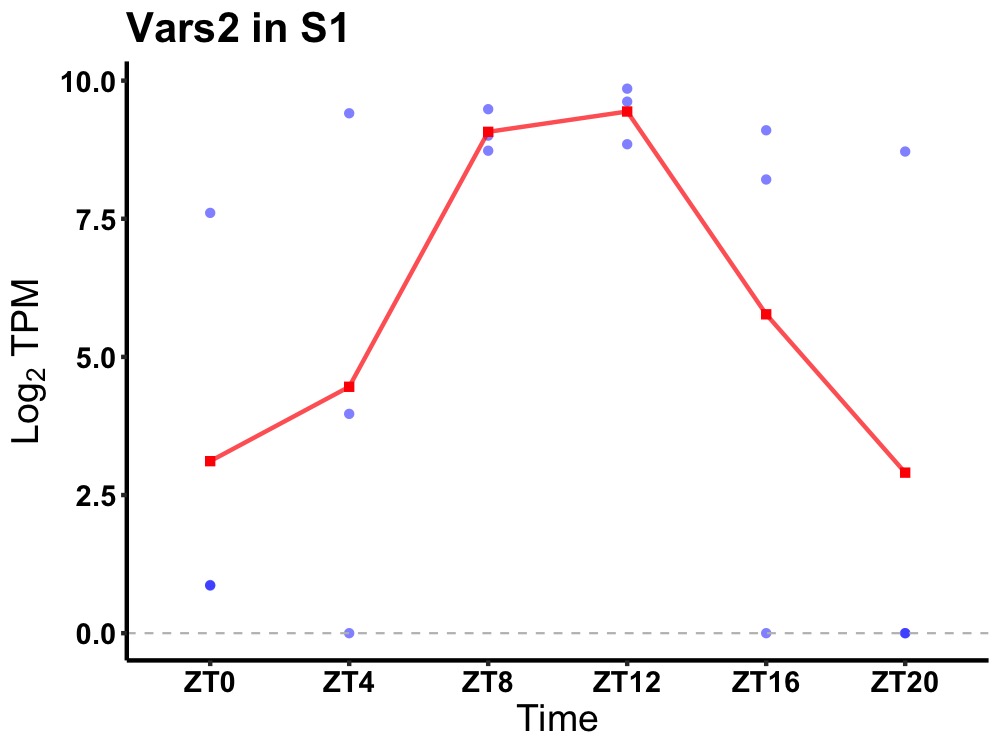 |
valine--tRNA ligase, mitochondrial | 0.043 | 20 | 12 | 3.56 |
| ENSMUSG00000013878 | Rnf170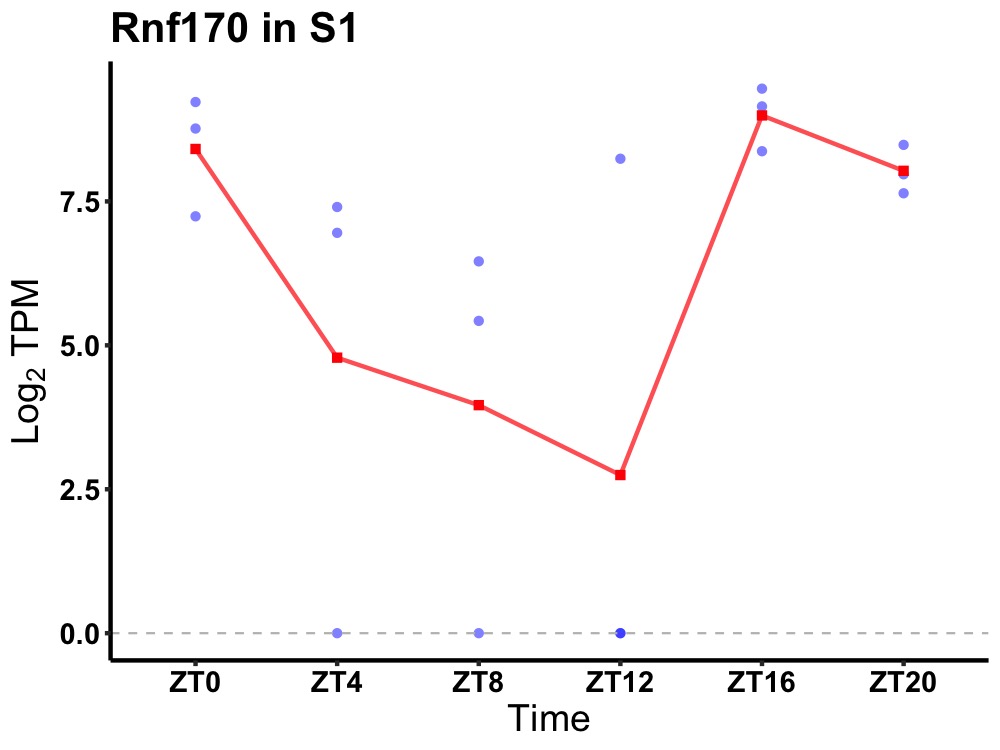 |
E3 ubiquitin-protein ligase RNF170 | 0.008 | 20 | 18 | 3.48 |
| ENSMUSG00000024869 | Gm49405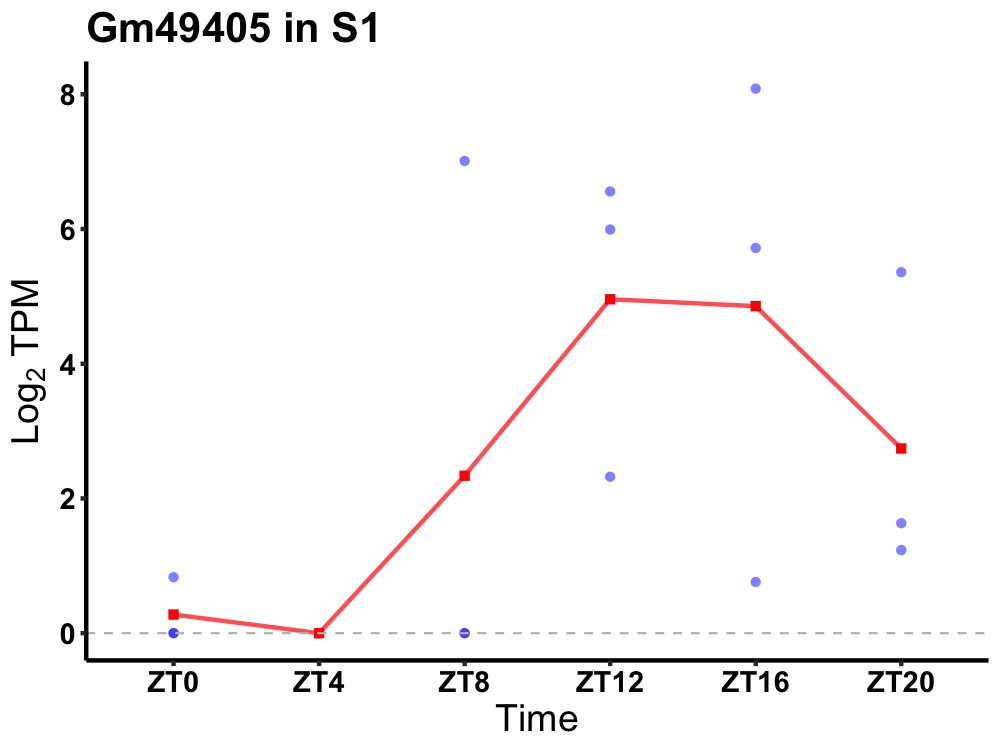 |
none | 0.036 | 24 | 16 | 3.46 |
| ENSMUSG00000054364 | Rhob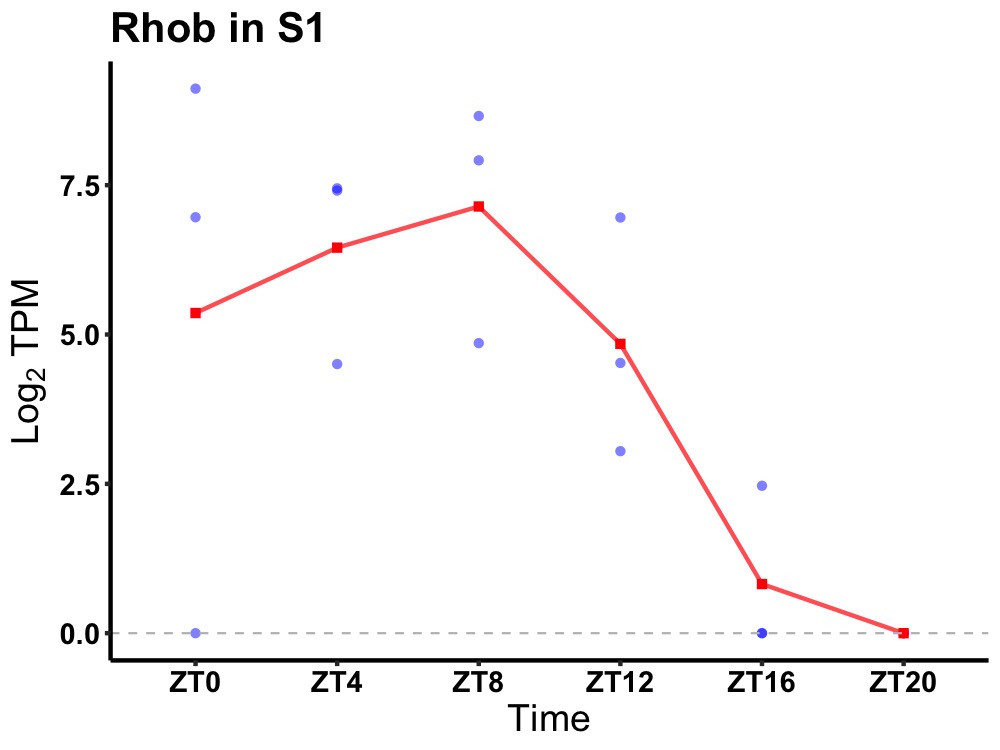 |
rho-related GTP-binding protein RhoB | 0.016 | 24 | 8 | 3.43 |
| ENSMUSG00000042675 | Ypel3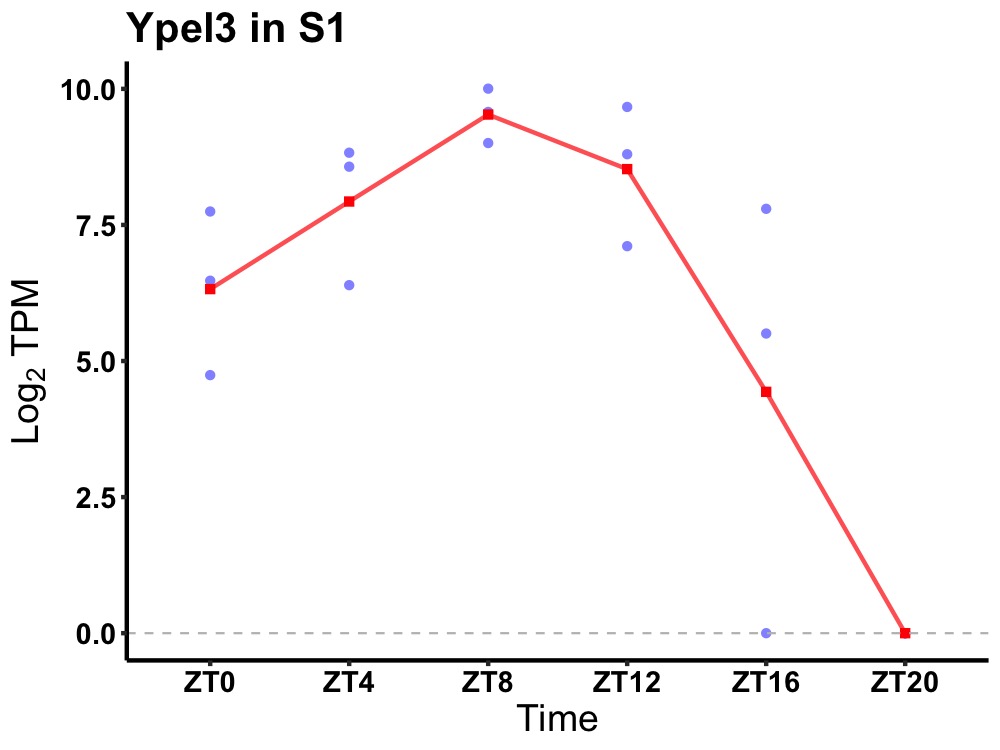 |
protein yippee-like 3 | 0.000 | 24 | 10 | 3.30 |
| ENSMUSG00000060121 | Gemin2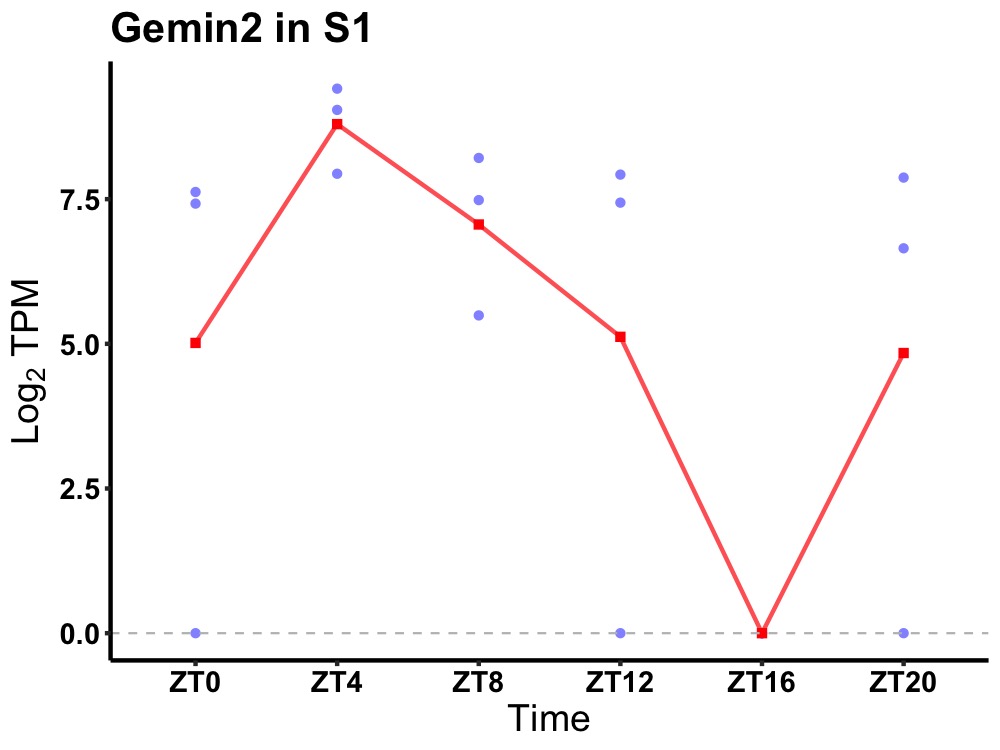 |
gem-associated protein 2 | 0.036 | 24 | 6 | 3.30 |
| ENSMUSG00000043866 | Taf10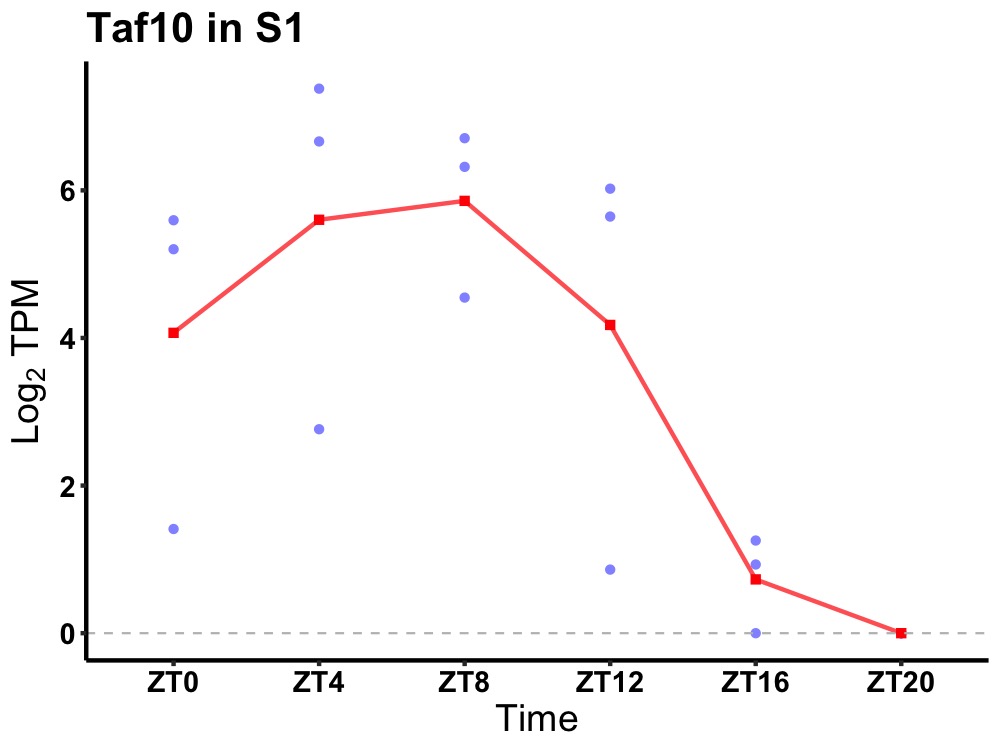 |
transcription initiation factor TFIID subunit 10 | 0.002 | 24 | 8 | 3.22 |
| ENSMUSG00000028010 | Gar1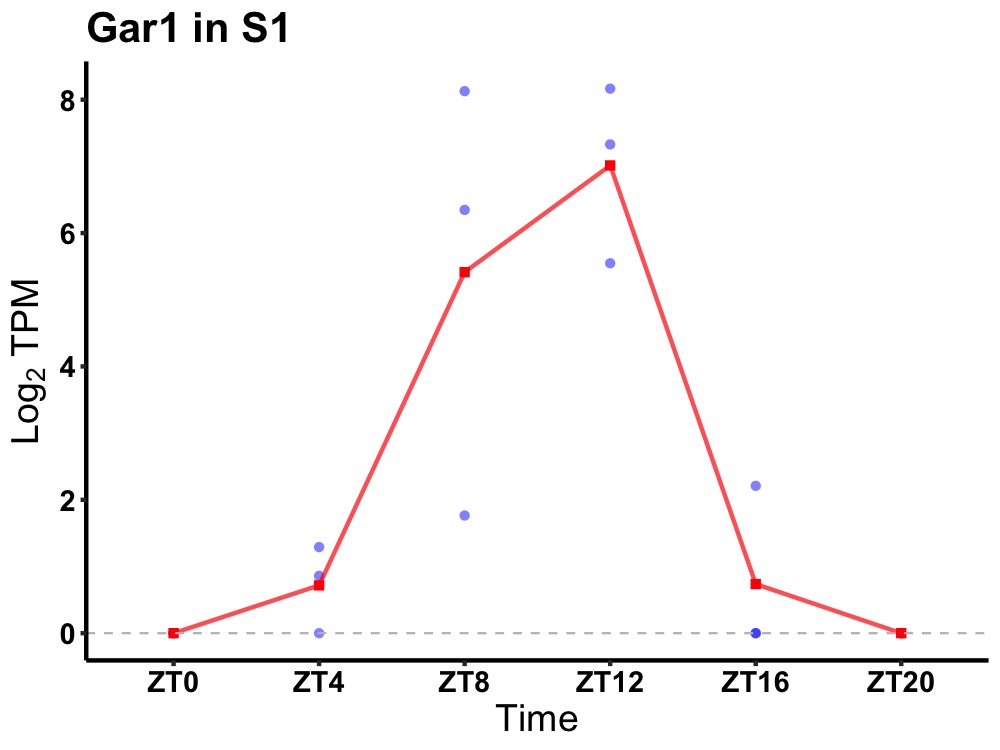 |
H/ACA ribonucleoprotein complex subunit 1 | 0.006 | 20 | 12 | 3.21 |
| ENSMUSG00000038611 | Phrf1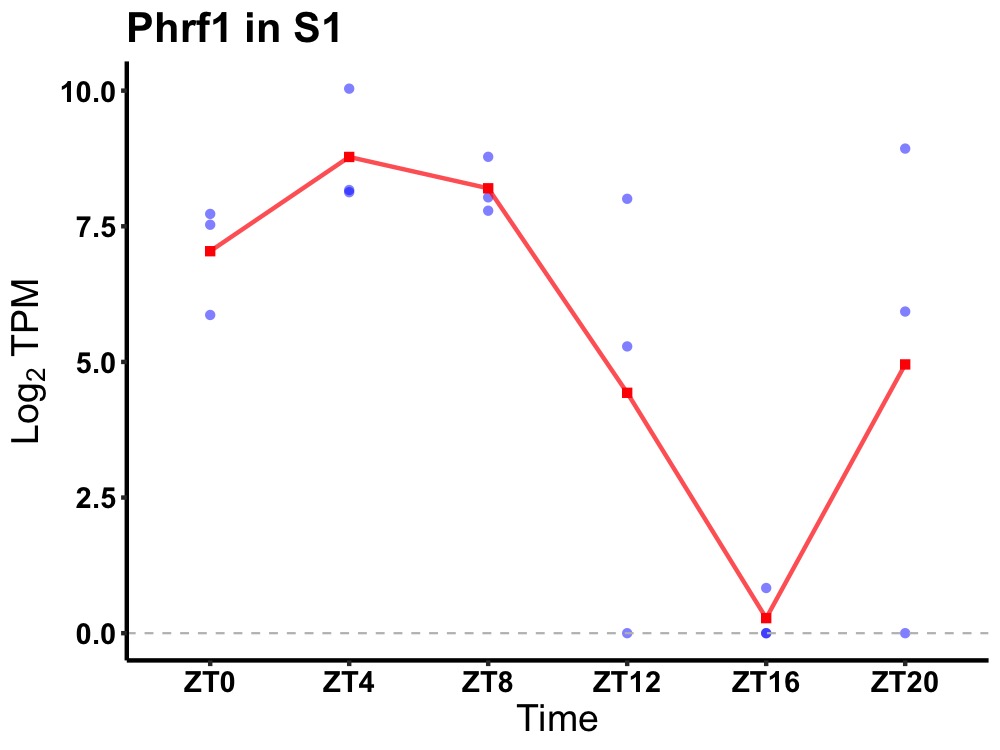 |
PHD and RING finger domain-containing protein 1 | 0.008 | 20 | 6 | 3.18 |
| ENSMUSG00000033703 | Fuk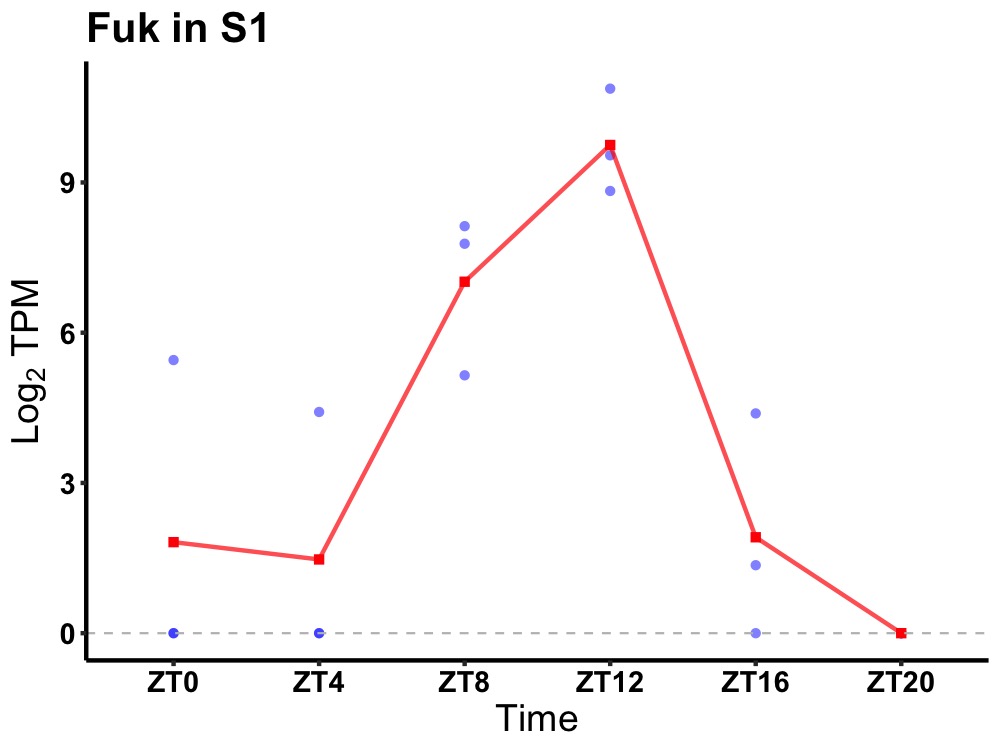 |
none | 0.004 | 20 | 12 | 3.12 |
| ENSMUSG00000008090 | Fgfrl1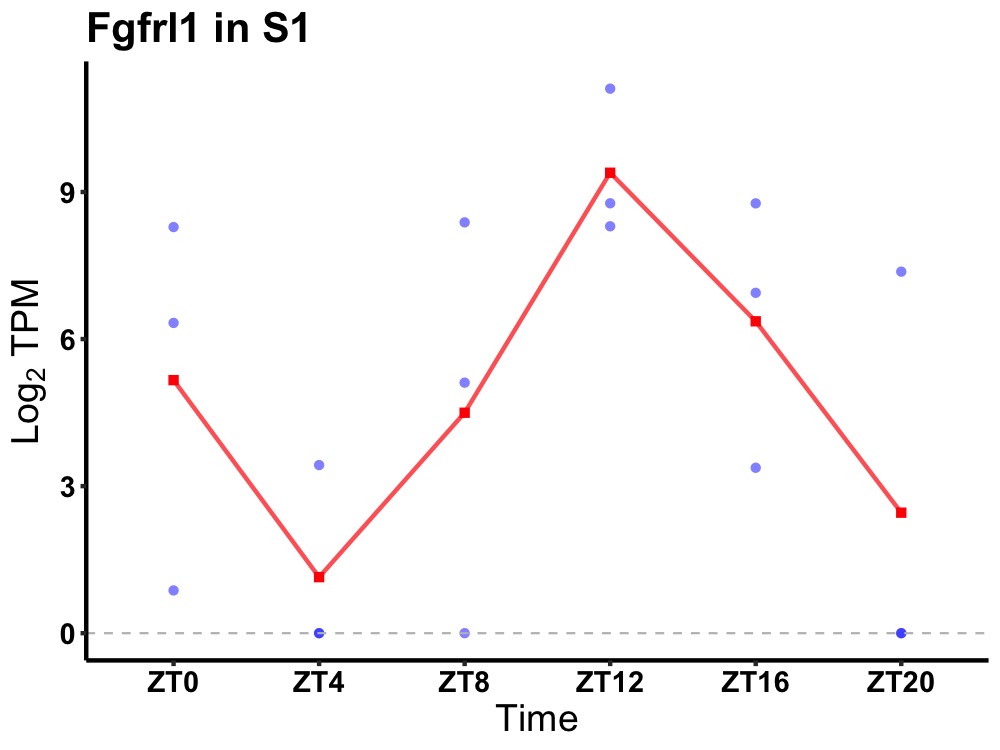 |
fibroblast growth factor receptor-like 1 | 0.036 | 20 | 14 | 3.12 |
| ENSMUSG00000029924 | Bcar1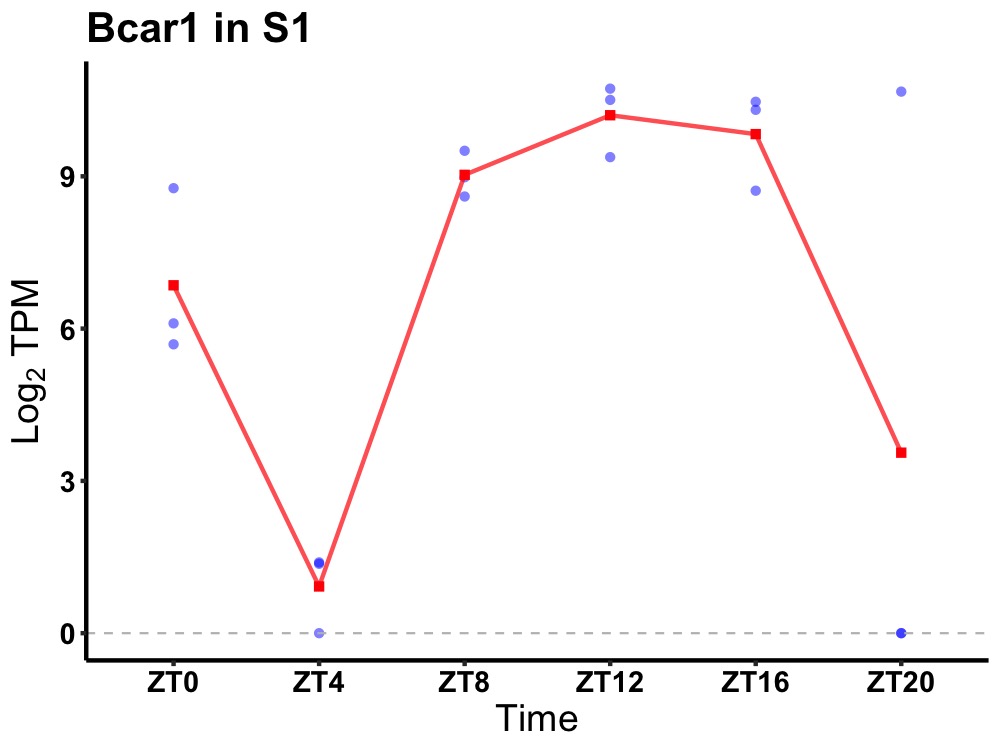 |
breast cancer anti-estrogen resistance protein 1 | 0.007 | 20 | 14 | 3.07 |
| ENSMUSG00000061393 | Acvr2b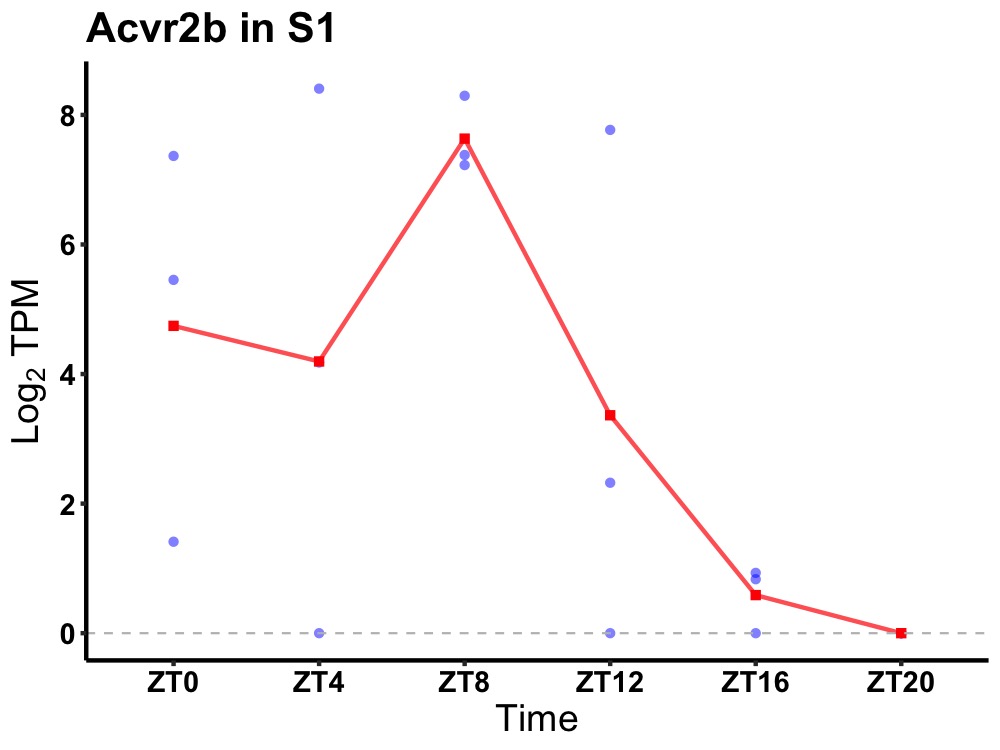 |
activin receptor type-2B | 0.049 | 24 | 8 | 2.95 |
| ENSMUSG00000094281 | Gm8034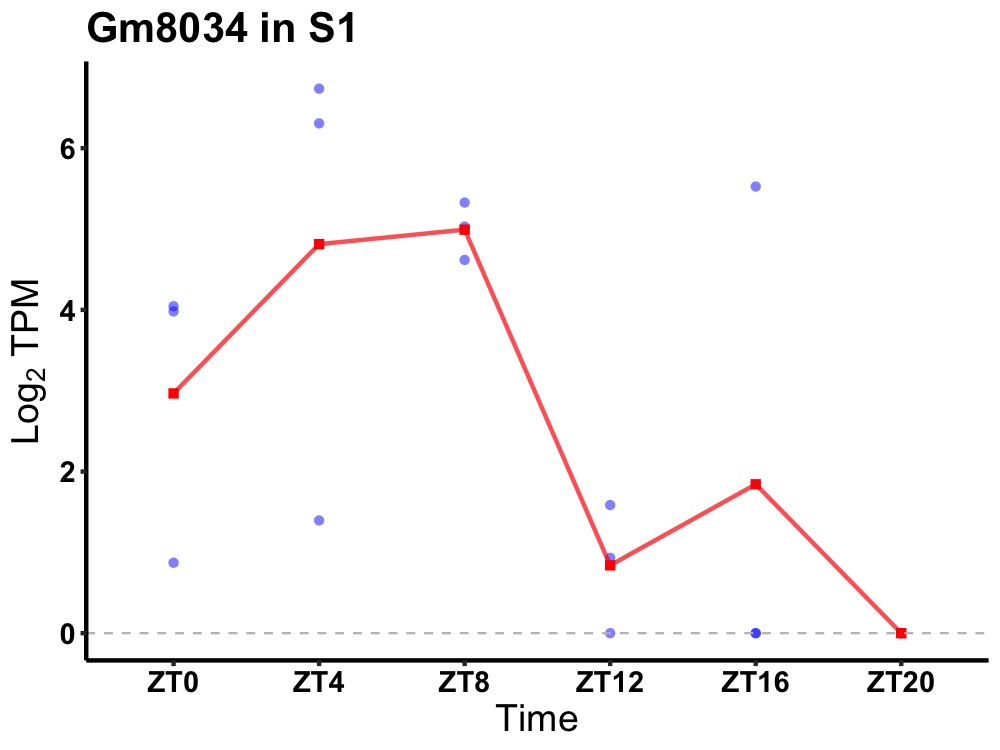 |
none | 0.023 | 24 | 6 | 2.86 |
| ENSMUSG00000053286 | Wdfy2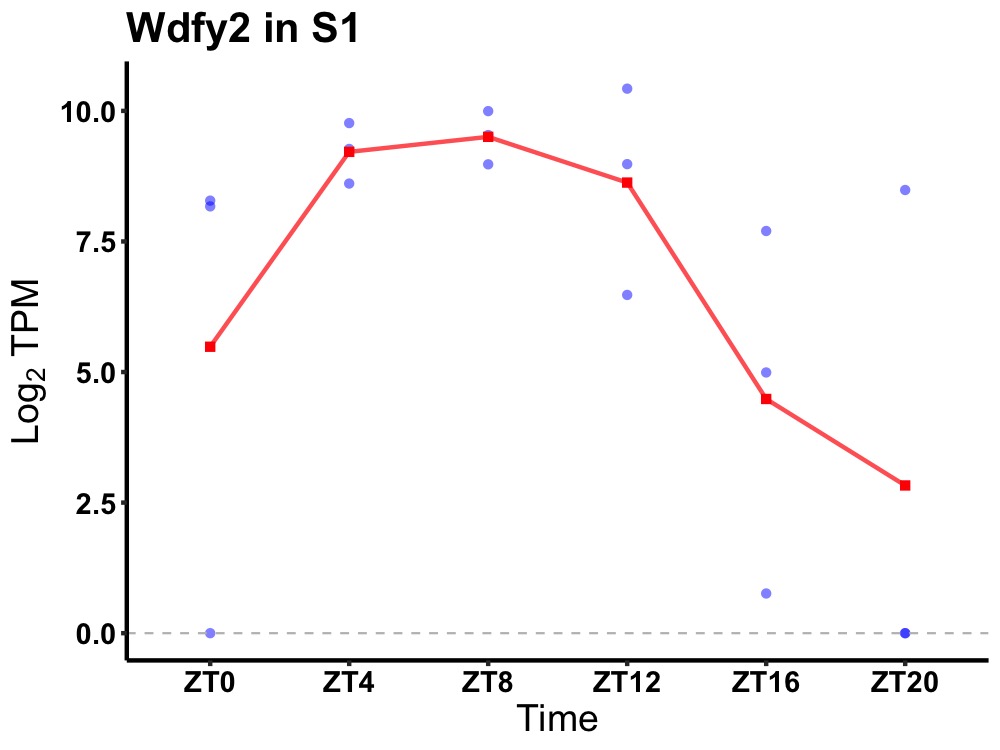 |
WD repeat and FYVE domain-containing protein 2 | 0.014 | 24 | 8 | 2.76 |
| ENSMUSG00000037344 | Slc12a9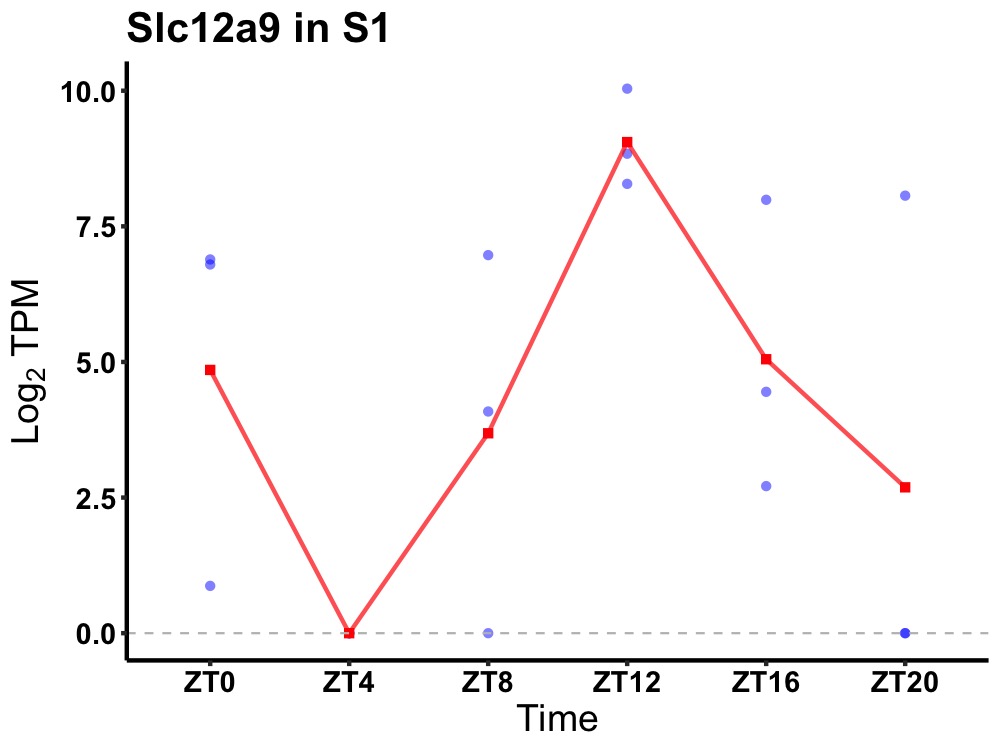 |
solute carrier family 12 member 9 | 0.023 | 20 | 14 | 2.76 |
| ENSMUSG00000035890 | Rnf126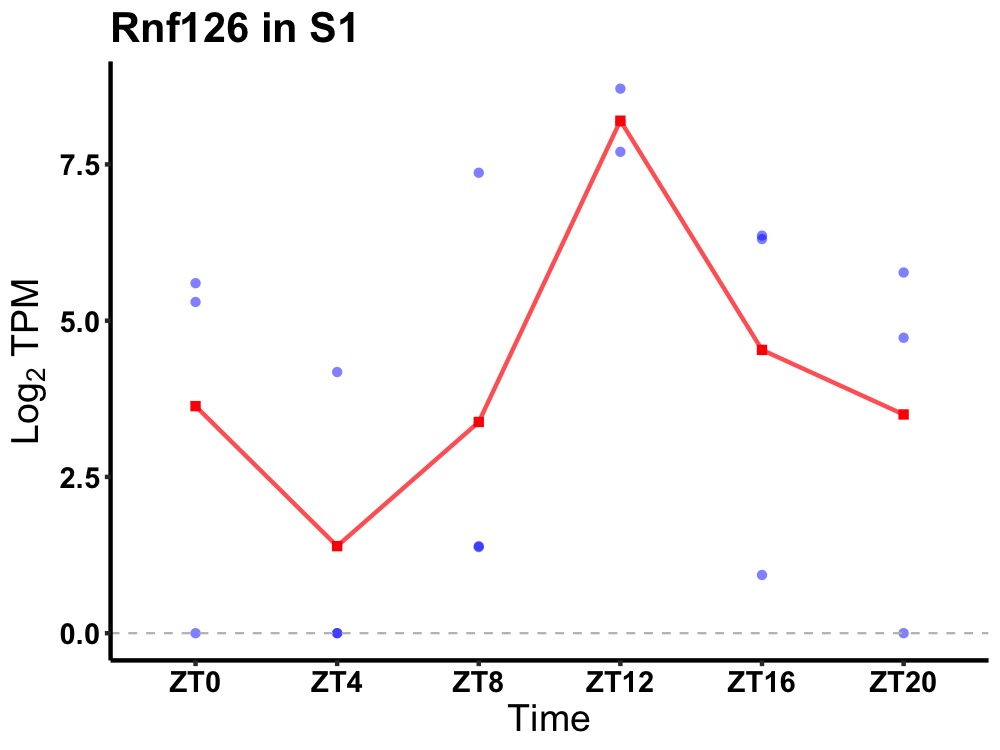 |
ring finger protein 126 | 0.043 | 20 | 14 | 2.66 |
| ENSMUSG00000030126 | Tmcc1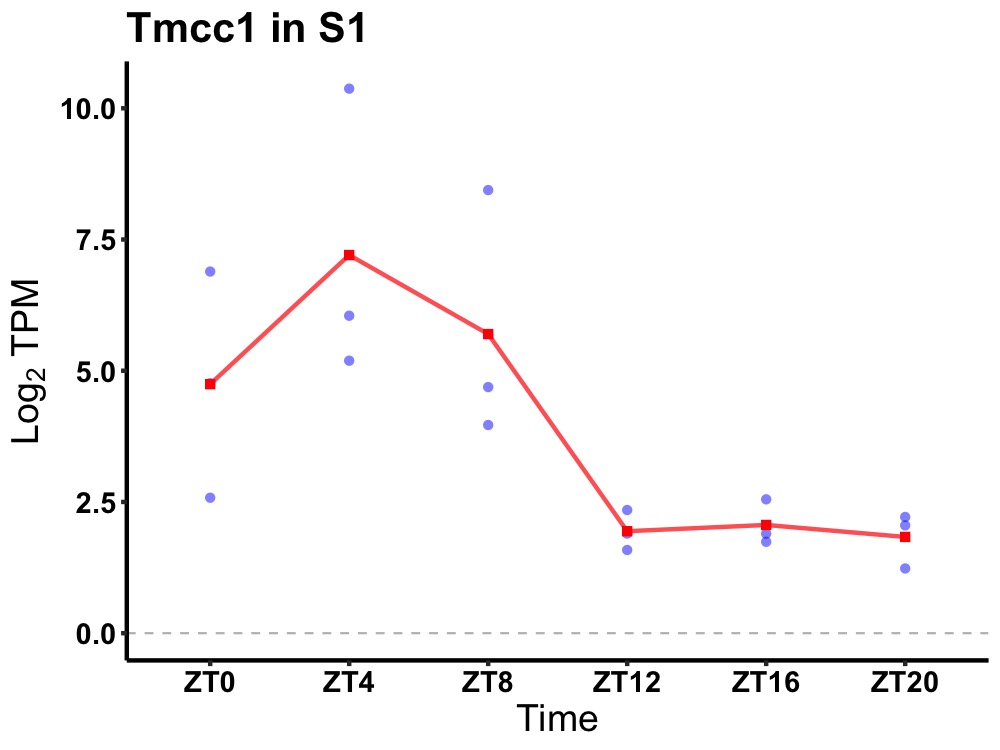 |
transmembrane and coiled coil domains 1 | 0.005 | 24 | 6 | 2.35 |
| ENSMUSG00000038745 | Baz1b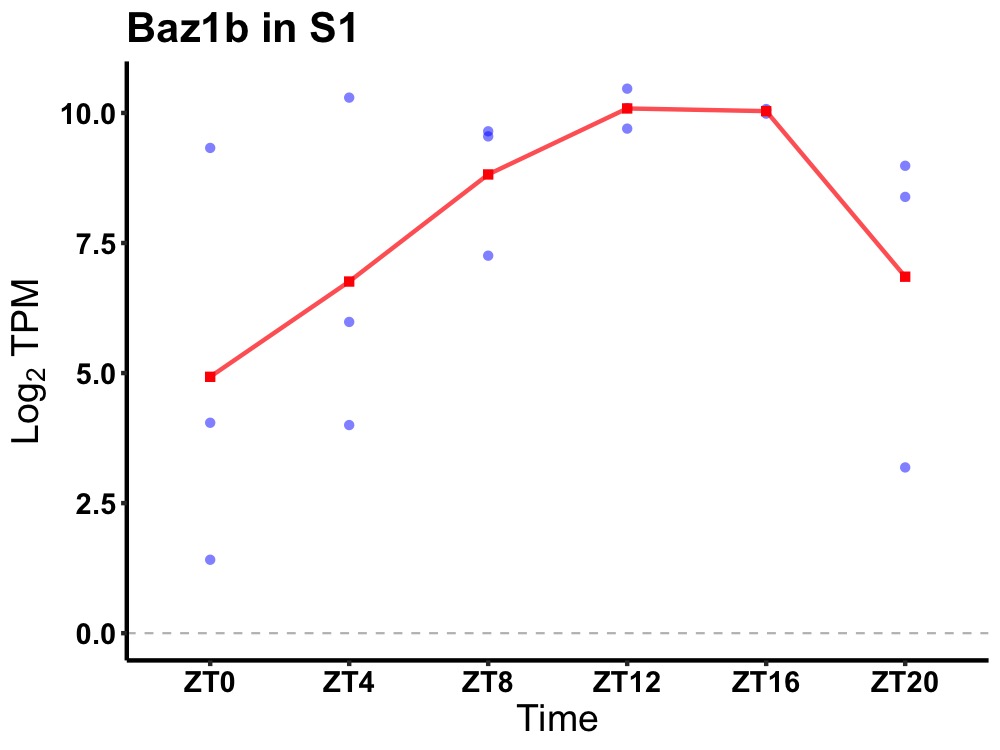 |
bromodomain adjacent to zinc finger domain, 1B | 0.019 | 24 | 14 | 2.30 |
| ENSMUSG00000020098 | Slc30a5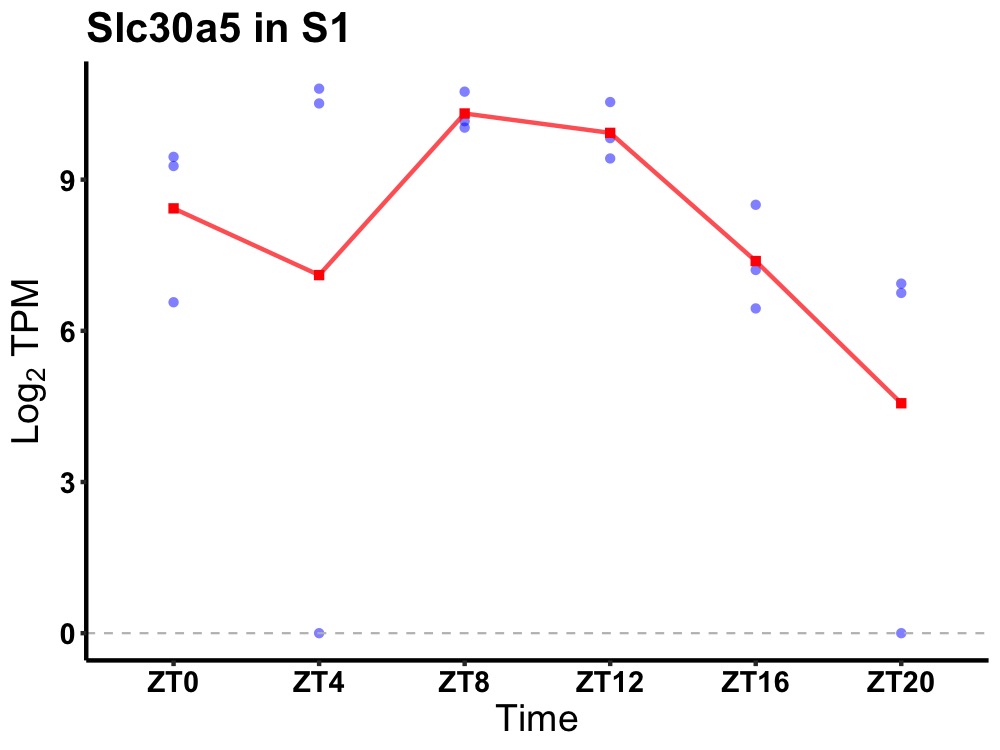 |
solute carrier family 30 (zinc transporter), member 5 | 0.023 | 24 | 8 | 2.18 |
| ENSMUSG00000085772 | D630024D03Rik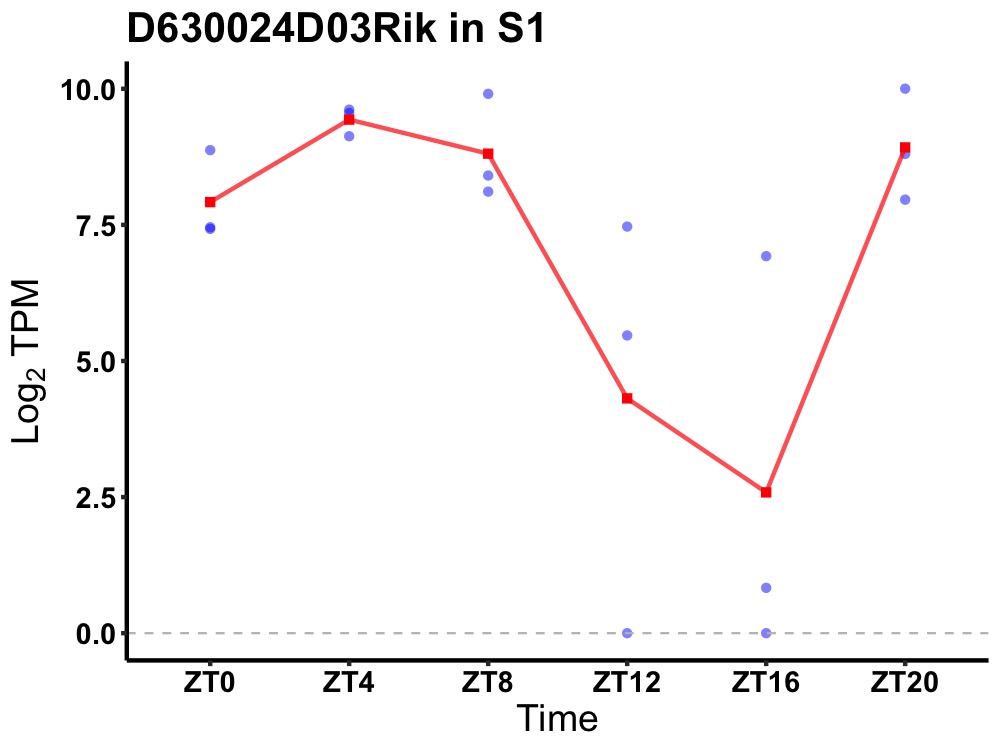 |
none | 0.003 | 20 | 6 | 2.16 |
| ENSMUSG00000046603 | Gnl1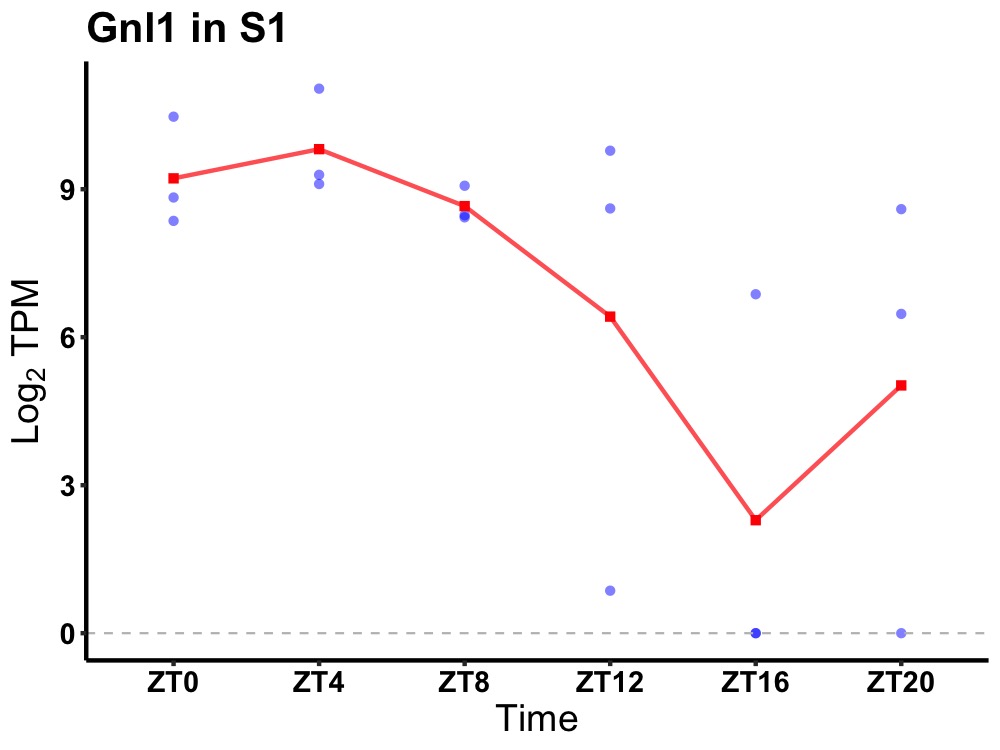 |
guanine nucleotide-binding protein-like 1 | 0.019 | 24 | 6 | 2.10 |
| ENSMUSG00000024014 | Pim1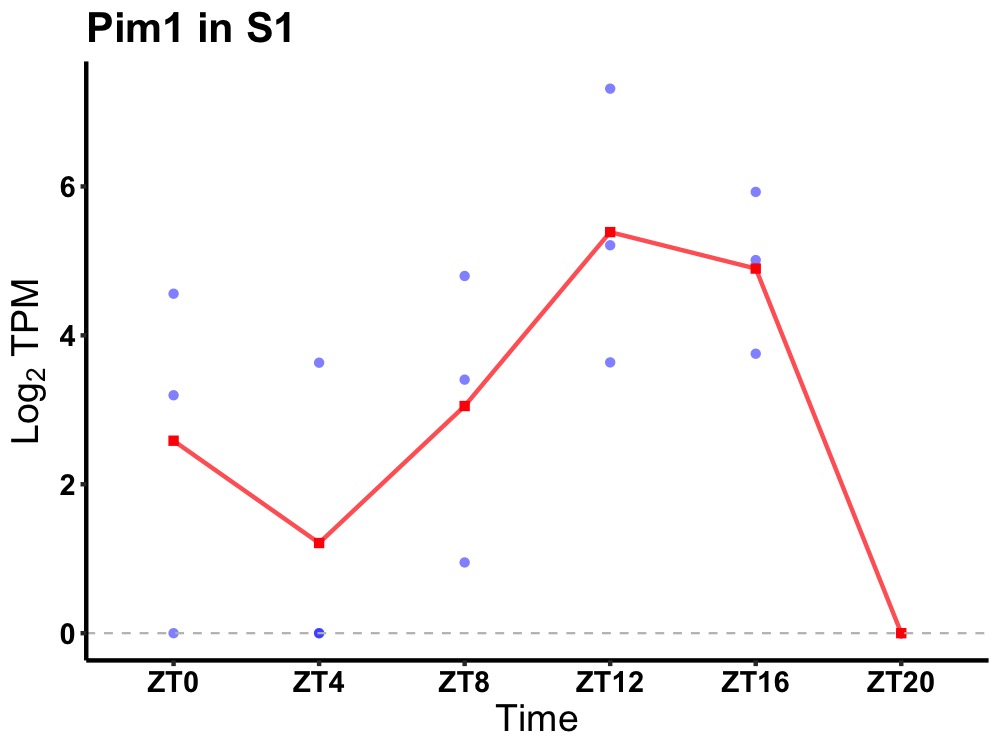 |
serine/threonine-protein kinase pim-1 | 0.008 | 20 | 14 | 1.94 |
| ENSMUSG00000041203 | Trir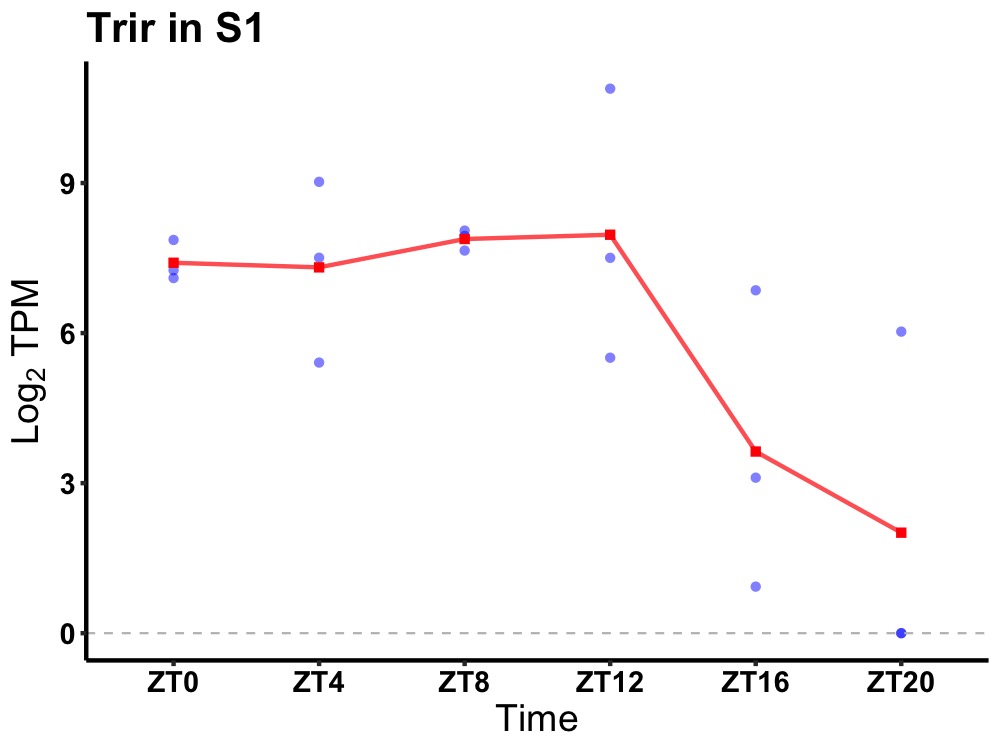 |
telomerase RNA component interacting RNase | 0.019 | 24 | 8 | 1.92 |
| ENSMUSG00000030298 | Cacul1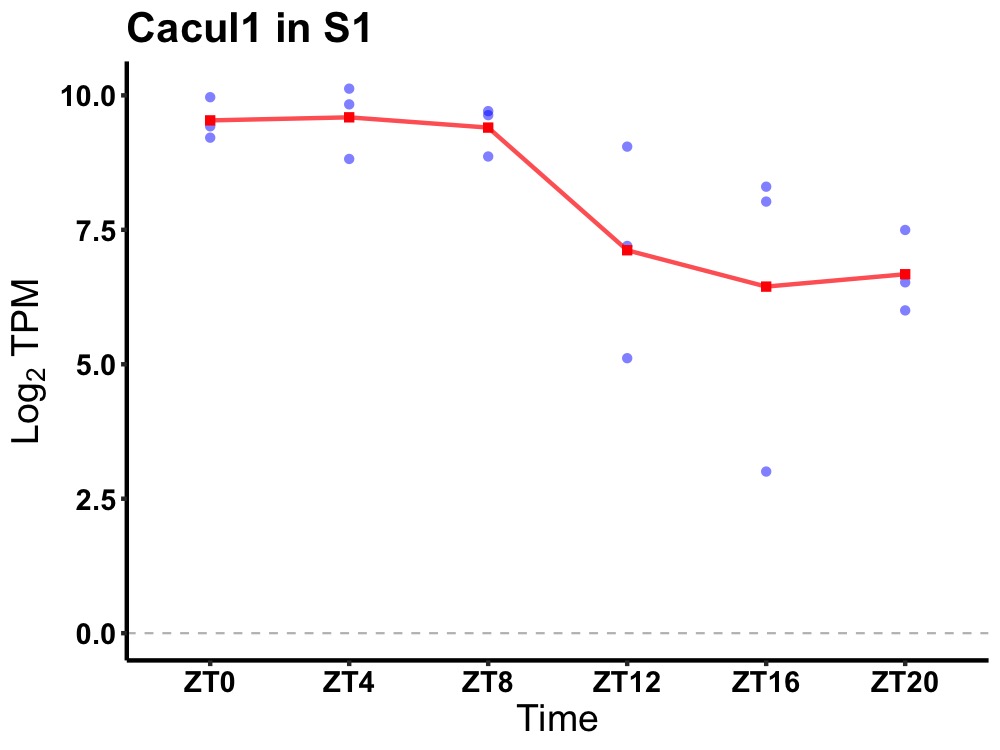 |
CDK2 associated, cullin domain 1 | 0.036 | 24 | 6 | 1.72 |
| ENSMUSG00000031604 | B3galnt2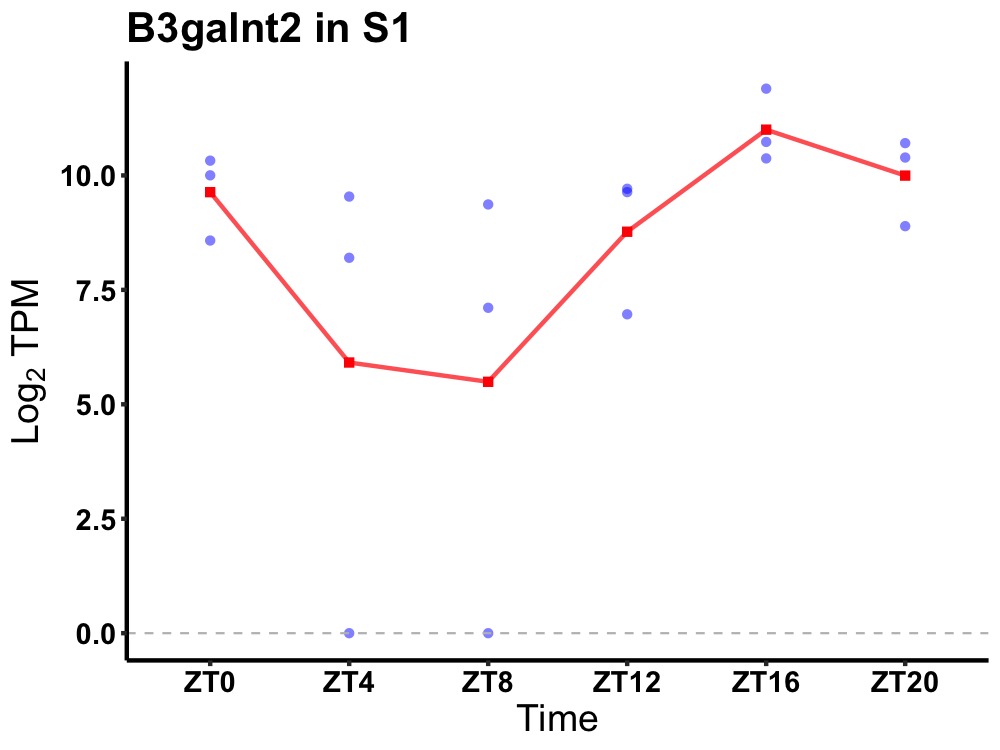 |
UDP-GalNAc:beta-1,3-N-acetylgalactosaminyltransferase 2 | 0.012 | 20 | 18 | 1.70 |
| ENSMUSG00000094664 | Shld2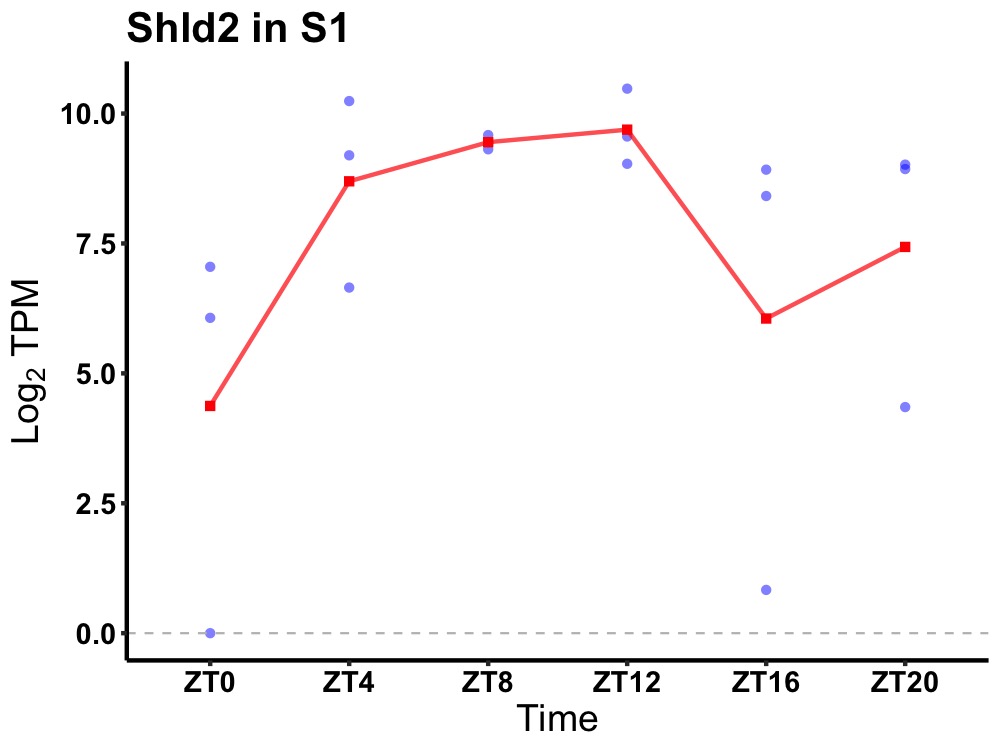 |
shieldin complex subunit 2 | 0.027 | 20 | 10 | 1.69 |
| ENSMUSG00000000934 | Jak2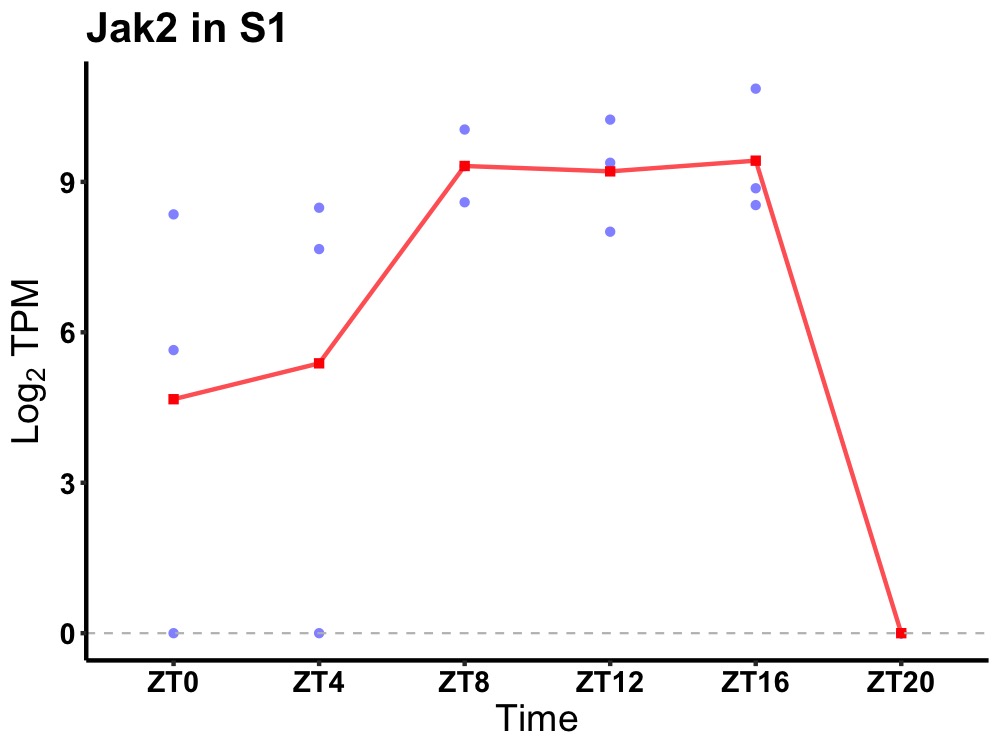 |
tyrosine-protein kinase JAK2 | 0.004 | 20 | 12 | 1.68 |
| ENSMUSG00000109324 | Med13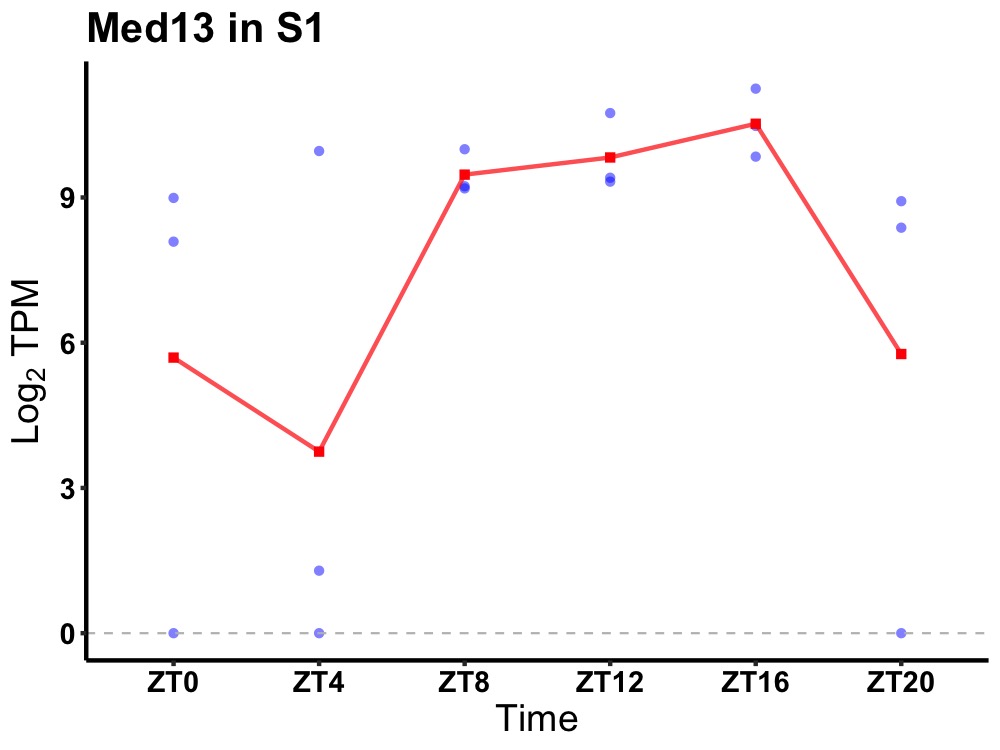 |
mediator of RNA polymerase II transcription subunit 13 | 0.043 | 24 | 14 | 1.67 |
| ENSMUSG00000038393 | Sema4g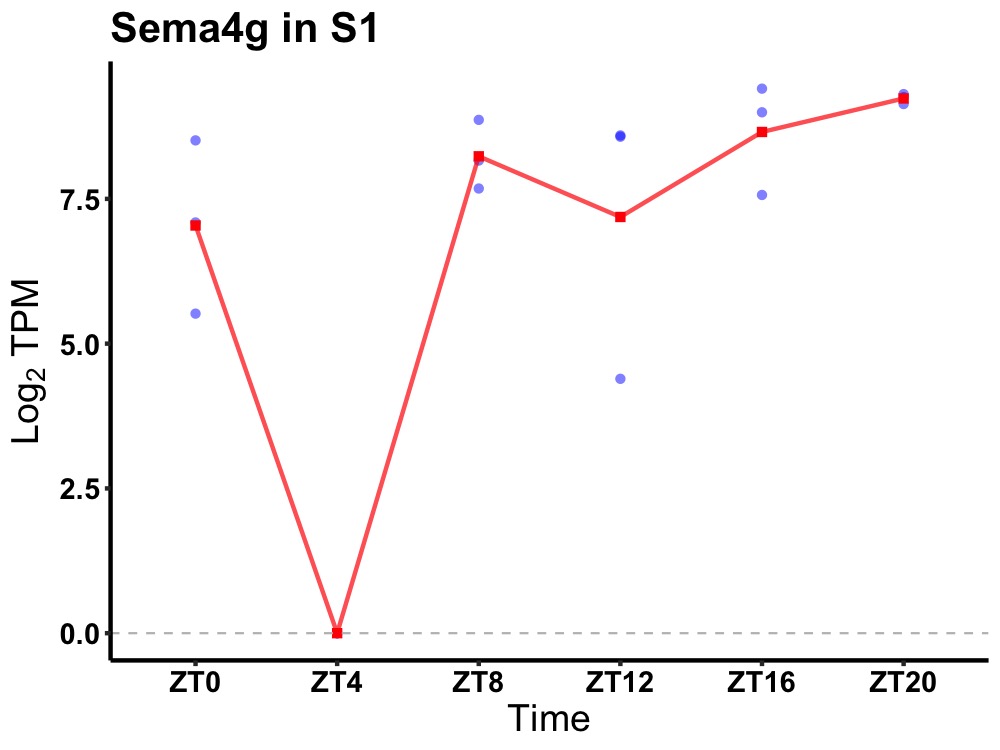 |
semaphorin-4G | 0.027 | 24 | 18 | 1.63 |
| ENSMUSG00000078592 | Gm4609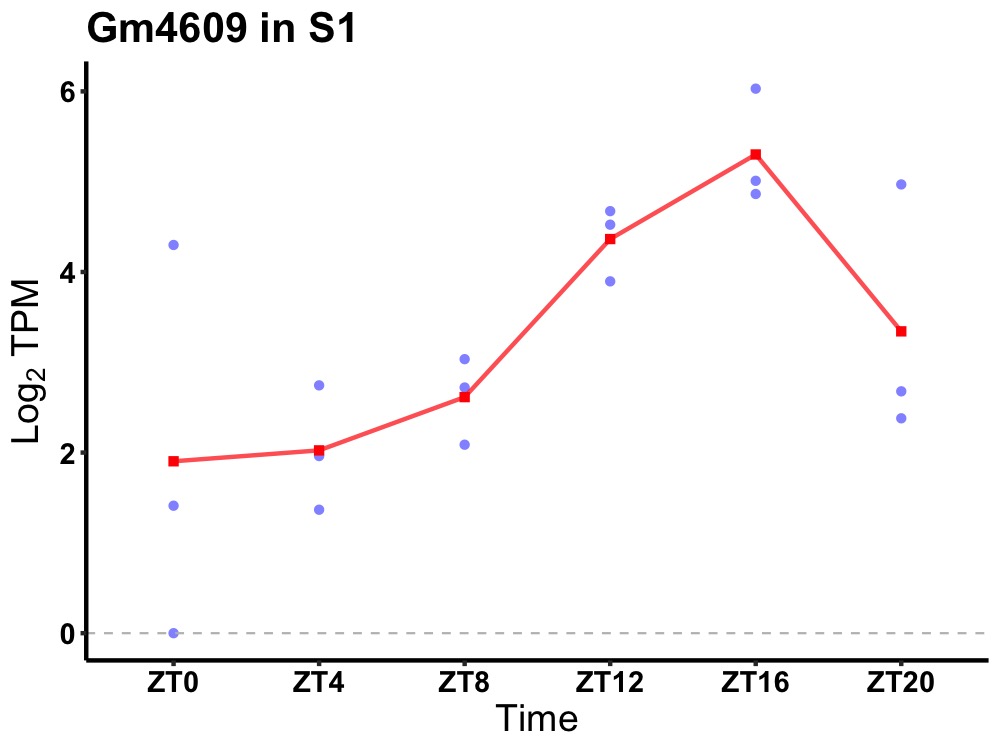 |
predicted gene 4609 | 0.007 | 24 | 16 | 1.60 |
| ENSMUSG00000024127 | 4833420G17Rik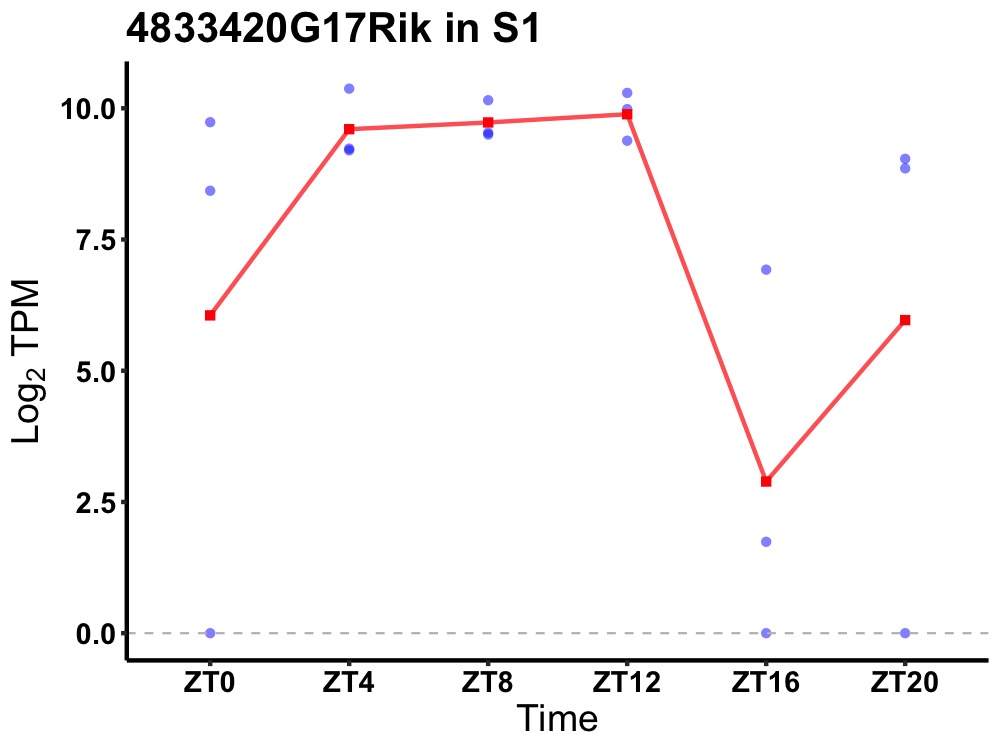 |
uncharacterized protein C5orf34 homolog | 0.043 | 20 | 8 | 1.58 |
| ENSMUSG00000043284 | Itpk1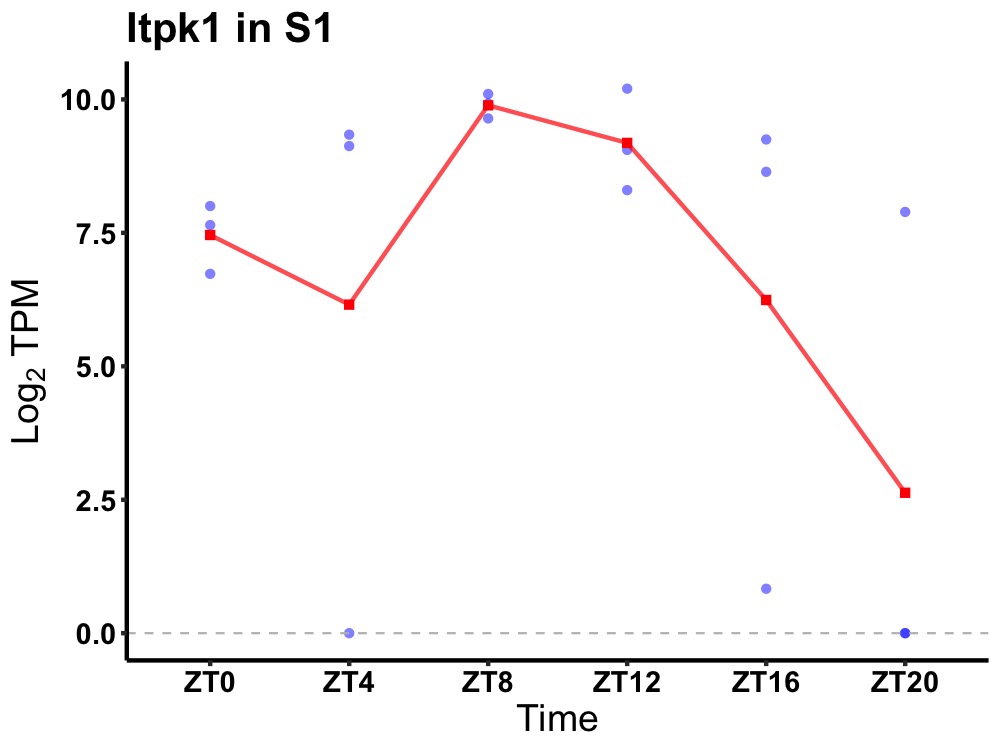 |
inositol-tetrakisphosphate 1-kinase | 0.014 | 24 | 10 | 1.56 |
| ENSMUSG00000039047 | Txnip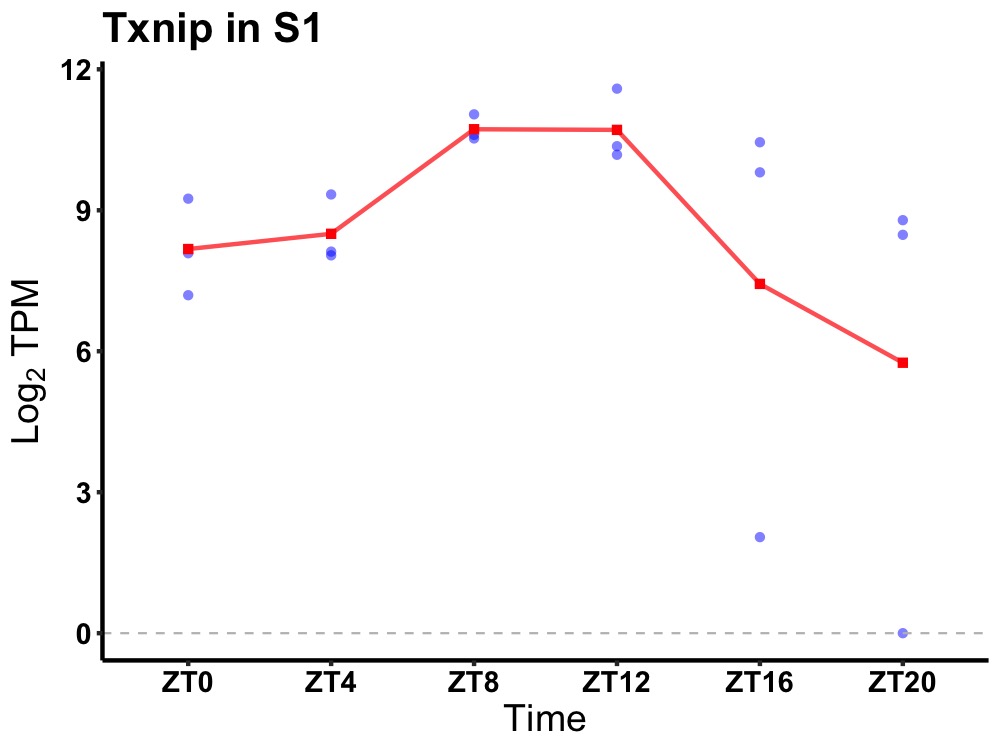 |
thioredoxin interacting protein | 0.027 | 20 | 12 | 1.55 |
| ENSMUSG00000024758 | Cyb5r4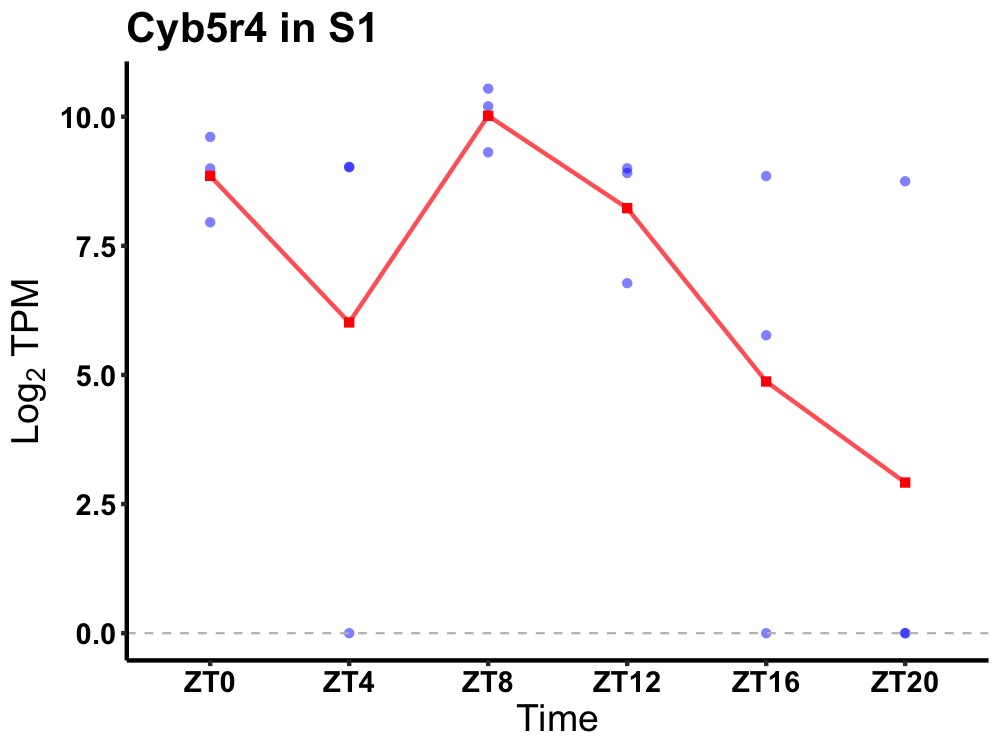 |
cytochrome b5 reductase 4 | 0.023 | 24 | 8 | 1.47 |
| ENSMUSG00000041852 | Tcf20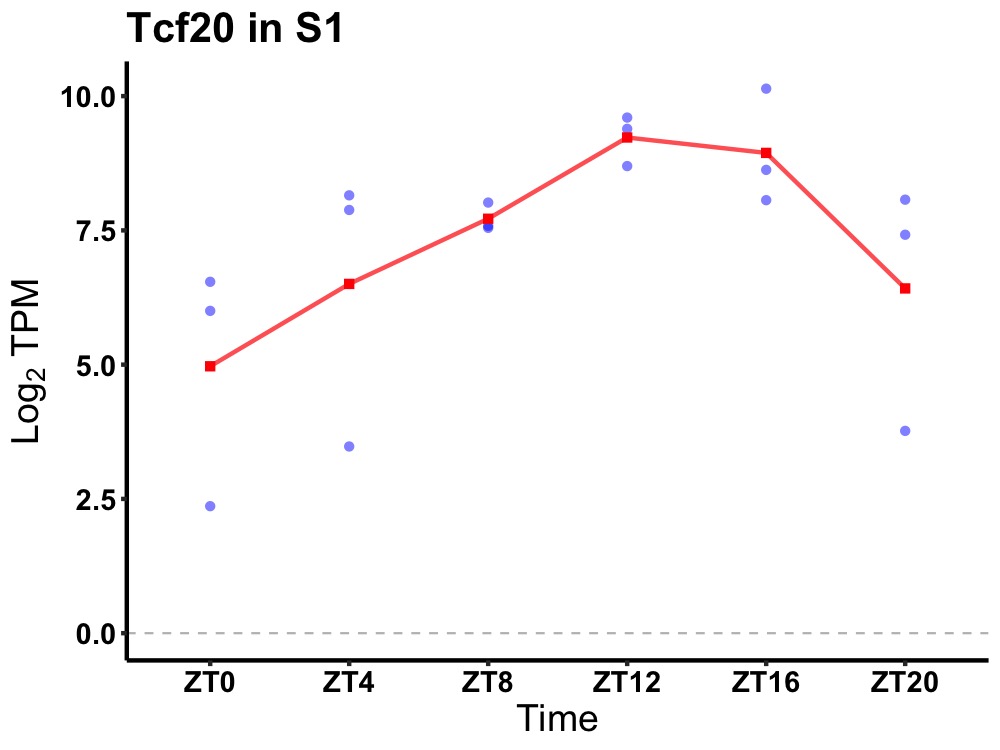 |
transcription factor 20 | 0.005 | 24 | 14 | 1.46 |
| ENSMUSG00000074521 | Gm14327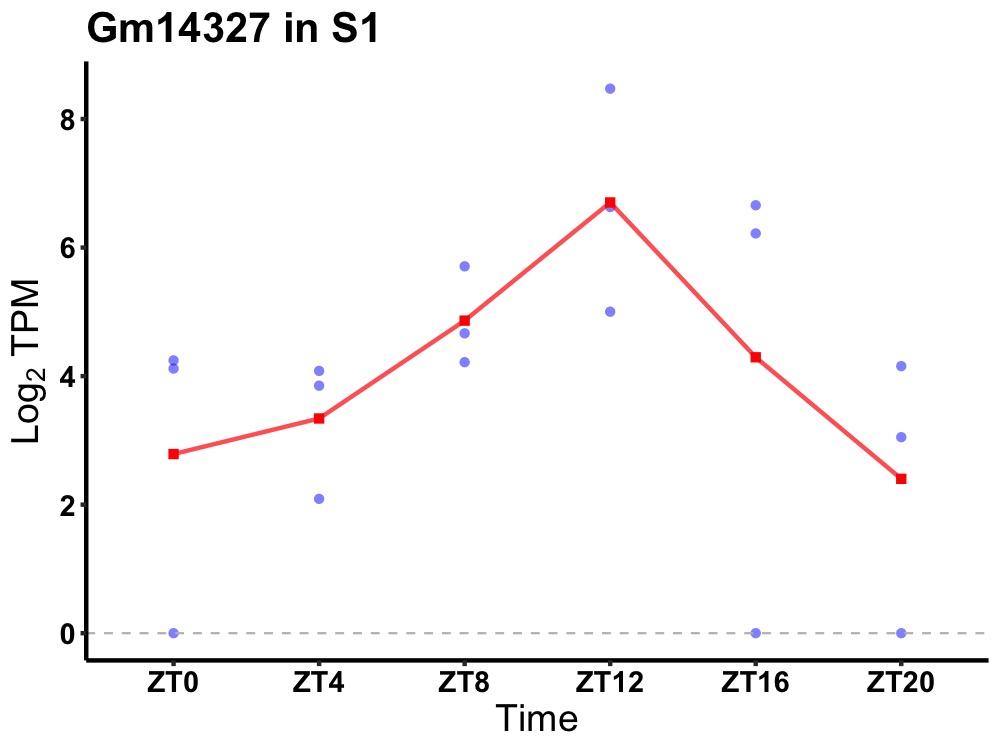 |
none | 0.031 | 20 | 14 | 1.45 |
| ENSMUSG00000021079 | Zbtb43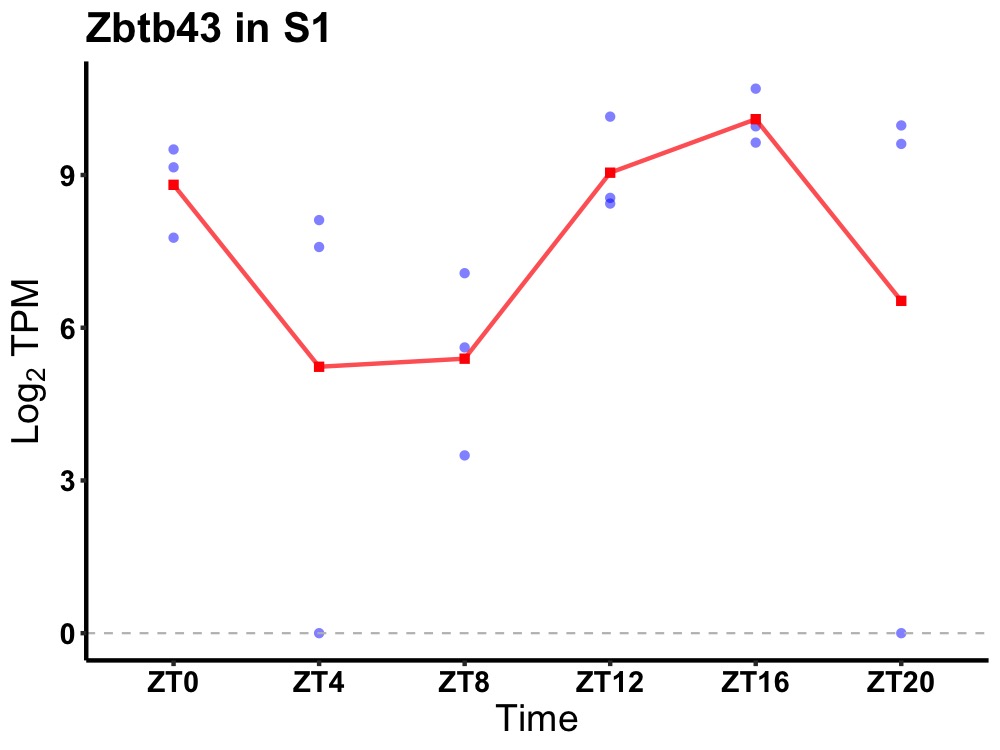 |
zinc finger and BTB domain containing 43 | 0.043 | 24 | 18 | 1.45 |
| ENSMUSG00000017288 | Wdr7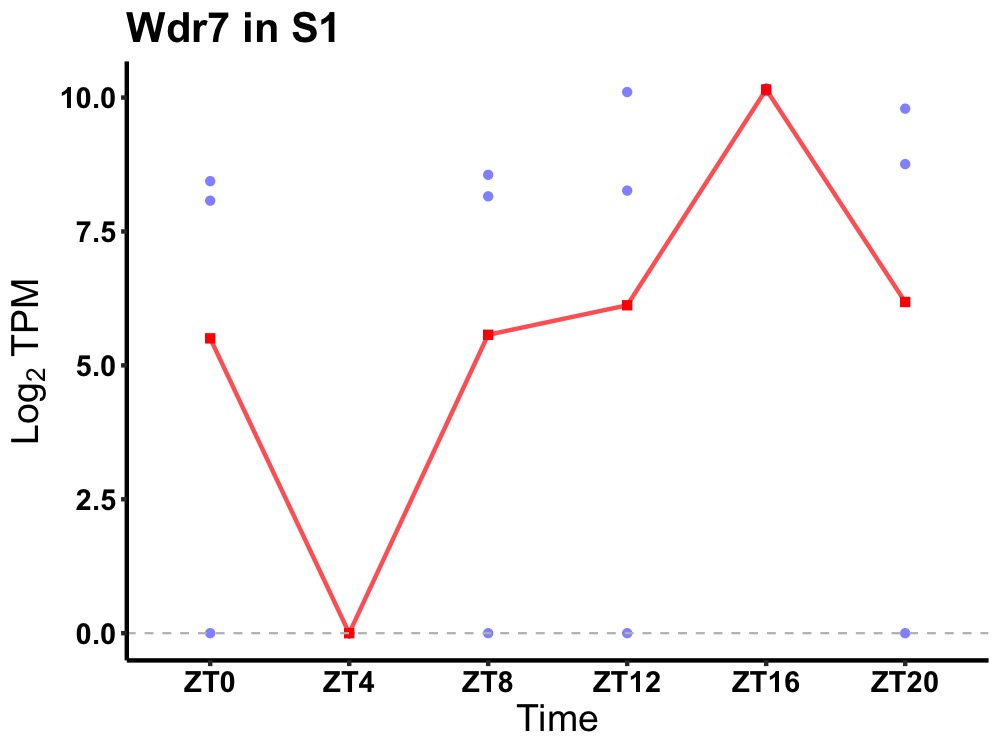 |
WD repeat-containing protein 7 | 0.019 | 24 | 16 | 1.45 |
| ENSMUSG00000096768 | Gm47283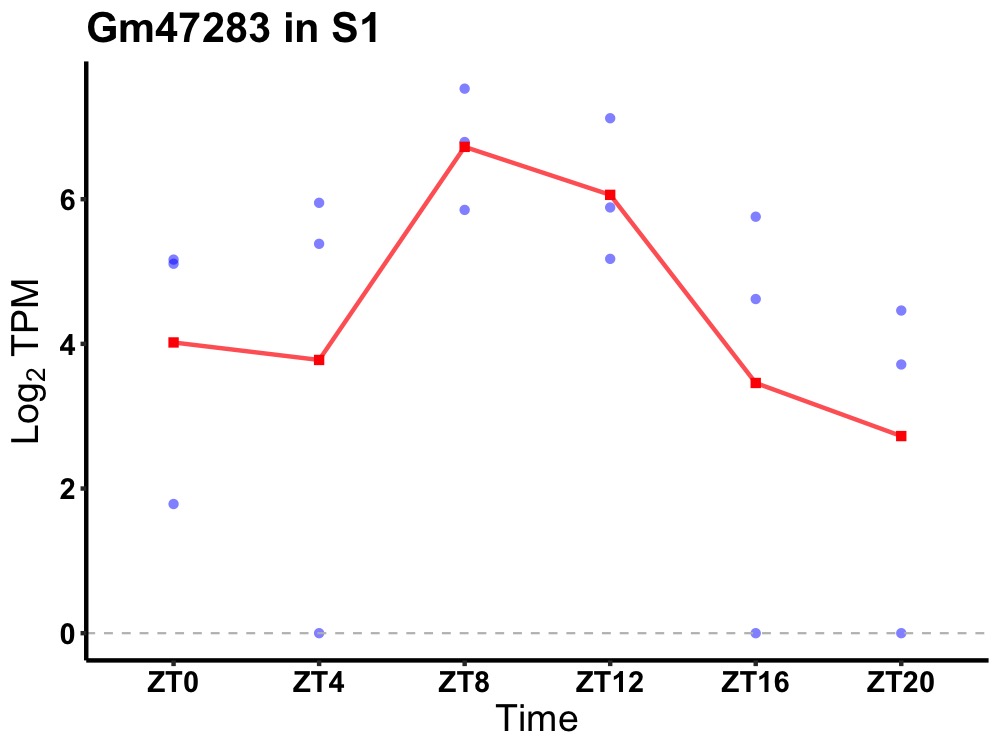 |
predicted gene, 47283 | 0.023 | 24 | 10 | 1.44 |
| ENSMUSG00000003458 | Cirbp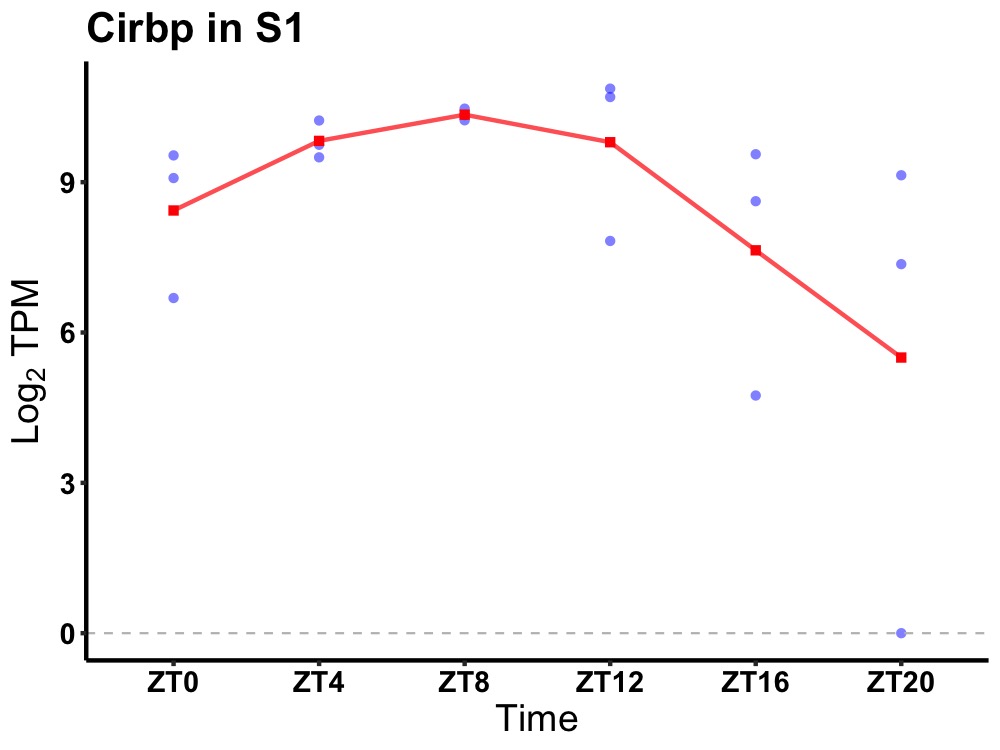 |
cold inducible RNA binding protein | 0.027 | 24 | 10 | 1.44 |
| ENSMUSG00000090264 | Eif4ebp3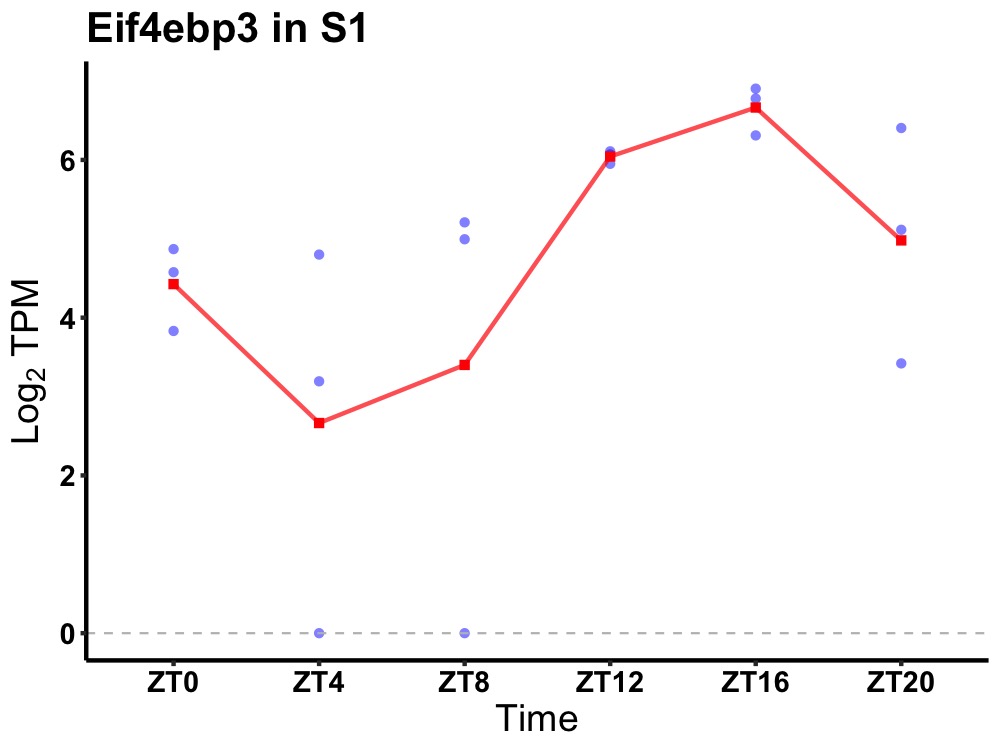 |
eukaryotic translation initiation factor 4E-binding protein 3 | 0.001 | 24 | 16 | 1.41 |
| ENSMUSG00000087361 | 0610043K17Rik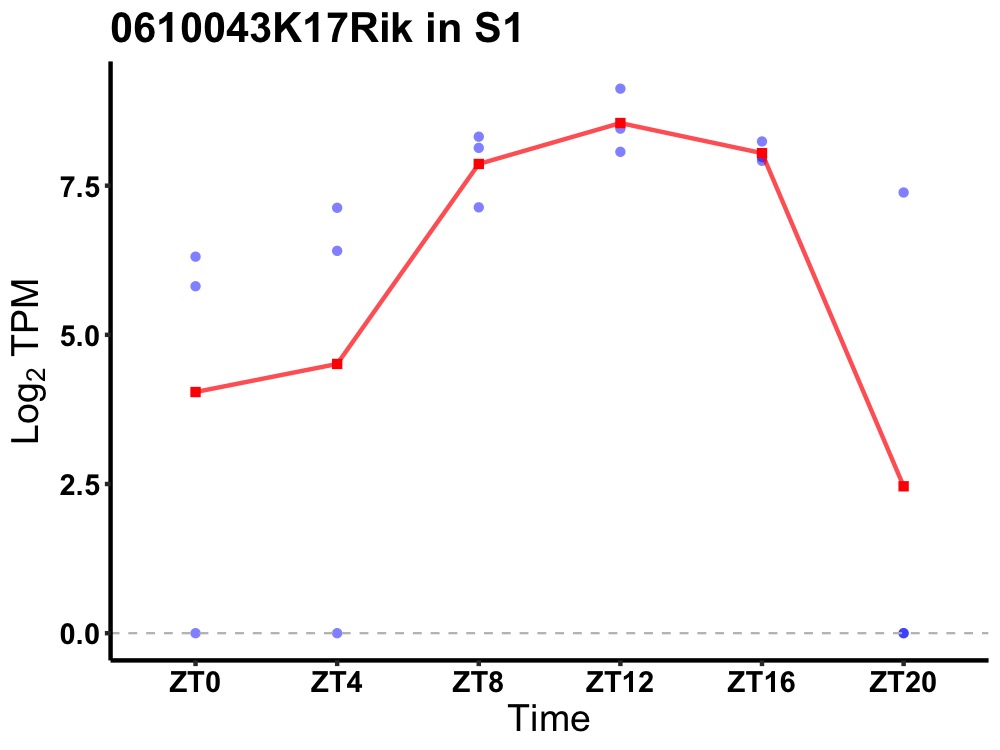 |
none | 0.001 | 20 | 12 | 1.39 |
| ENSMUSG00000020225 | Tbcel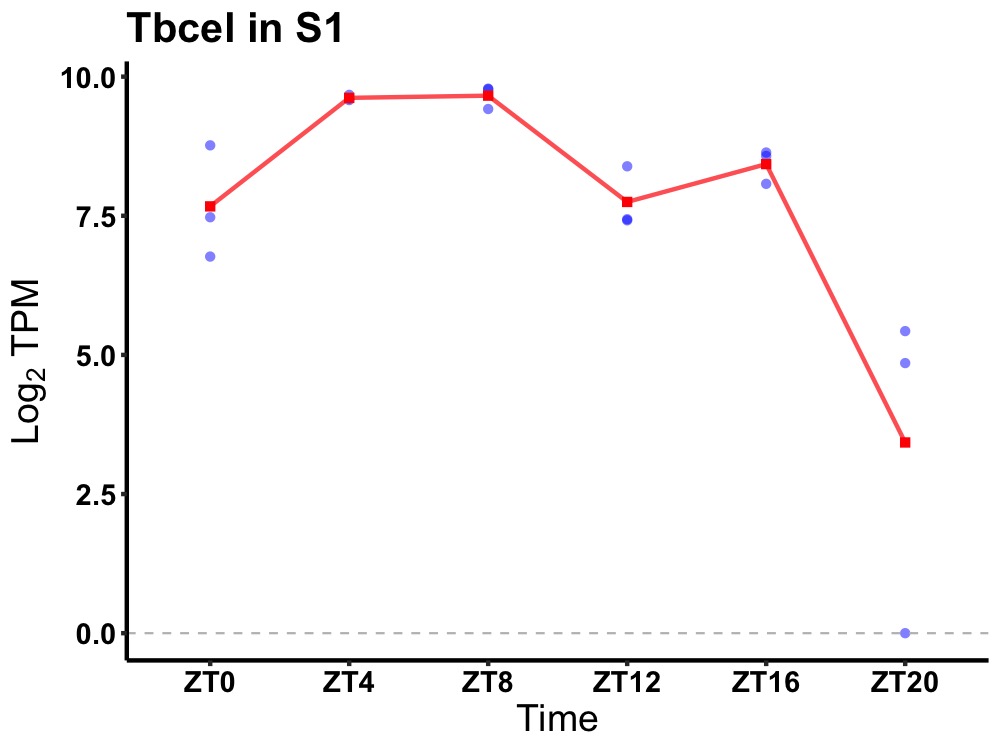 |
tubulin folding cofactor E-like | 0.005 | 24 | 8 | 1.38 |
| ENSMUSG00000022292 | Zc3h7b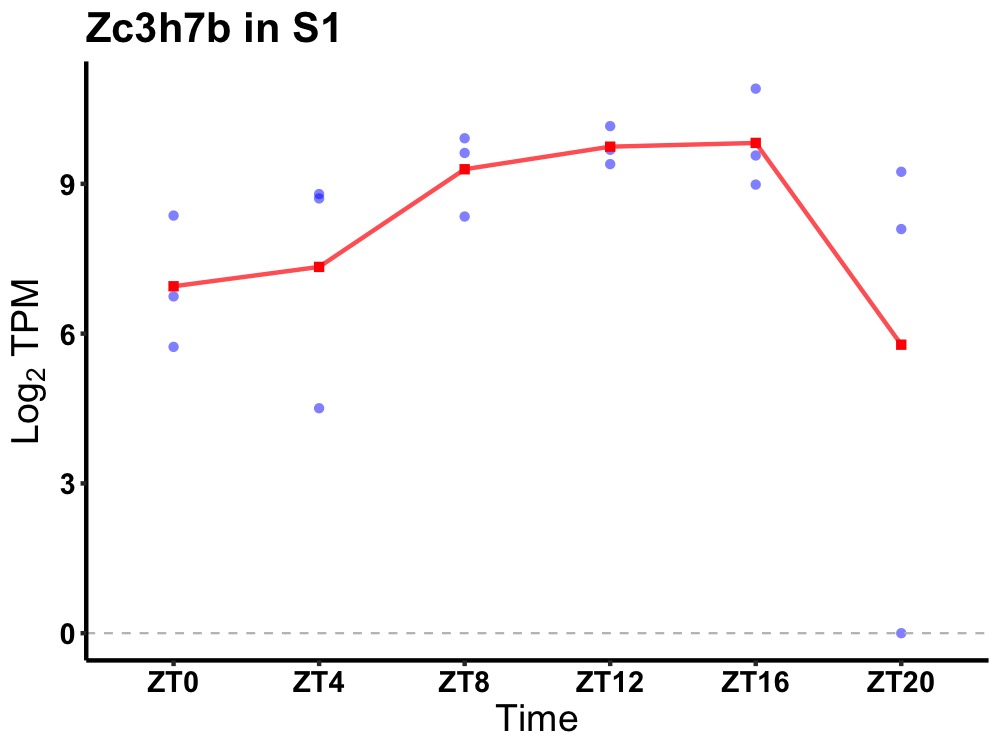 |
zinc finger CCCH type containing 7B | 0.036 | 24 | 12 | 1.37 |
| ENSMUSG00000058873 | Gm5582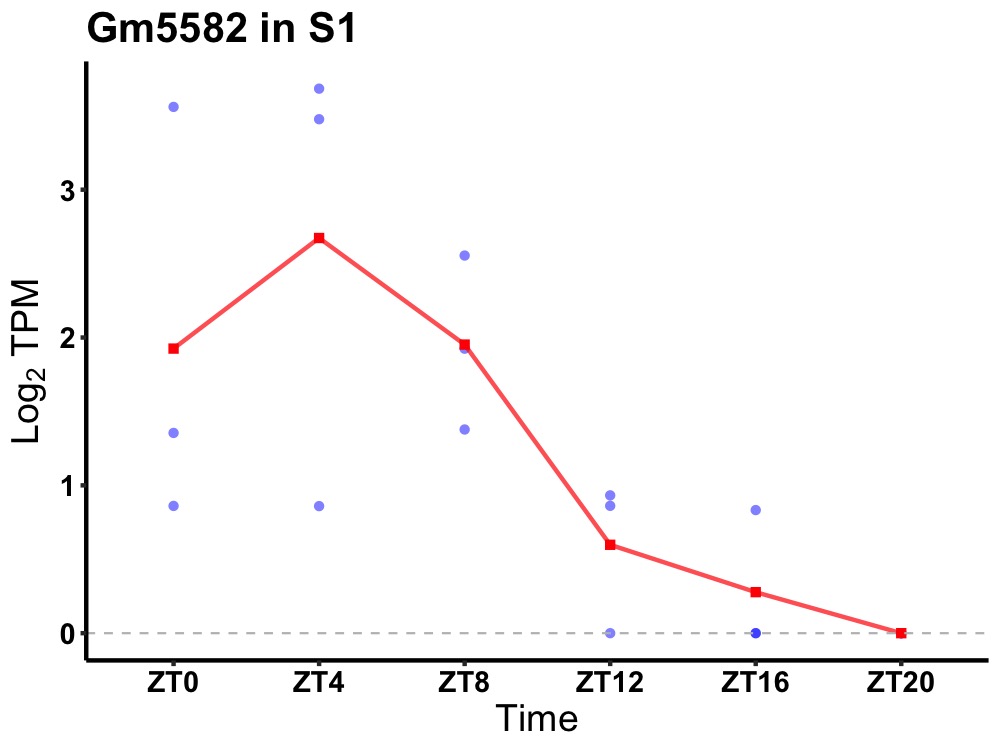 |
none | 0.008 | 24 | 6 | 1.36 |
| ENSMUSG00000032418 | Rnf123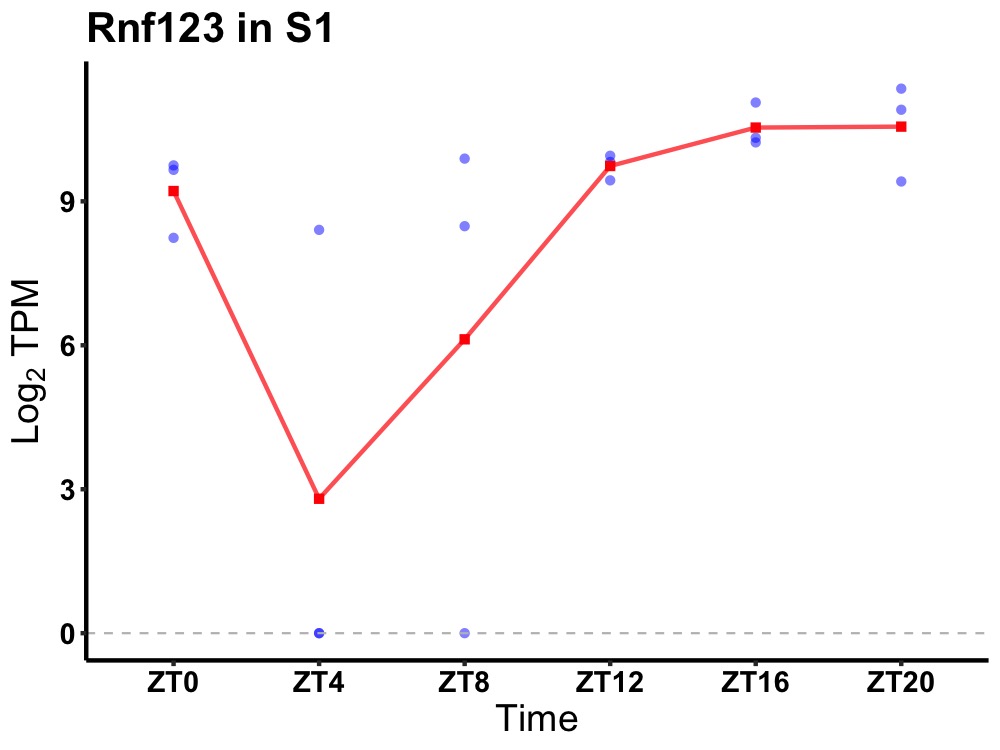 |
E3 ubiquitin-protein ligase RNF123 | 0.005 | 24 | 18 | 1.36 |
| ENSMUSG00000035104 | Rab3ip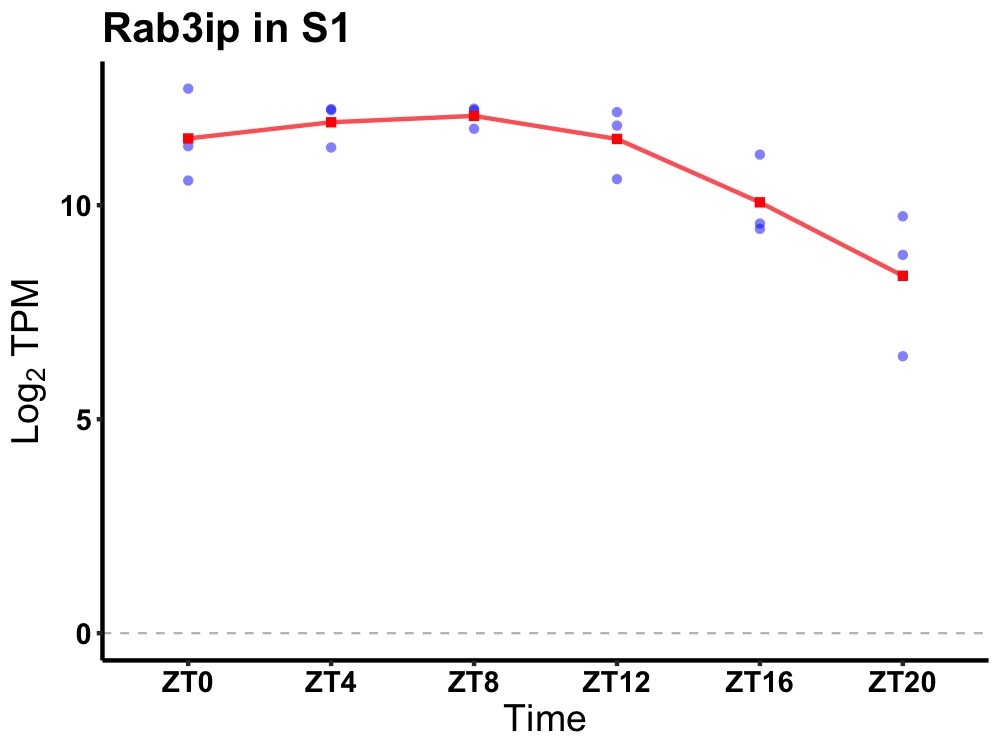 |
RAB3A interacting protein | 0.007 | 24 | 8 | 1.35 |
| ENSMUSG00000020922 | Fbxo40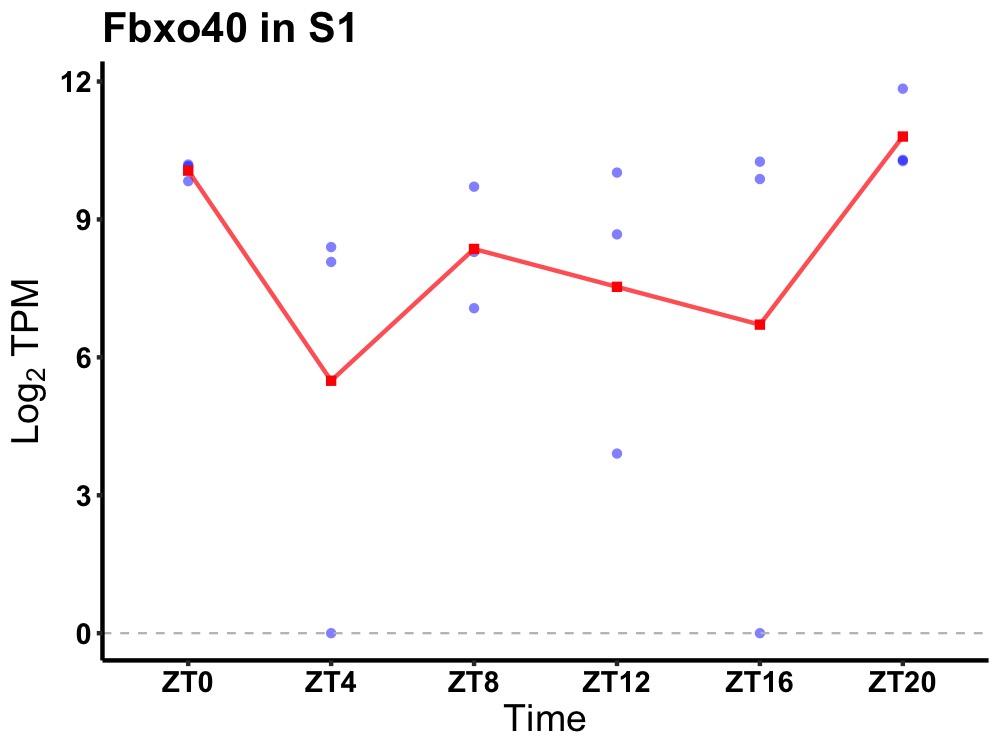 |
F-box only protein 40 | 0.031 | 24 | 20 | 1.34 |
| ENSMUSG00000054387 | Mdm4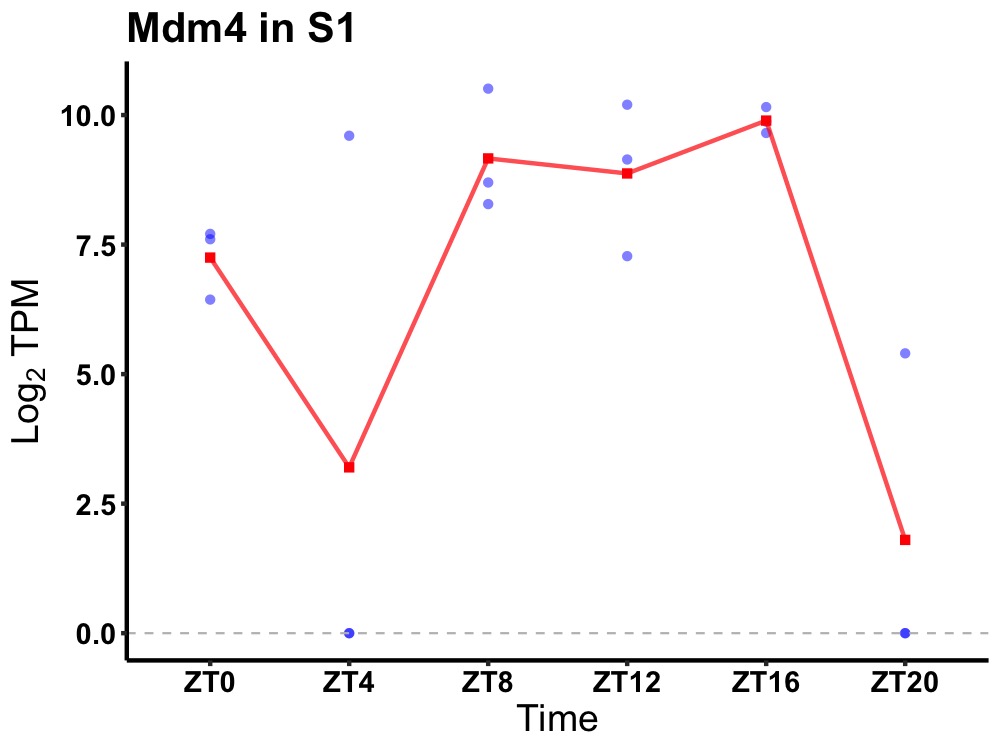 |
protein Mdm4 | 0.036 | 20 | 14 | 1.34 |
| ENSMUSG00000025982 | Marveld3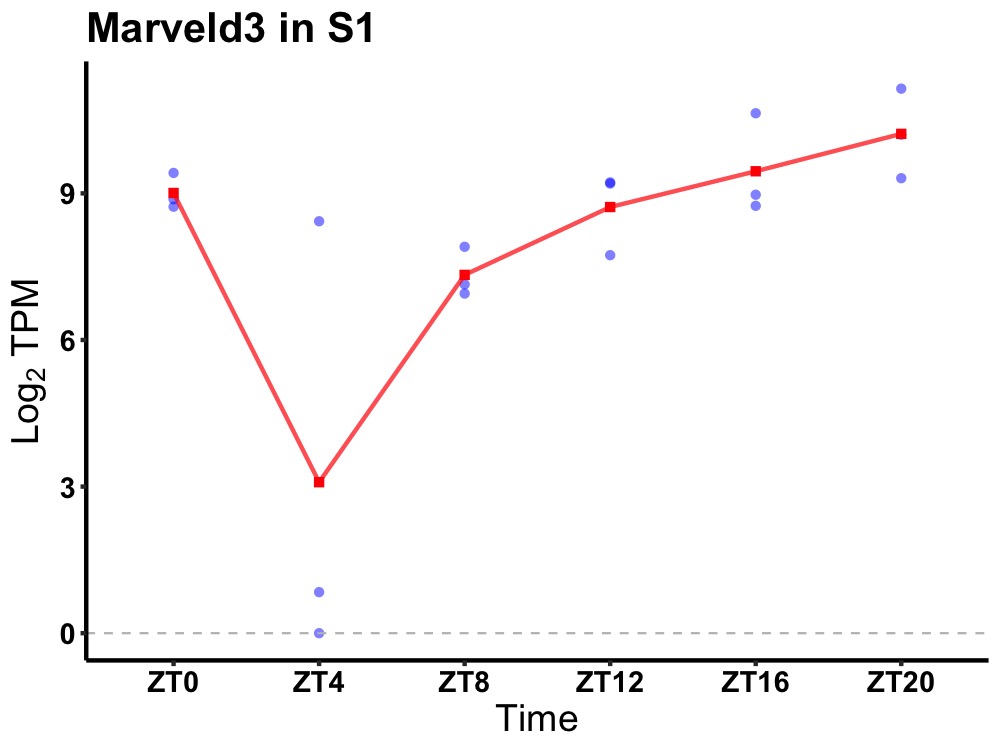 |
MARVEL (membrane-associating) domain containing 3 | 0.005 | 24 | 20 | 1.31 |
| ENSMUSG00000025487 | Otud7b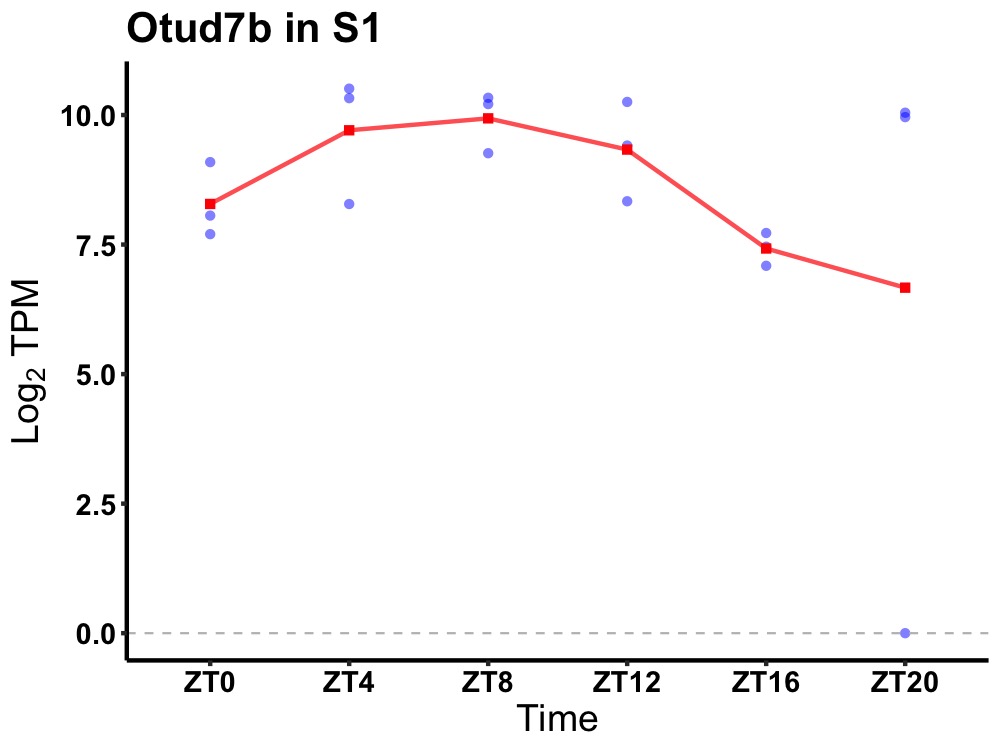 |
OTU domain containing 7B | 0.036 | 20 | 8 | 1.28 |
| ENSMUSG00000050199 | Sgk1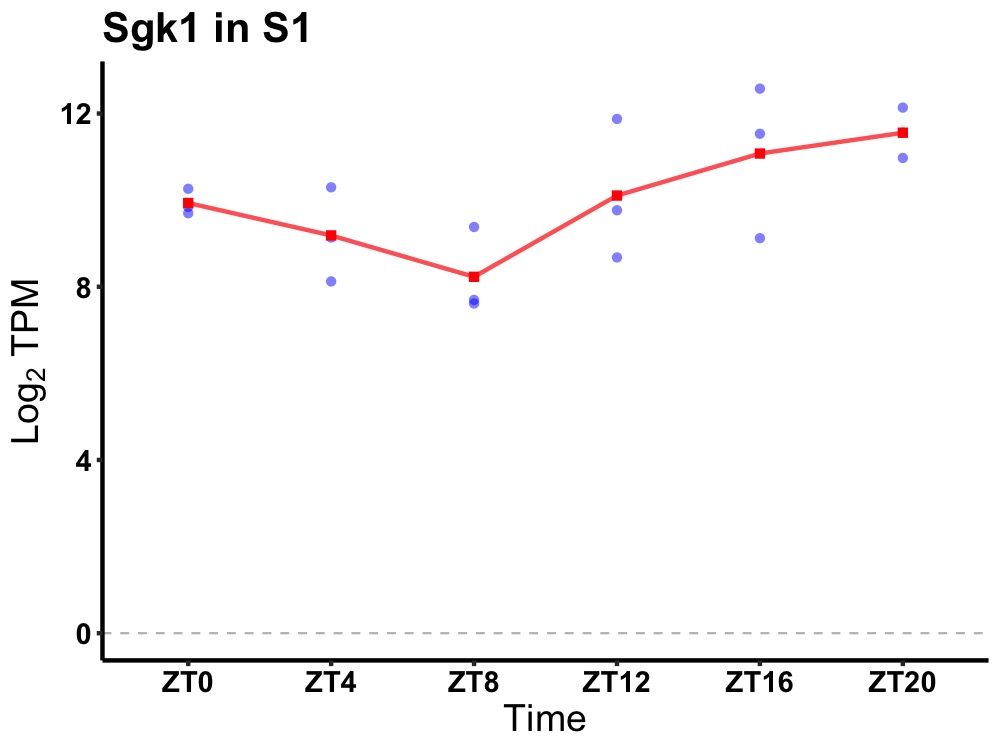 |
serum/glucocorticoid regulated kinase 1 | 0.036 | 24 | 20 | 1.27 |
| ENSMUSG00000089785 | Nvl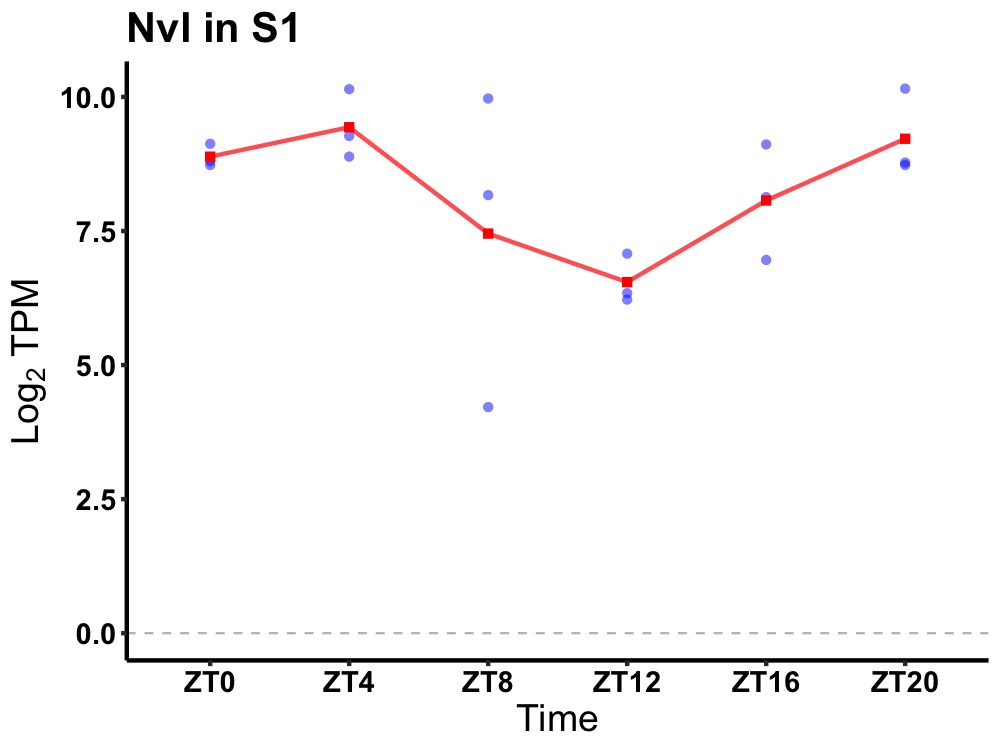 |
nuclear VCP-like | 0.049 | 20 | 4 | 1.27 |
| ENSMUSG00000083392 | Cse1l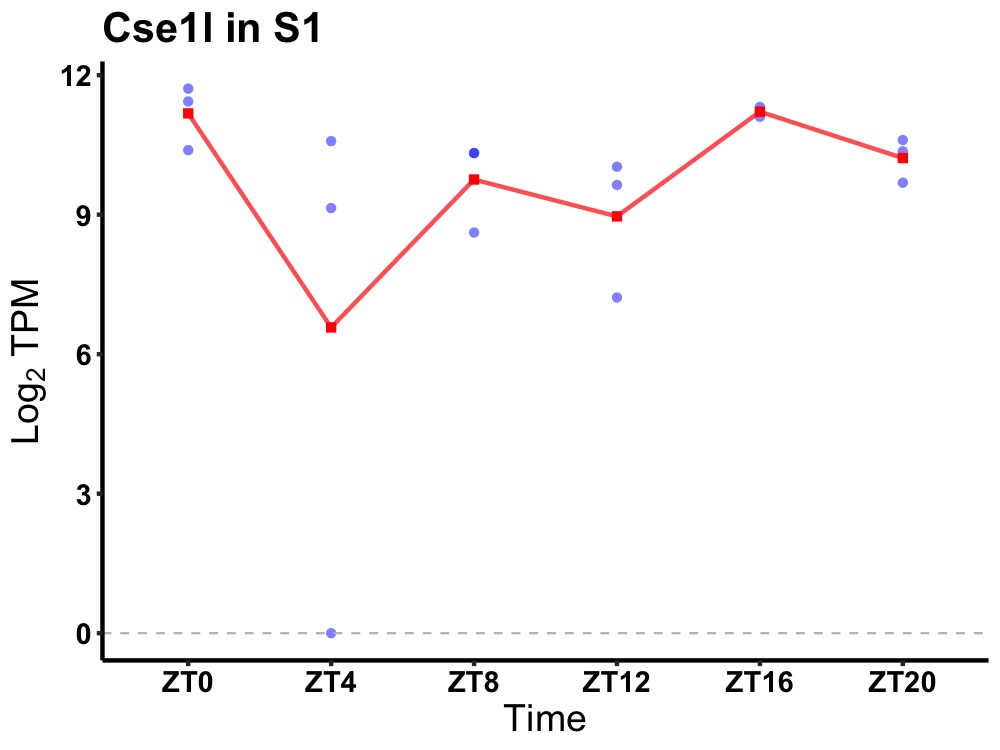 |
chromosome segregation 1-like (S. cerevisiae) | 0.049 | 20 | 18 | 1.26 |
| ENSMUSG00000115049 | Gm49172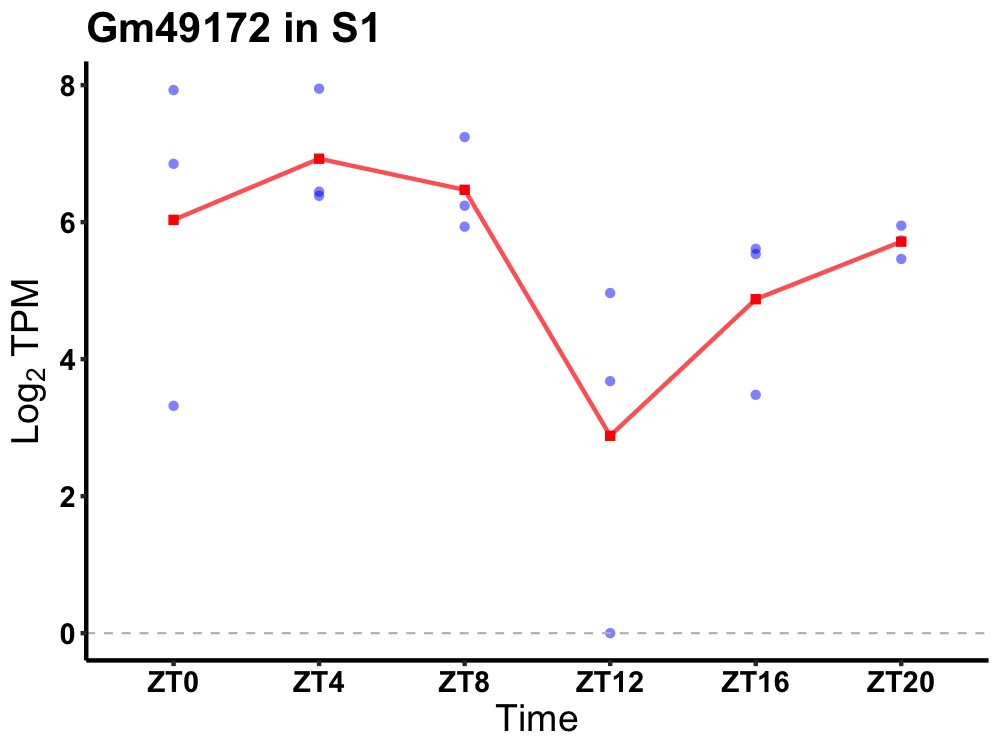 |
predicted gene, 49172 | 0.036 | 24 | 4 | 1.26 |
| ENSMUSG00000033427 | Mknk2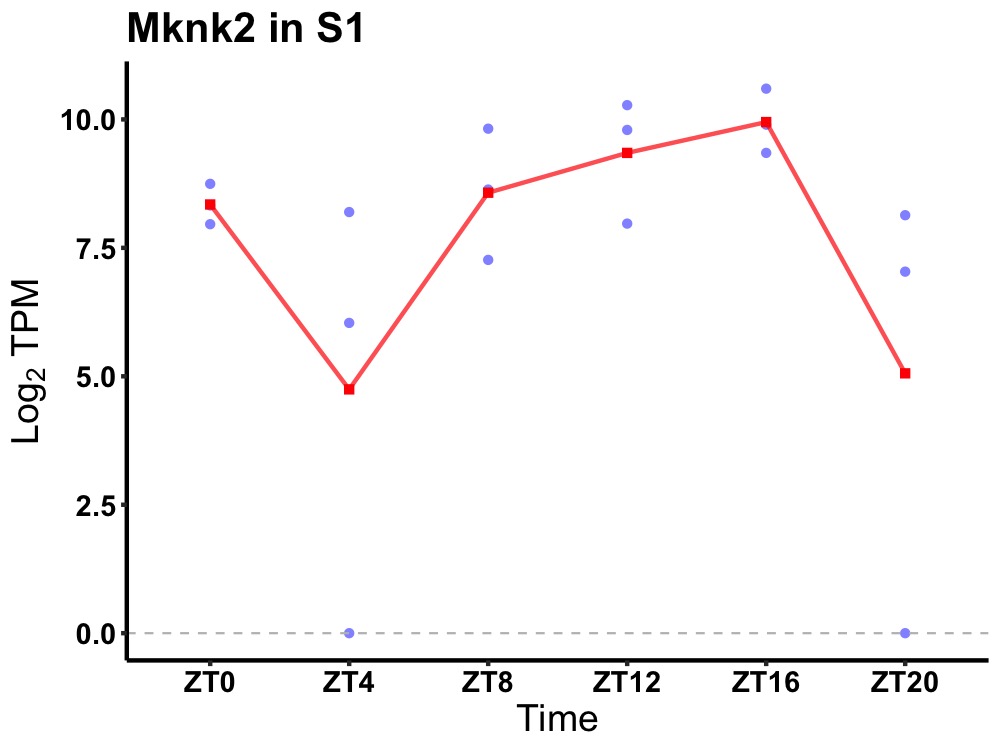 |
MAP kinase-interacting serine/threonine kinase 2 | 0.023 | 20 | 14 | 1.26 |
| ENSMUSG00000039361 | Ggps1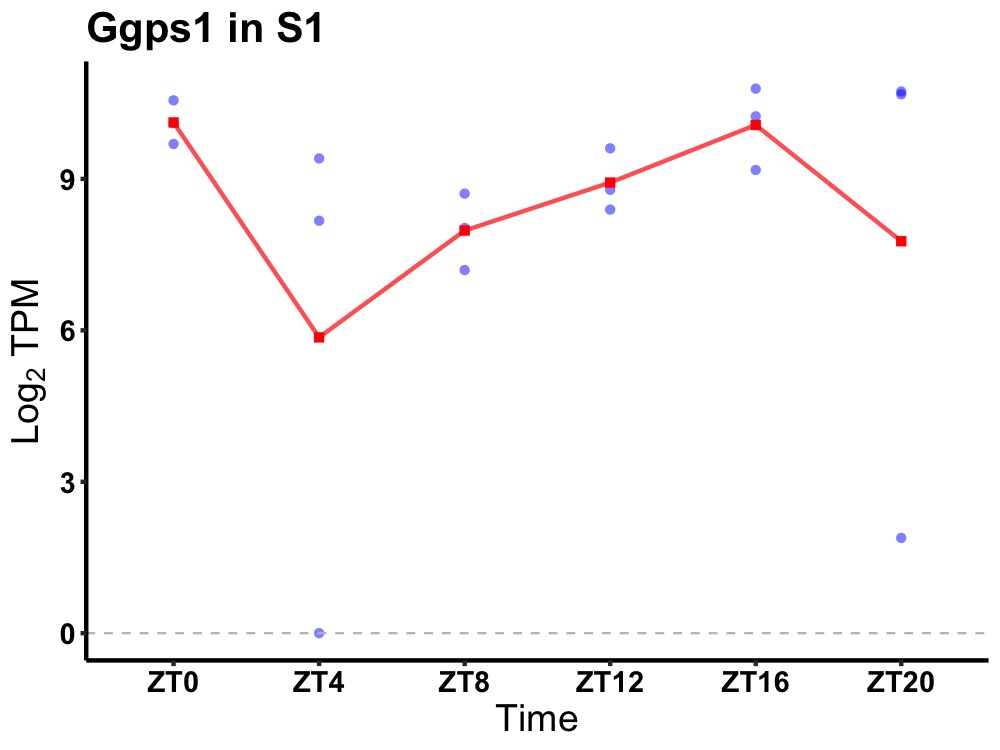 |
geranylgeranyl diphosphate synthase 1 | 0.049 | 24 | 20 | 1.25 |
| ENSMUSG00000115593 | Tsc22d3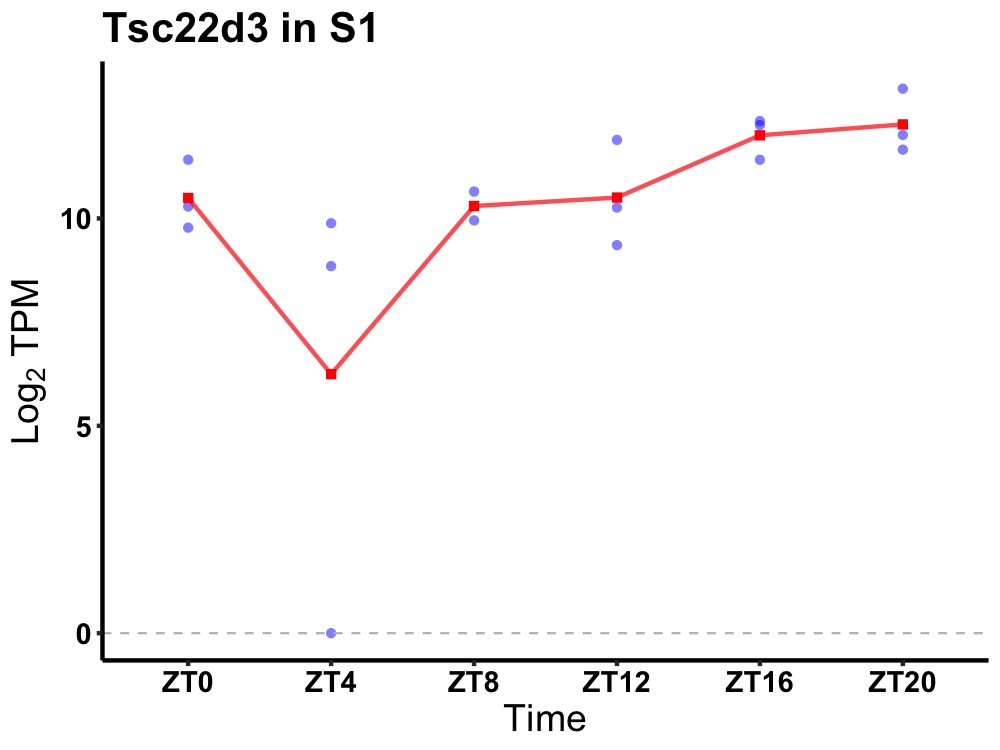 |
TSC22 domain family, member 3 | 0.014 | 24 | 18 | 1.24 |
| ENSMUSG00000040998 | Sephs1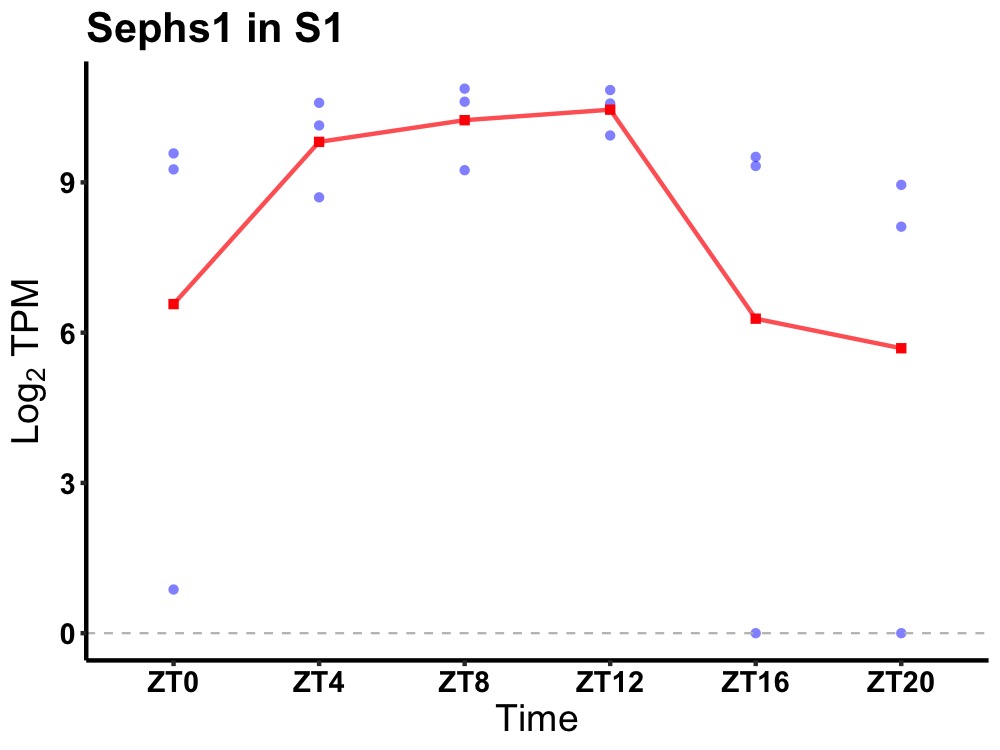 |
selenophosphate synthetase 1 | 0.031 | 24 | 10 | 1.23 |
| ENSMUSG00000032745 | Metap1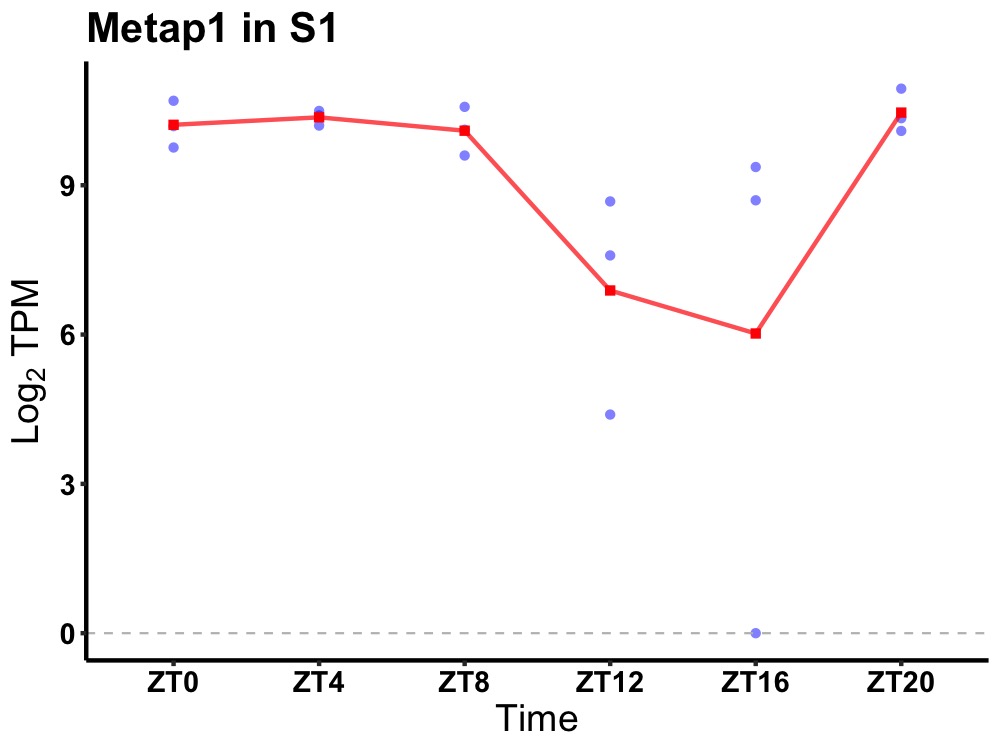 |
methionyl aminopeptidase 1 | 0.005 | 20 | 4 | 1.23 |
| ENSMUSG00000115503 | Nat8f5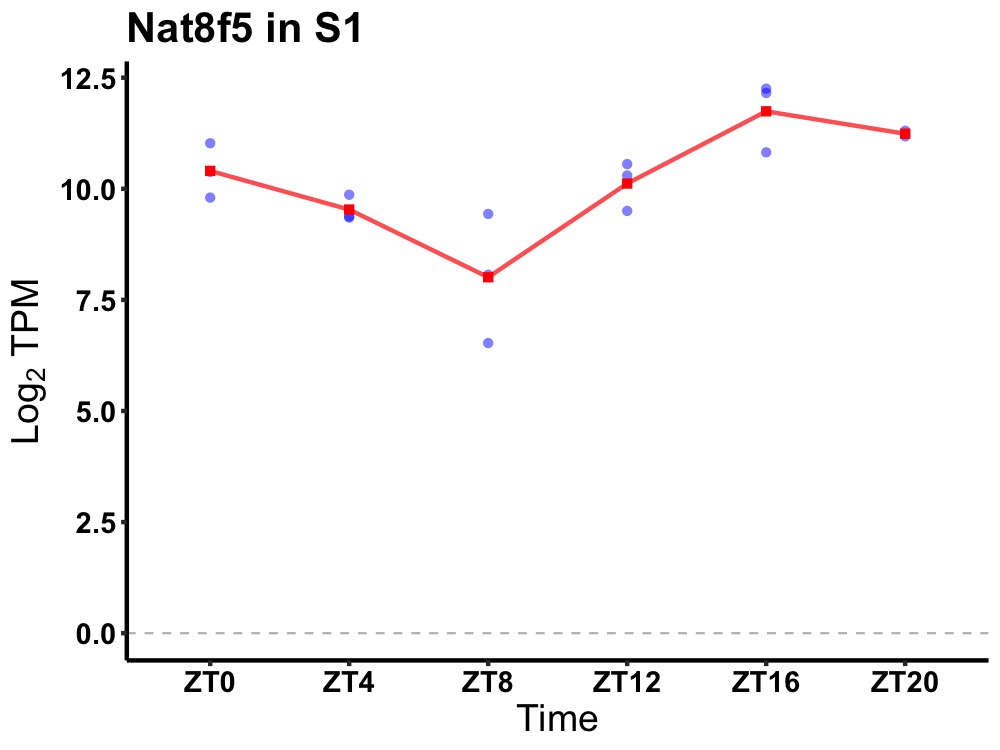 |
N-acetyltransferase 8 (GCN5-related) family member 5 | 0.000 | 24 | 20 | 1.22 |
| ENSMUSG00000032399 | Fbxo6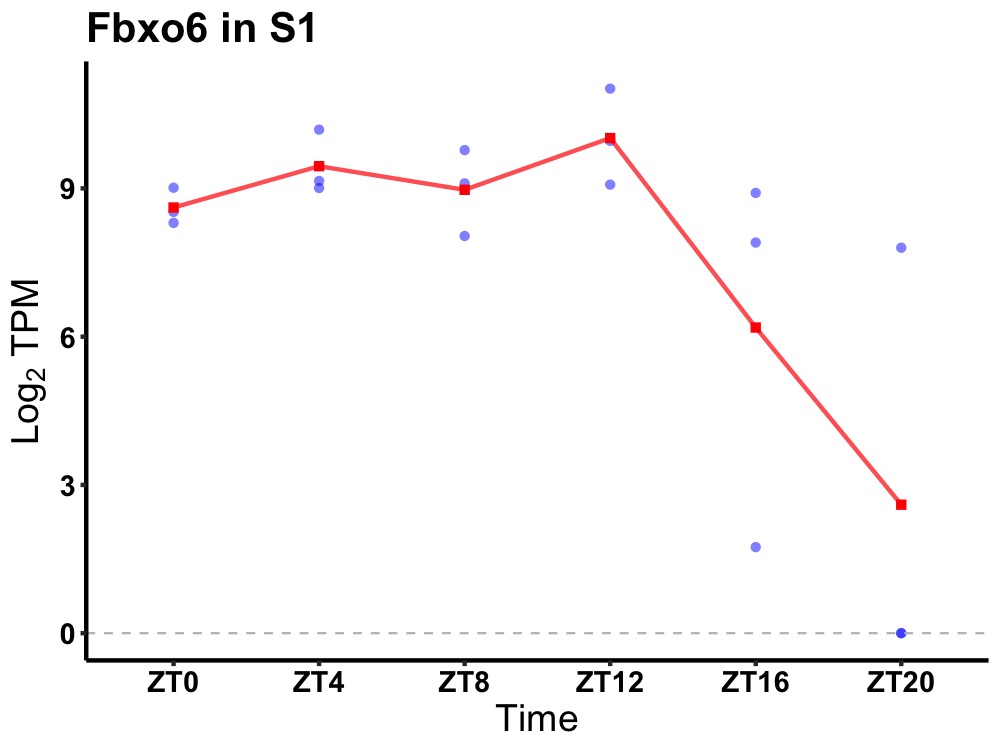 |
F-box protein 6 | 0.027 | 24 | 8 | 1.22 |
| ENSMUSG00000028150 | Tbrg4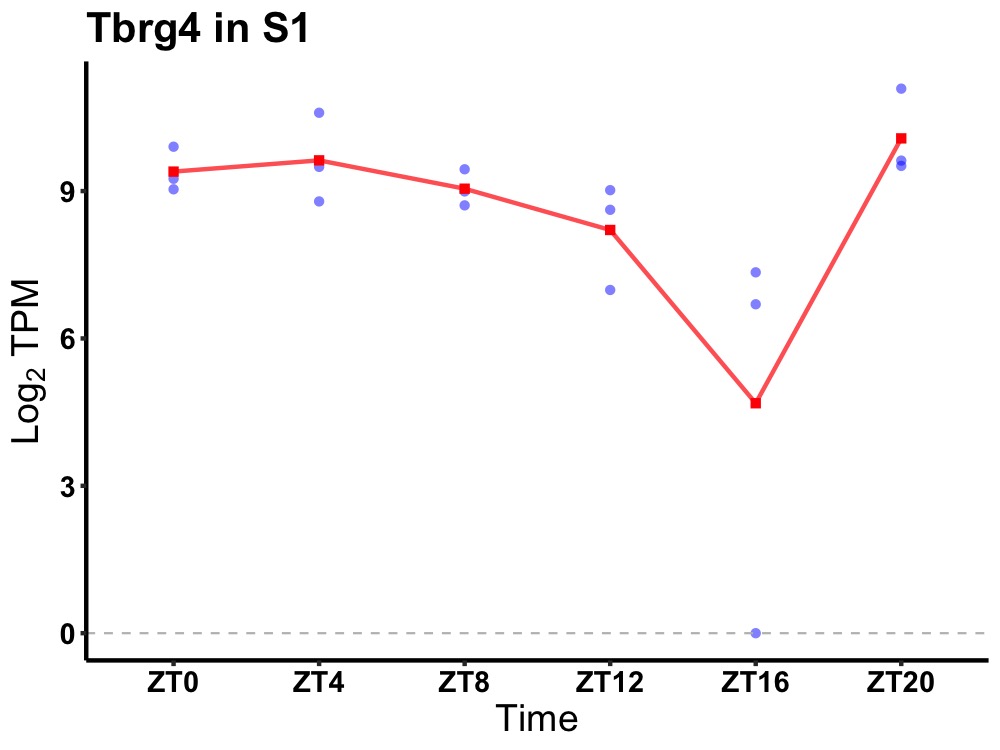 |
transforming growth factor beta regulated gene 4 | 0.036 | 20 | 4 | 1.19 |
| ENSMUSG00000022951 | Lactb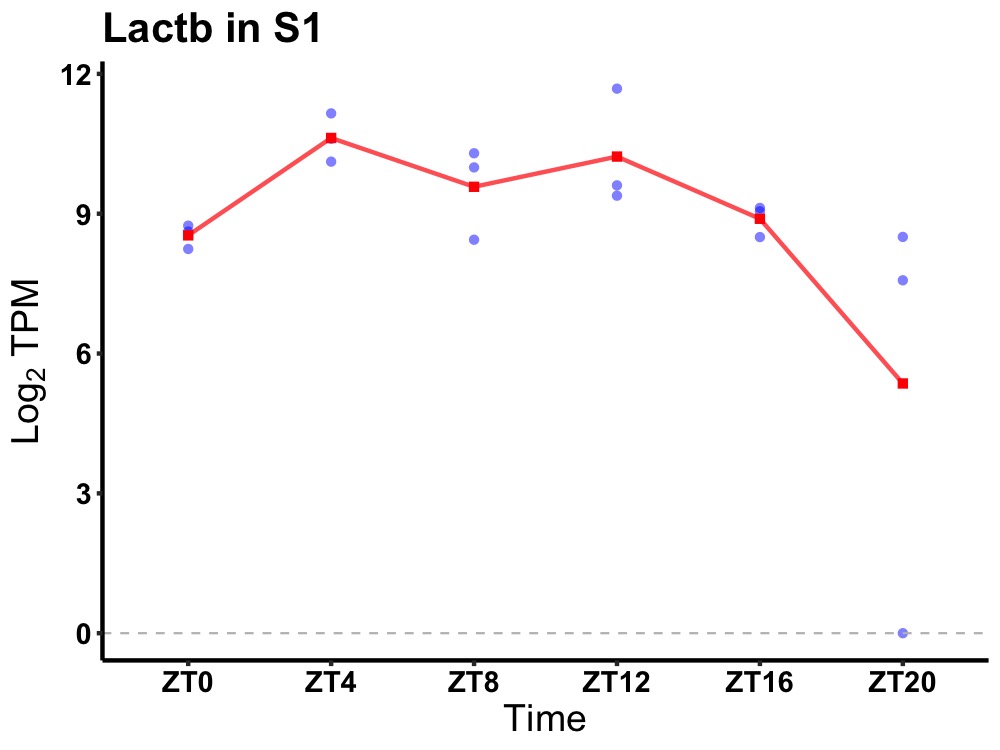 |
lactamase, beta | 0.049 | 24 | 10 | 1.19 |
| ENSMUSG00000081046 | Gm12846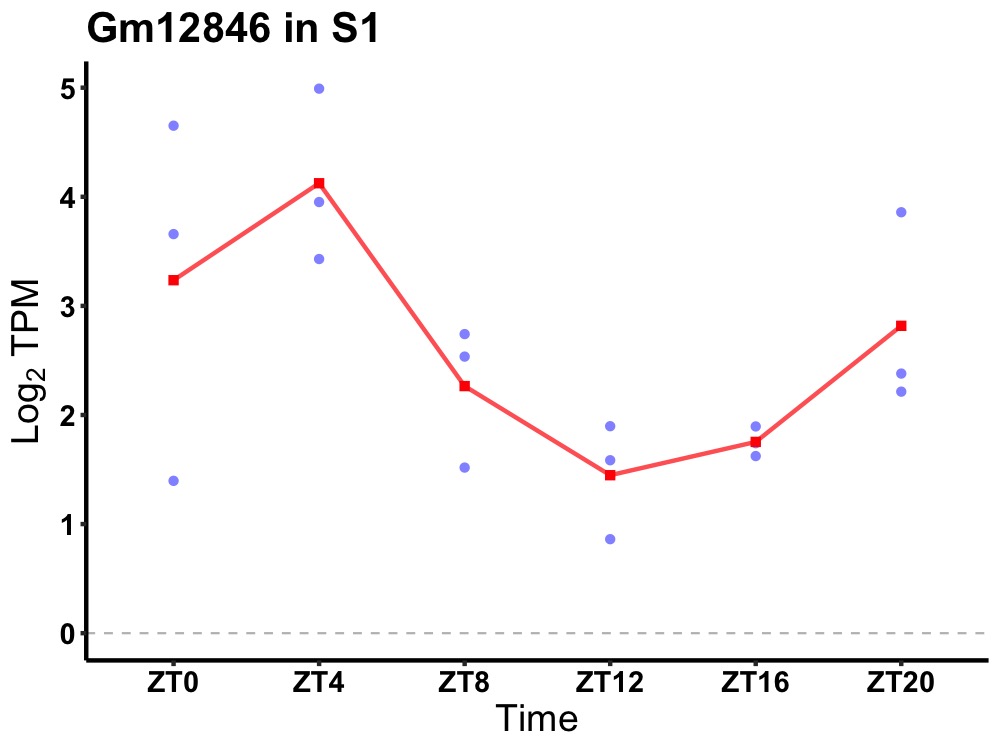 |
predicted gene 12846 | 0.049 | 20 | 4 | 1.17 |
| ENSMUSG00000111108 | Gm17795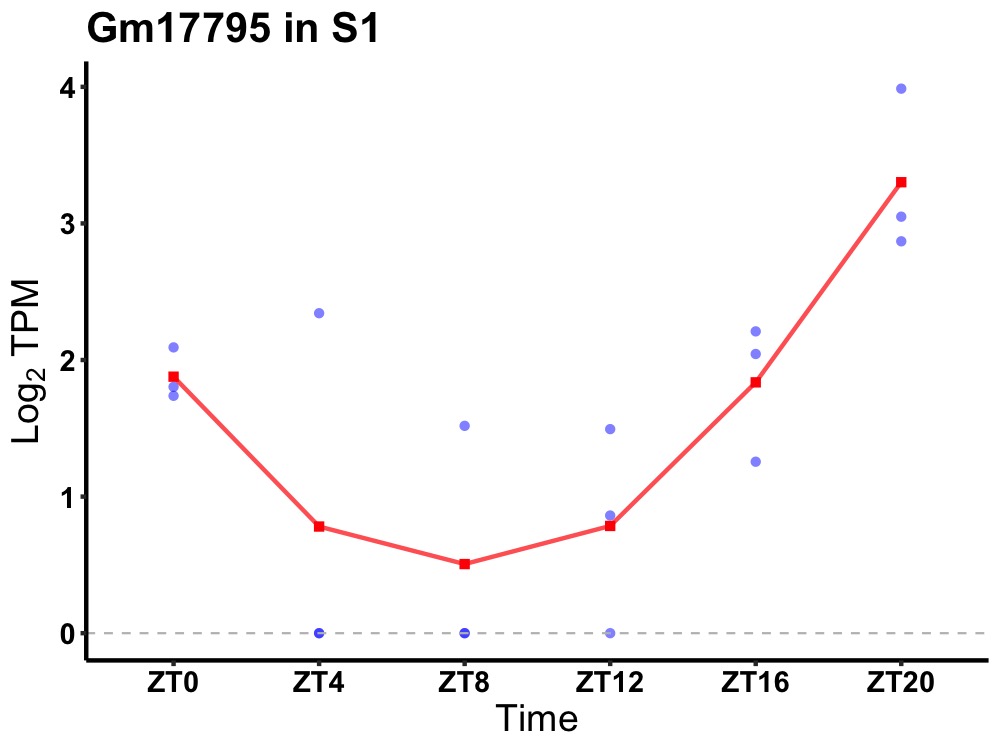 |
none | 0.010 | 24 | 20 | 1.16 |
| ENSMUSG00000054426 | A930005H10Rik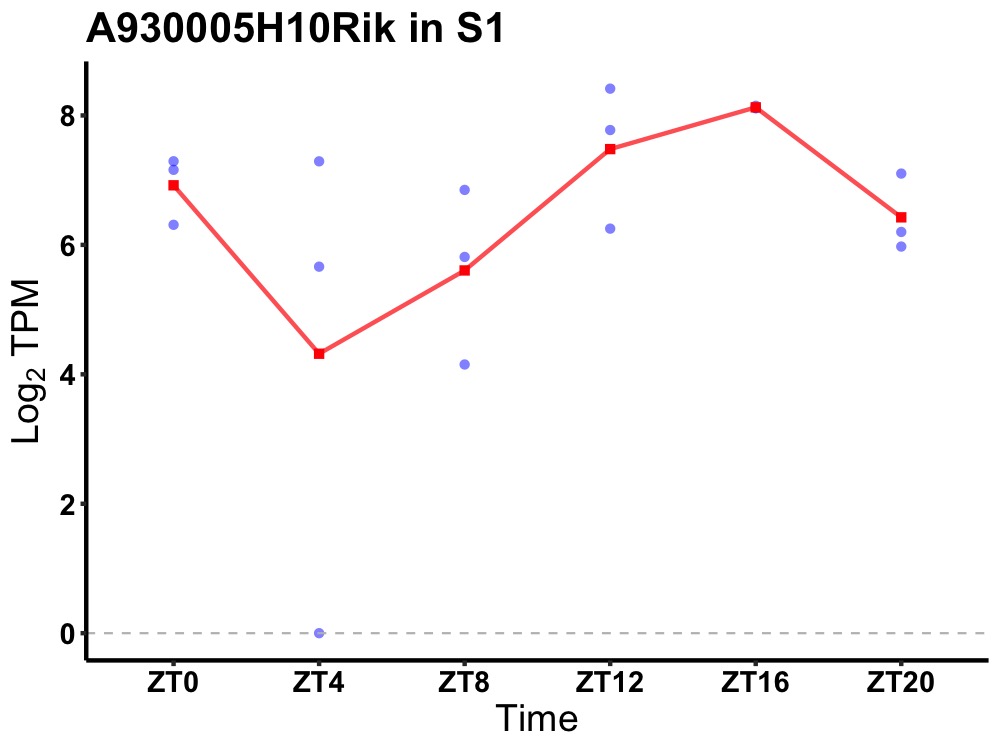 |
none | 0.005 | 20 | 16 | 1.16 |
| ENSMUSG00000029804 | Asb9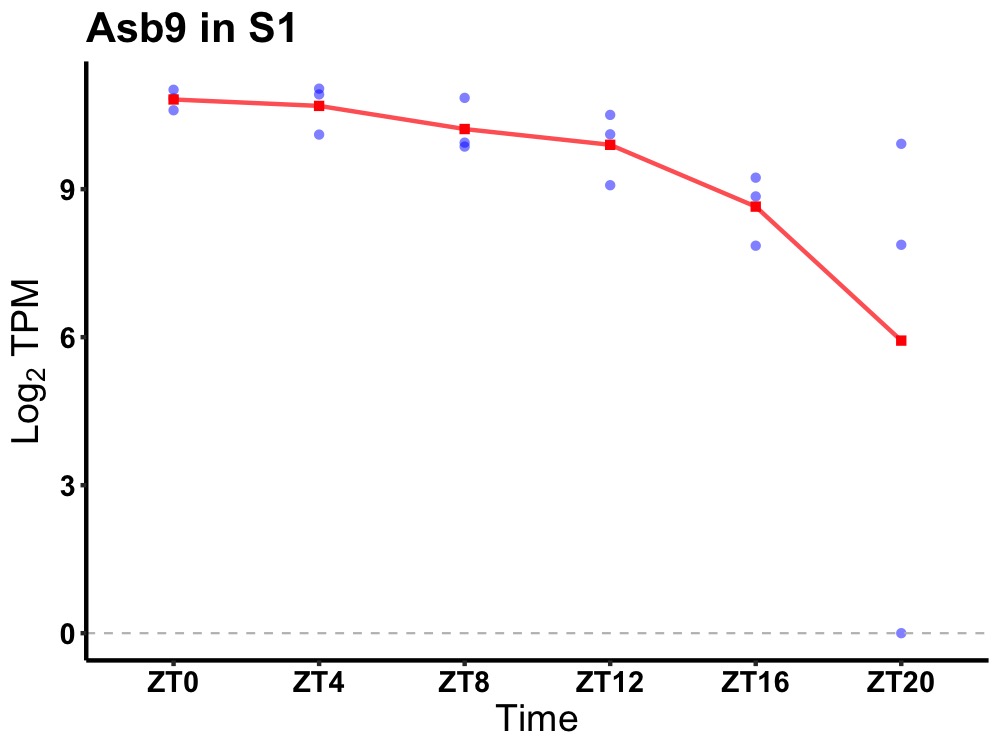 |
ankyrin repeat and SOCS box-containing 9 | 0.019 | 24 | 6 | 1.14 |
| ENSMUSG00000034875 | Mga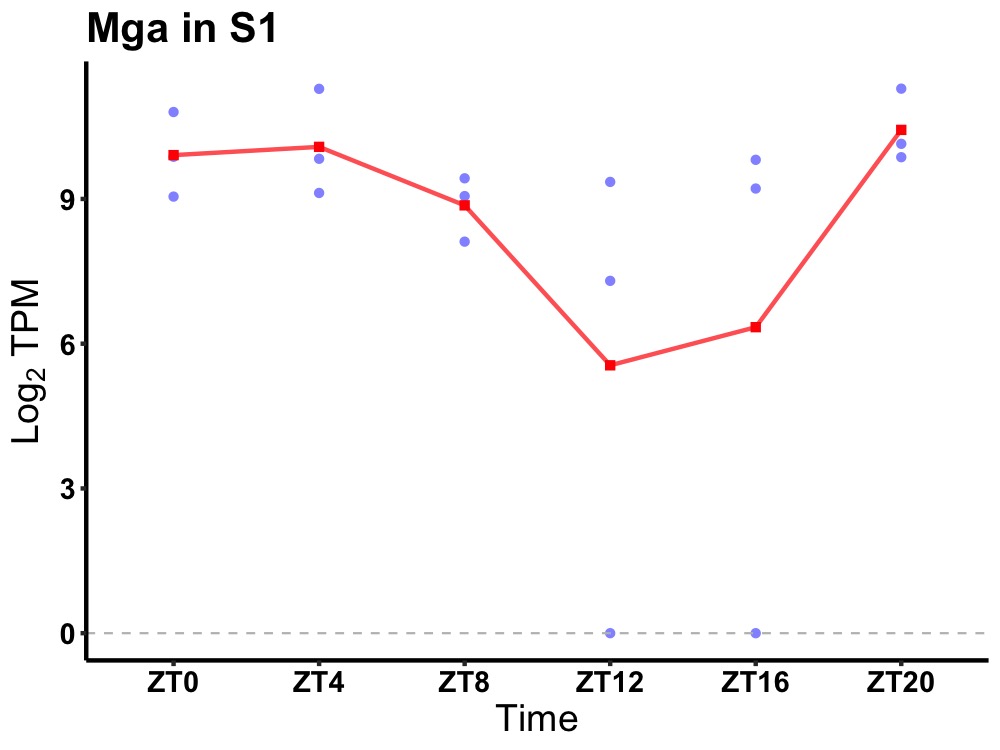 |
MAX gene-associated protein | 0.043 | 20 | 2 | 1.14 |
| ENSMUSG00000079942 | 2210408F21Rik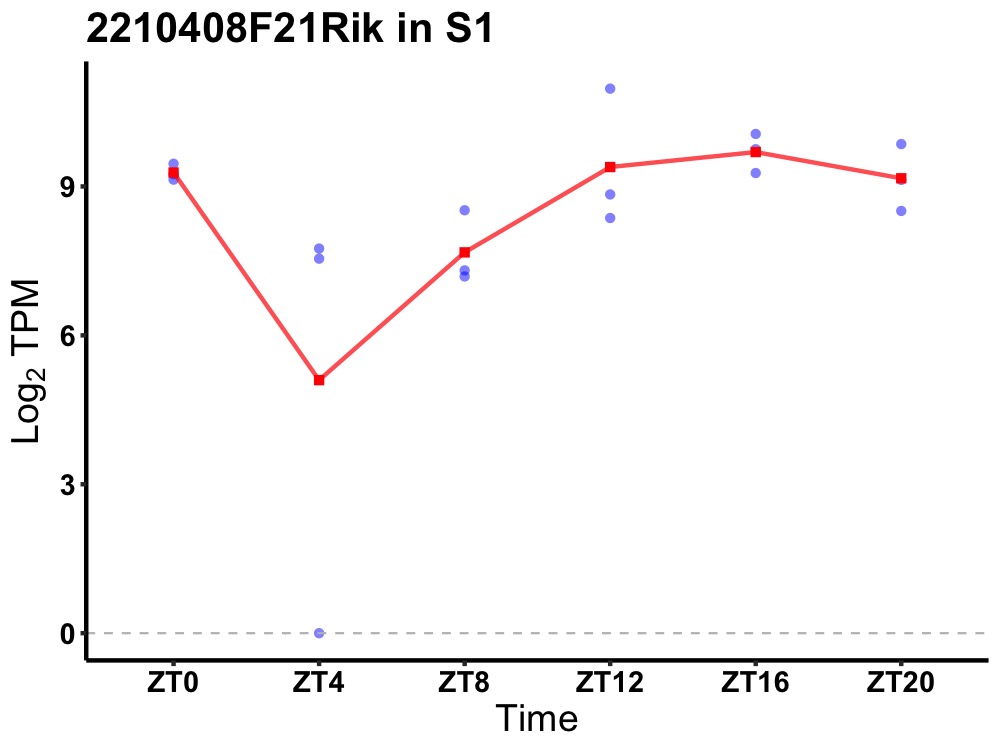 |
RIKEN cDNA 2210408F21 gene | 0.007 | 20 | 18 | 1.13 |
| ENSMUSG00000024570 | Wsb2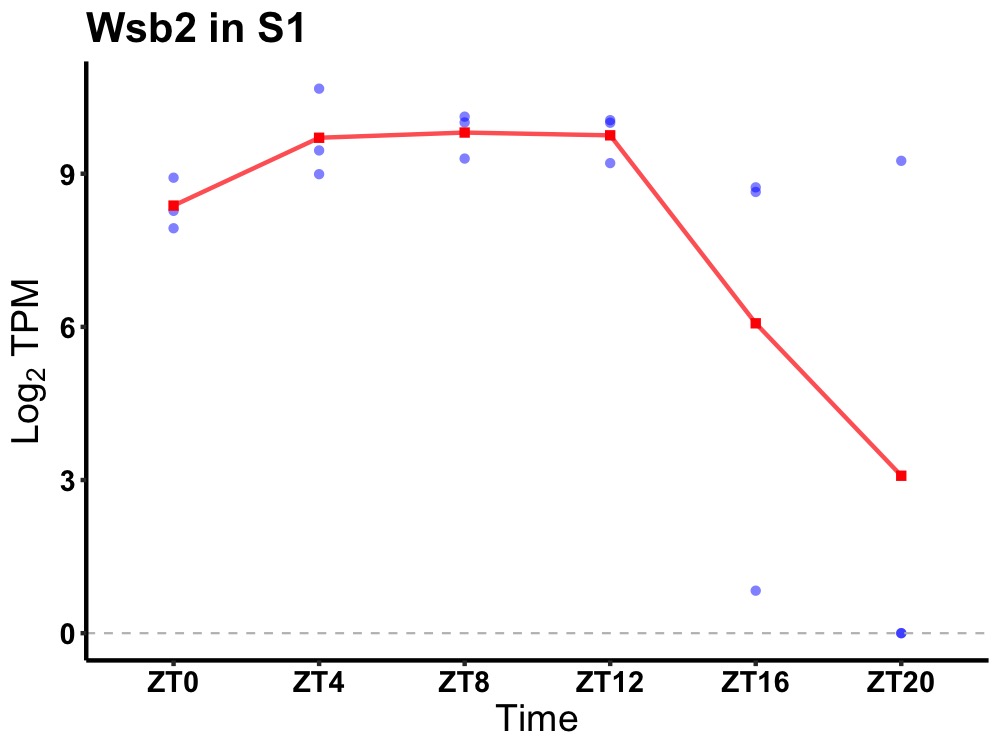 |
WD repeat and SOCS box-containing protein 2 | 0.010 | 24 | 10 | 1.13 |
| ENSMUSG00000037754 | Eif3b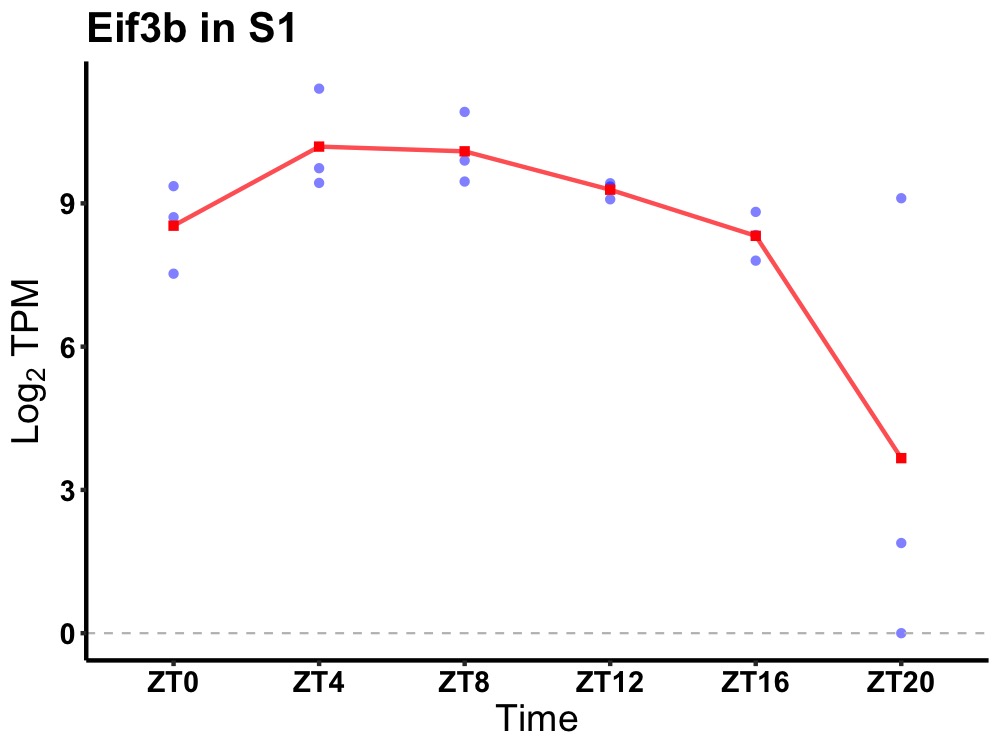 |
eukaryotic translation initiation factor 3, subunit B | 0.000 | 24 | 8 | 1.10 |
| ENSMUSG00000034729 | Prok1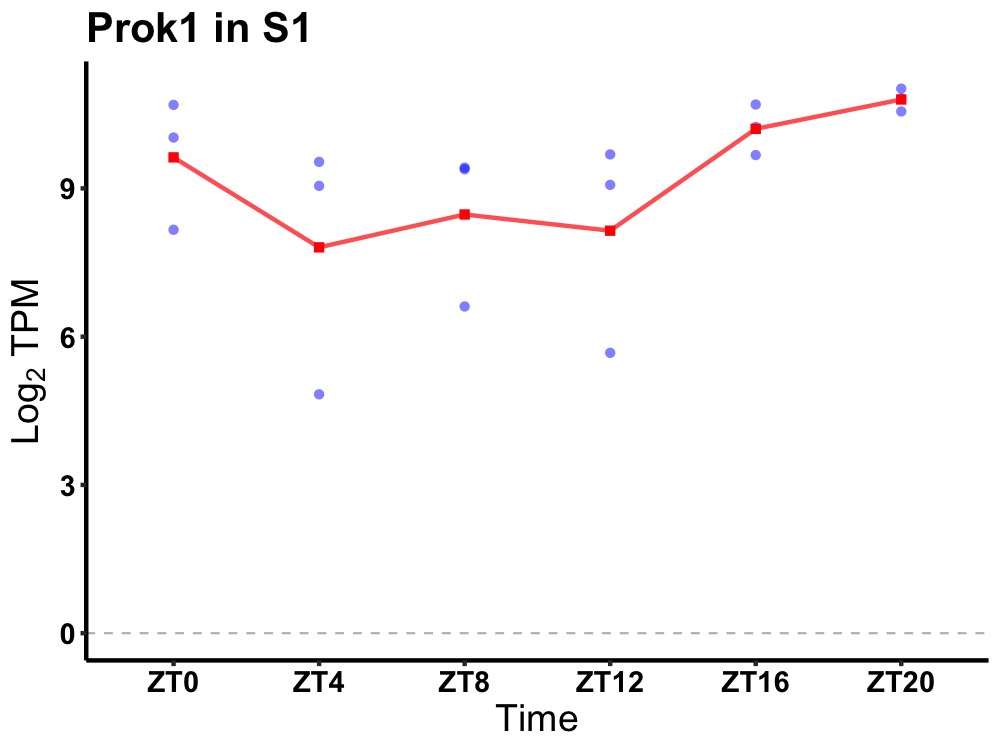 |
prokineticin 1 | 0.014 | 24 | 20 | 1.10 |
| ENSMUSG00000019920 | Stra6l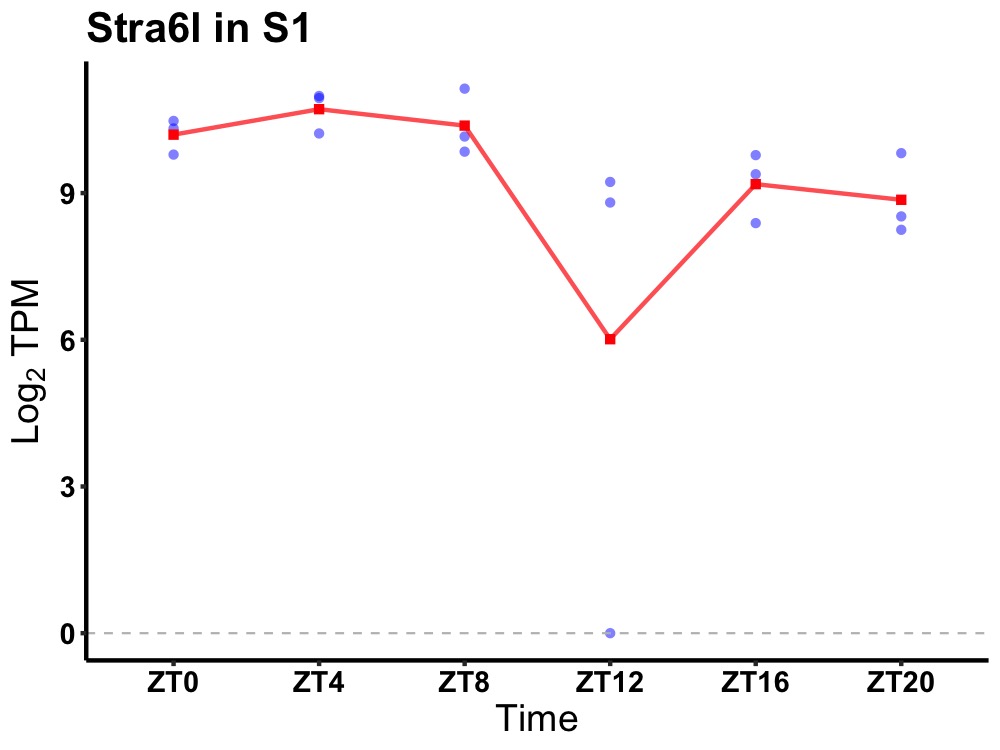 |
STRA6-like | 0.027 | 24 | 6 | 1.09 |
| ENSMUSG00000026281 | Osgin1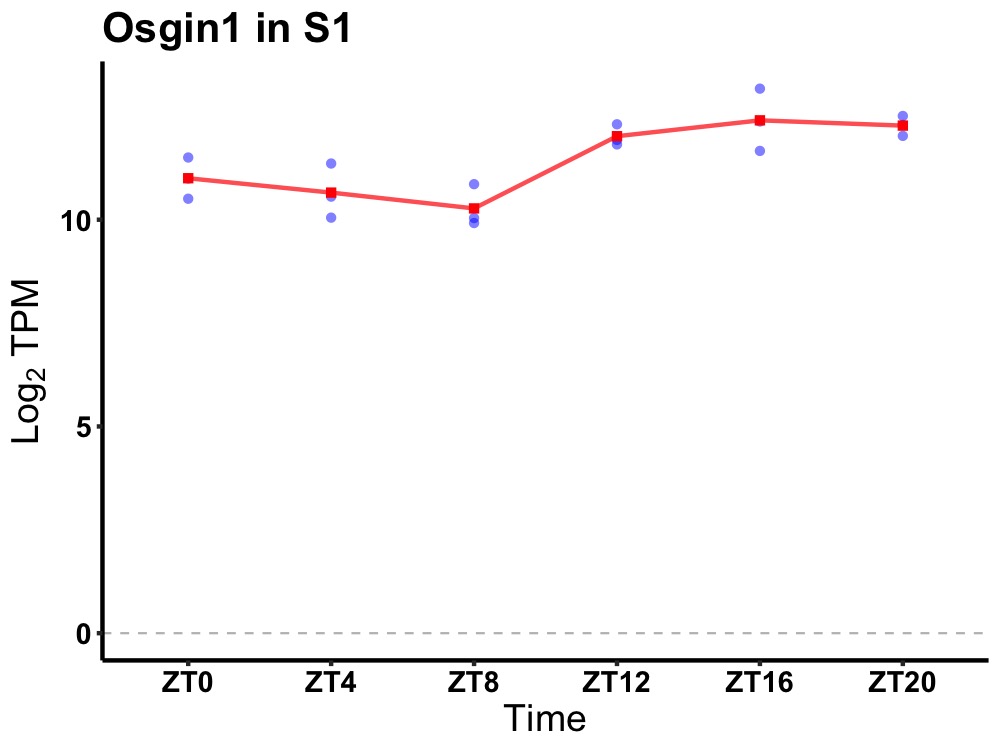 |
oxidative stress induced growth inhibitor 1 | 0.002 | 24 | 18 | 1.07 |
| ENSMUSG00000050270 | Tmem220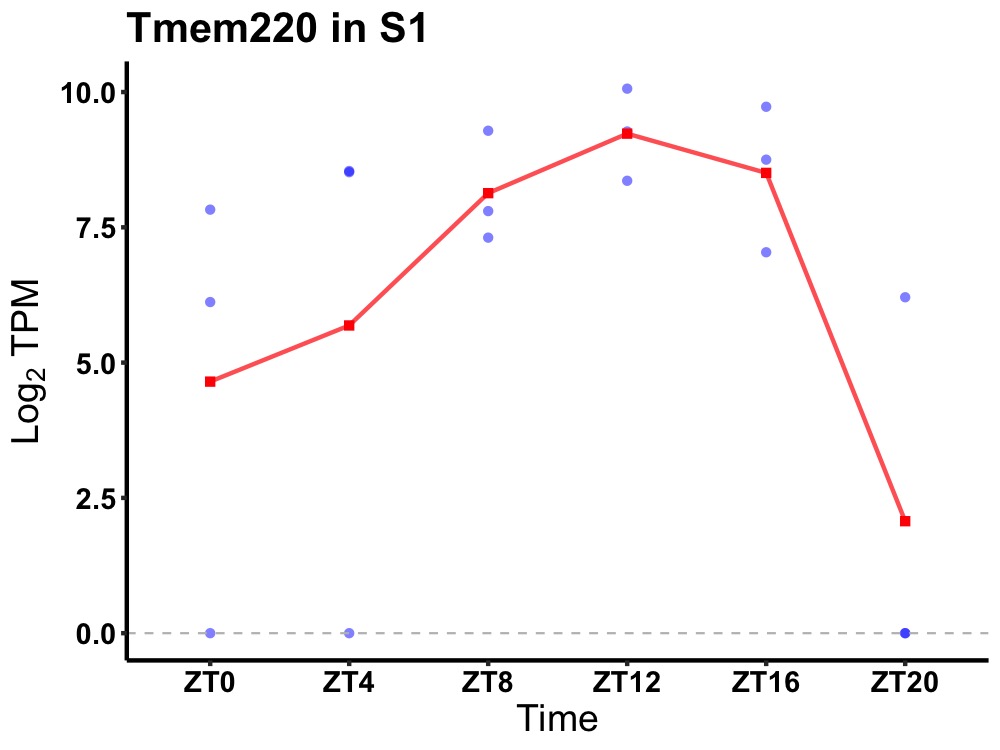 |
transmembrane protein 220 | 0.043 | 20 | 12 | 1.07 |
| ENSMUSG00000029551 | Psmg3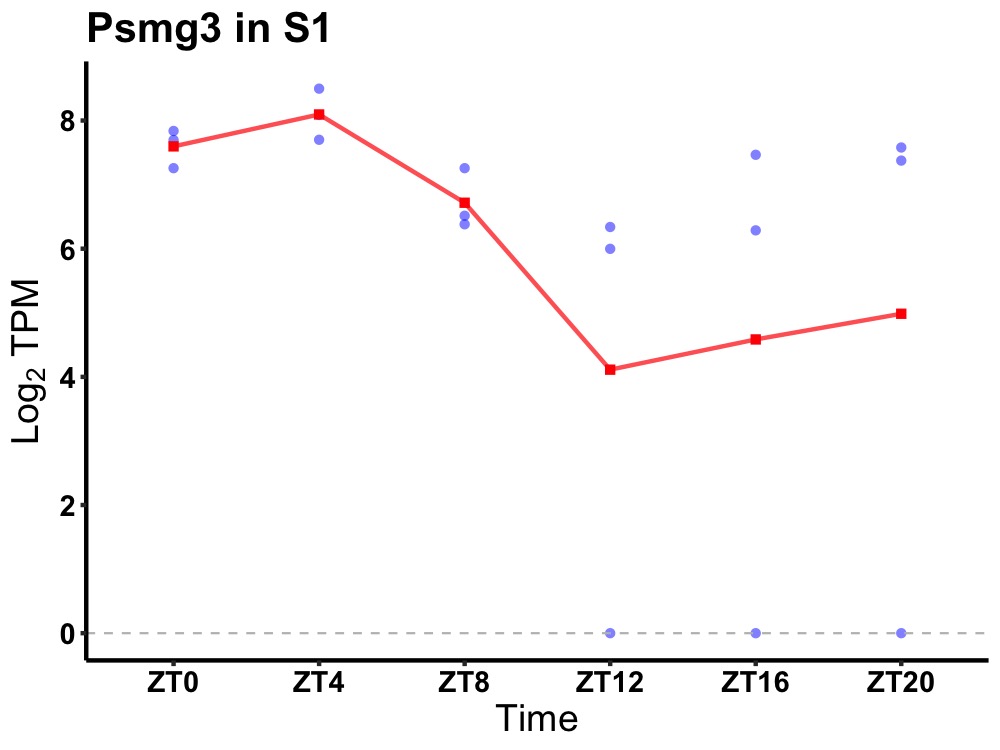 |
proteasome assembly chaperone 3 | 0.012 | 20 | 4 | 1.06 |
| ENSMUSG00000028423 | Herc3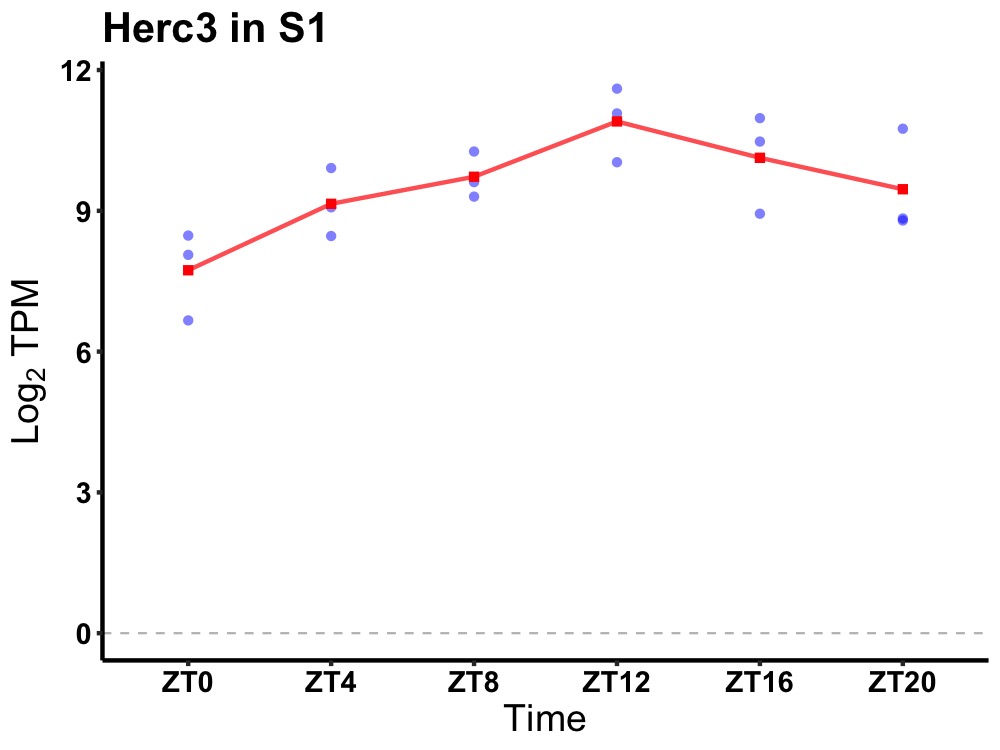 |
hect domain and RLD 3 | 0.003 | 24 | 14 | 1.05 |
| ENSMUSG00000062906 | Ctps2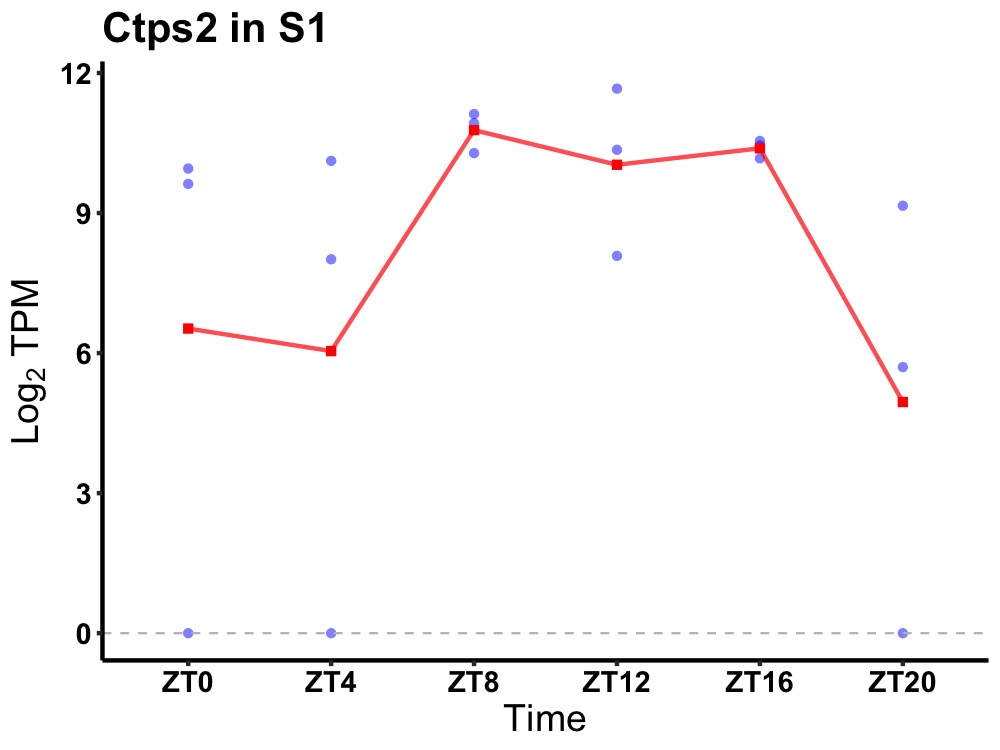 |
cytidine 5'-triphosphate synthase 2 | 0.023 | 20 | 12 | 1.04 |
| ENSMUSG00000050628 | Entpd7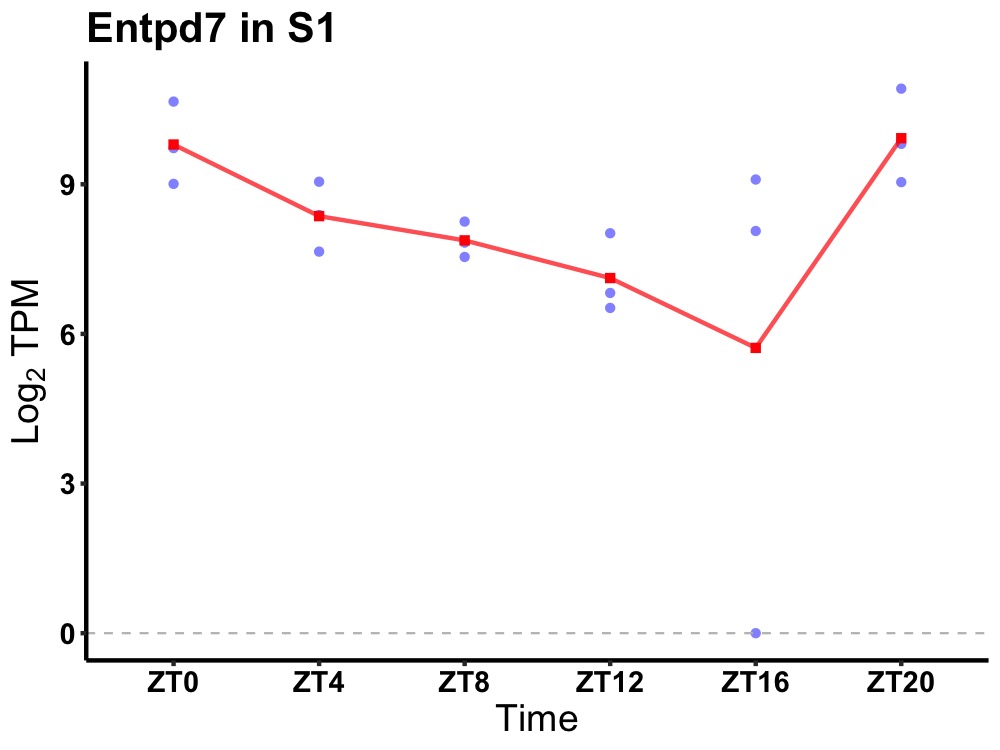 |
ectonucleoside triphosphate diphosphohydrolase 7 | 0.007 | 20 | 2 | 1.04 |
| ENSMUSG00000027804 | Gtf2e2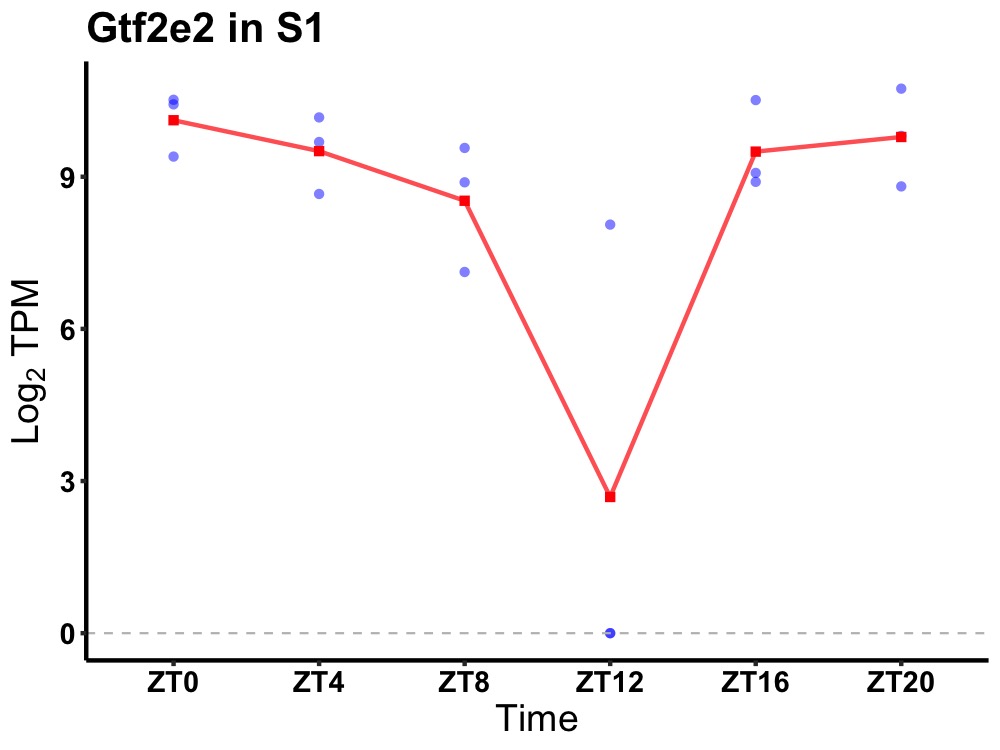 |
general transcription factor II E, polypeptide 2 (beta subunit) | 0.036 | 24 | 0 | 1.02 |
| ENSMUSG00000001783 | Zfyve27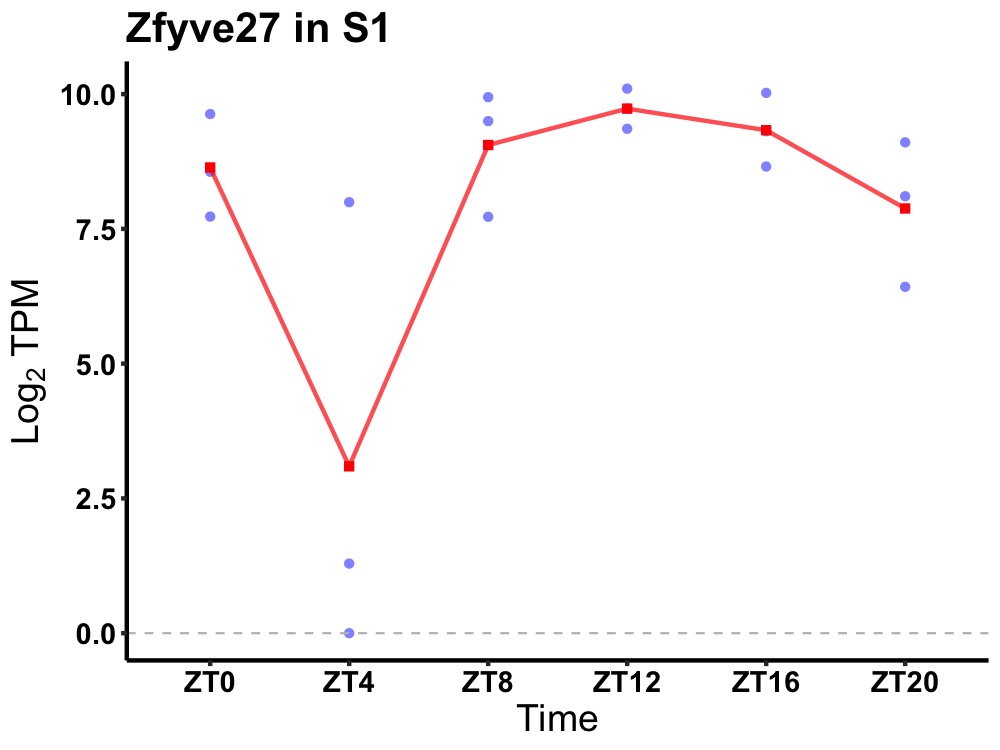 |
protrudin | 0.014 | 20 | 14 | 1.02 |
| ENSMUSG00000082253 | Gm14639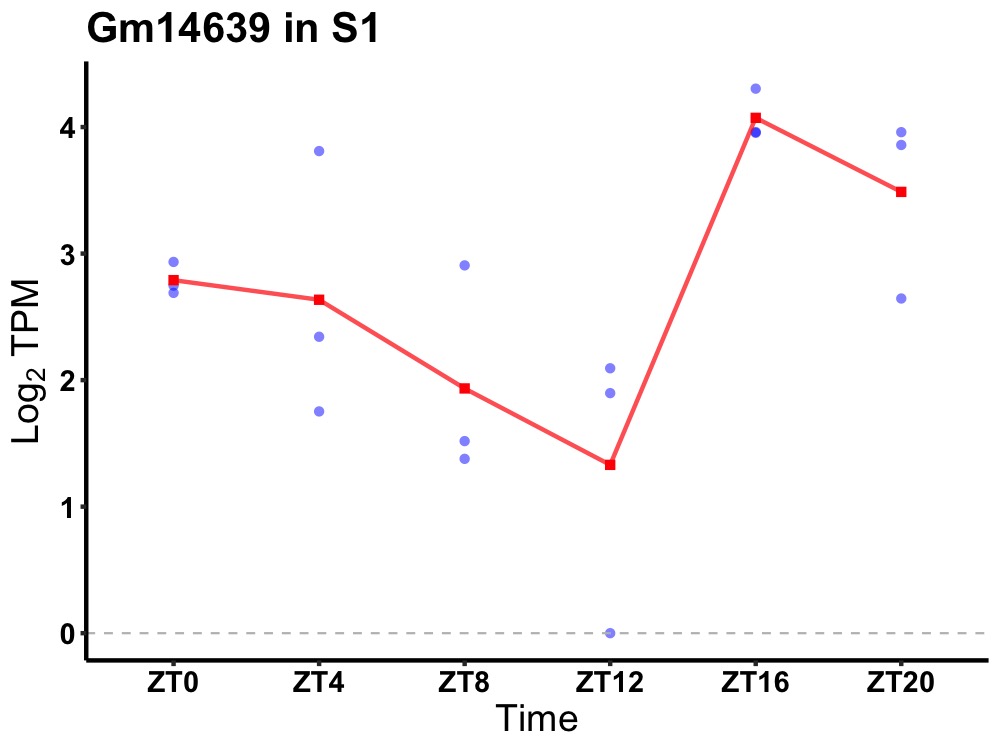 |
predicted gene 14639 | 0.036 | 20 | 18 | 1.01 |
| ENSMUSG00000099627 | Gm6757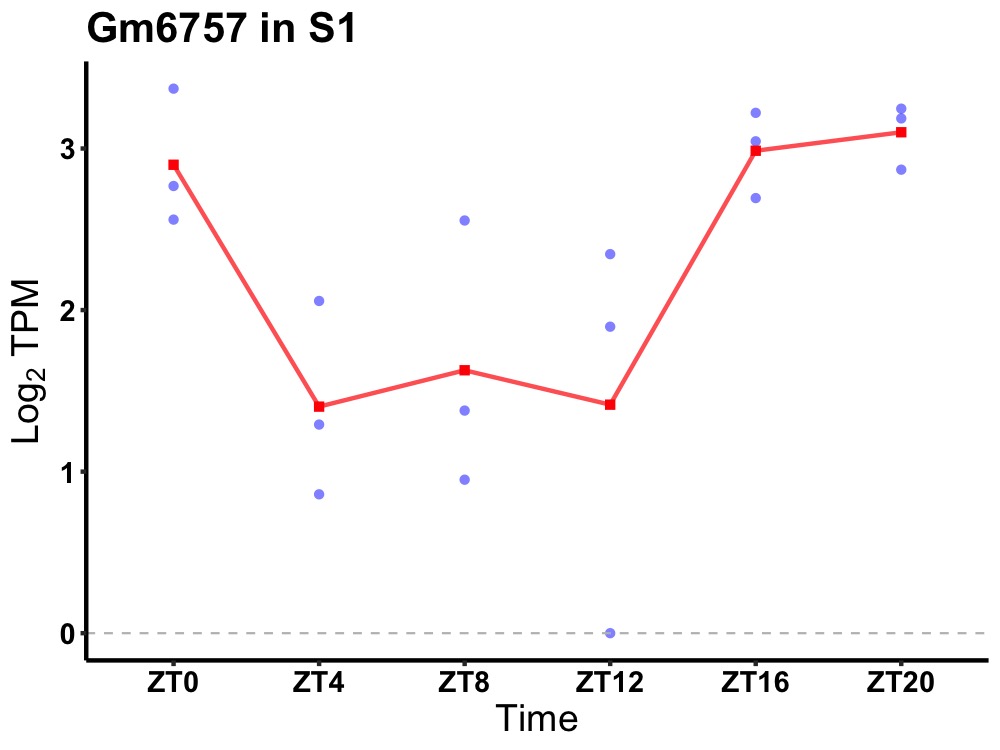 |
predicted gene 6757 | 0.014 | 24 | 20 | 1.01 |
| ENSMUSG00000003363 | Gm19696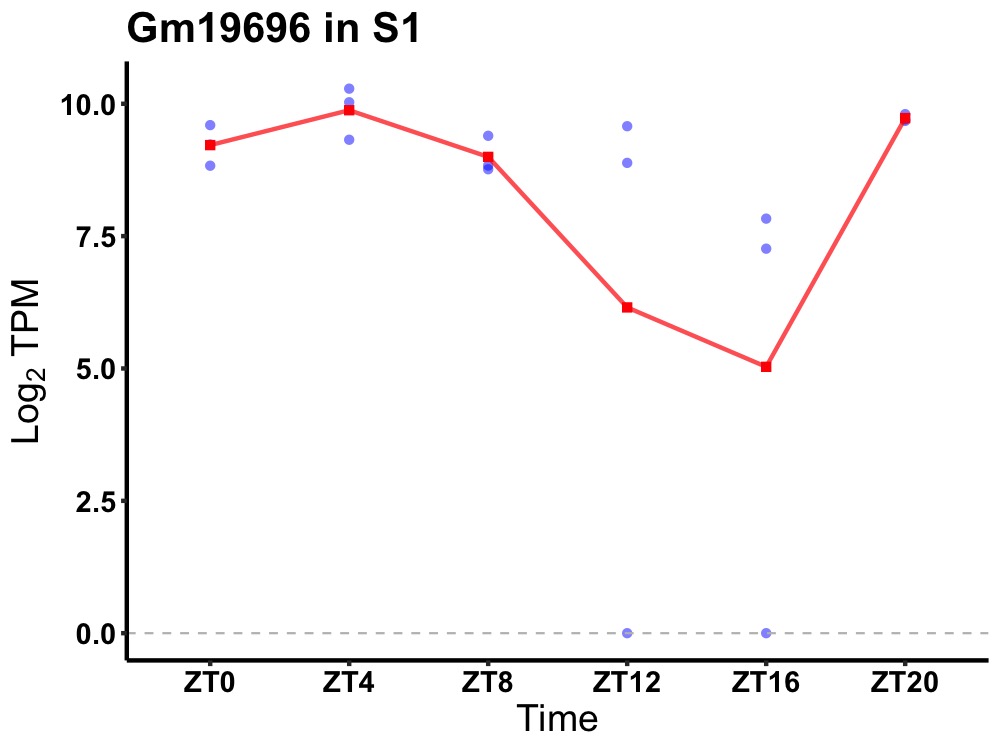 |
none | 0.008 | 20 | 4 | 0.99 |
| ENSMUSG00000083289 | Gm8812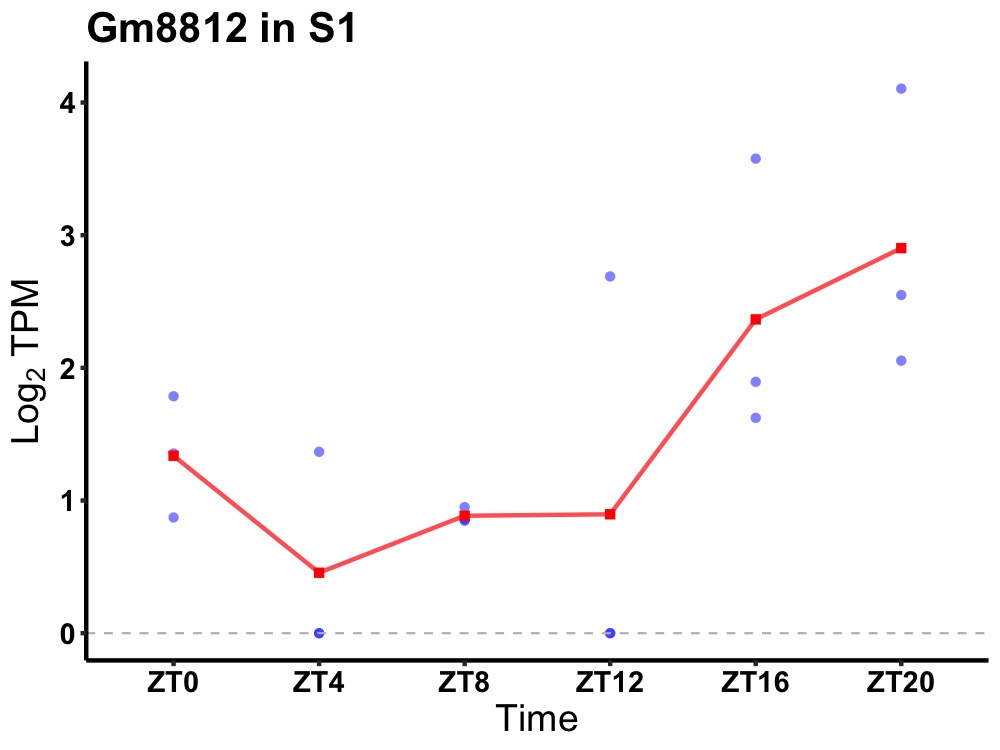 |
none | 0.049 | 24 | 20 | 0.97 |
| ENSMUSG00000111446 | Rorc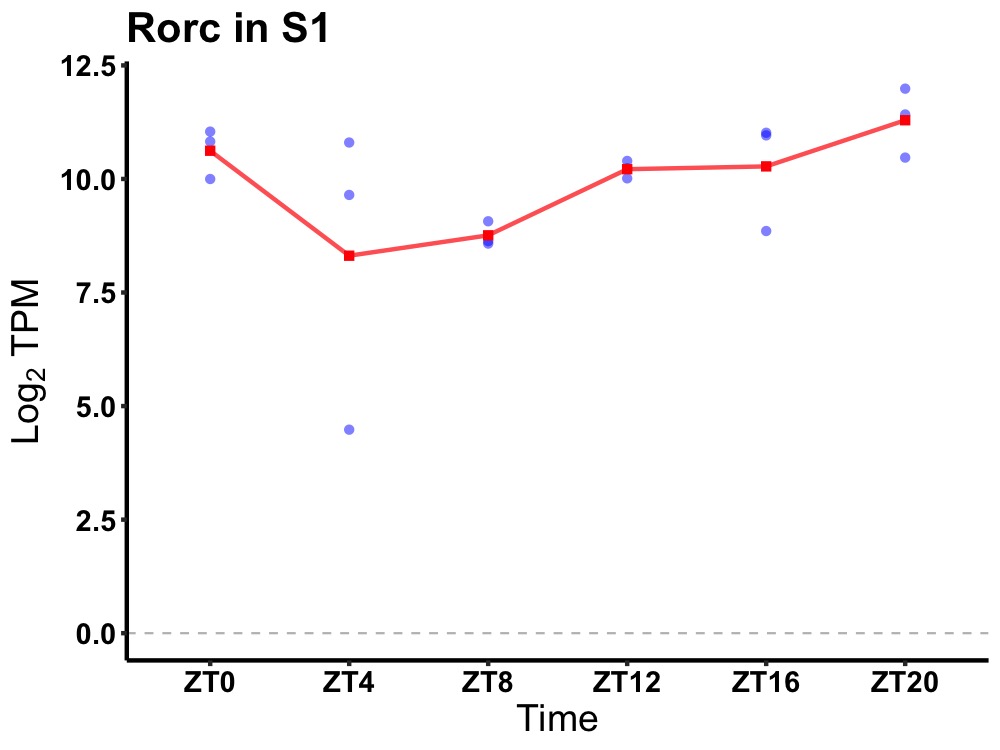 |
RAR-related orphan receptor gamma | 0.019 | 24 | 20 | 0.97 |
| ENSMUSG00000024064 | Upp2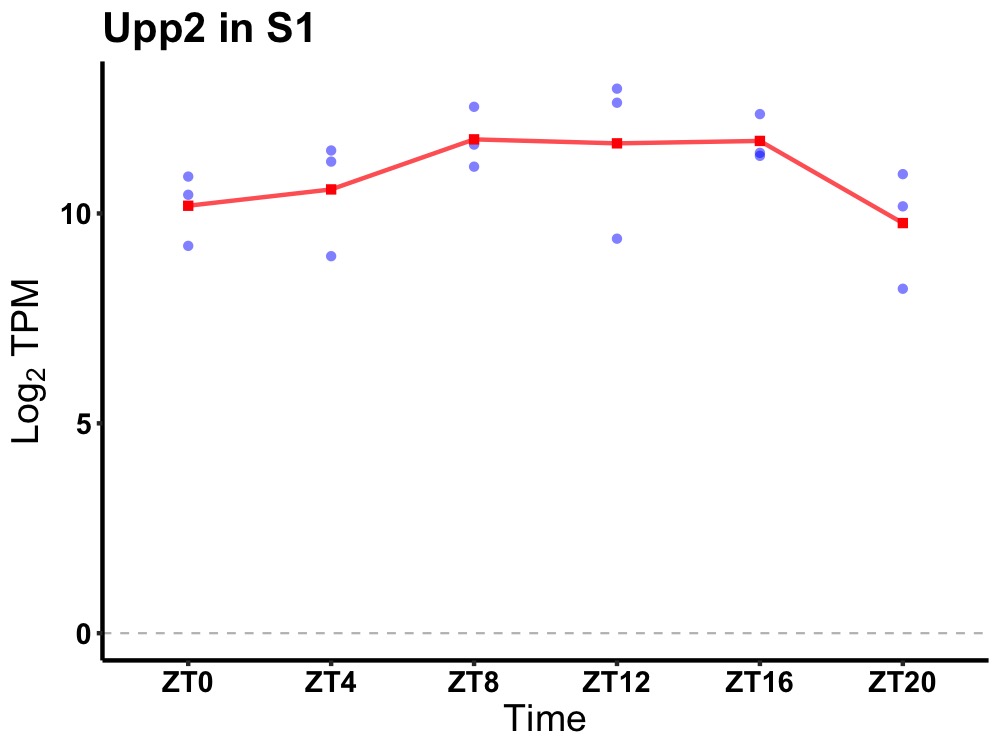 |
uridine phosphorylase 2 | 0.049 | 20 | 12 | 0.96 |
| ENSMUSG00000044211 | Gm7887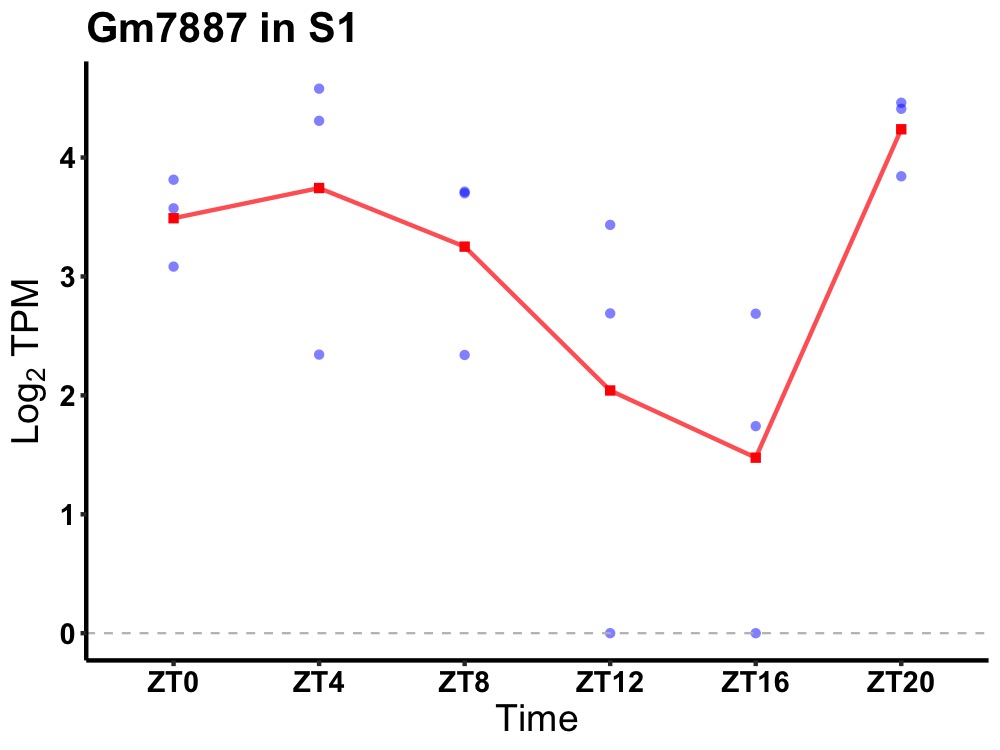 |
predicted gene 7887 | 0.031 | 20 | 4 | 0.95 |
| ENSMUSG00000026915 | Plrg1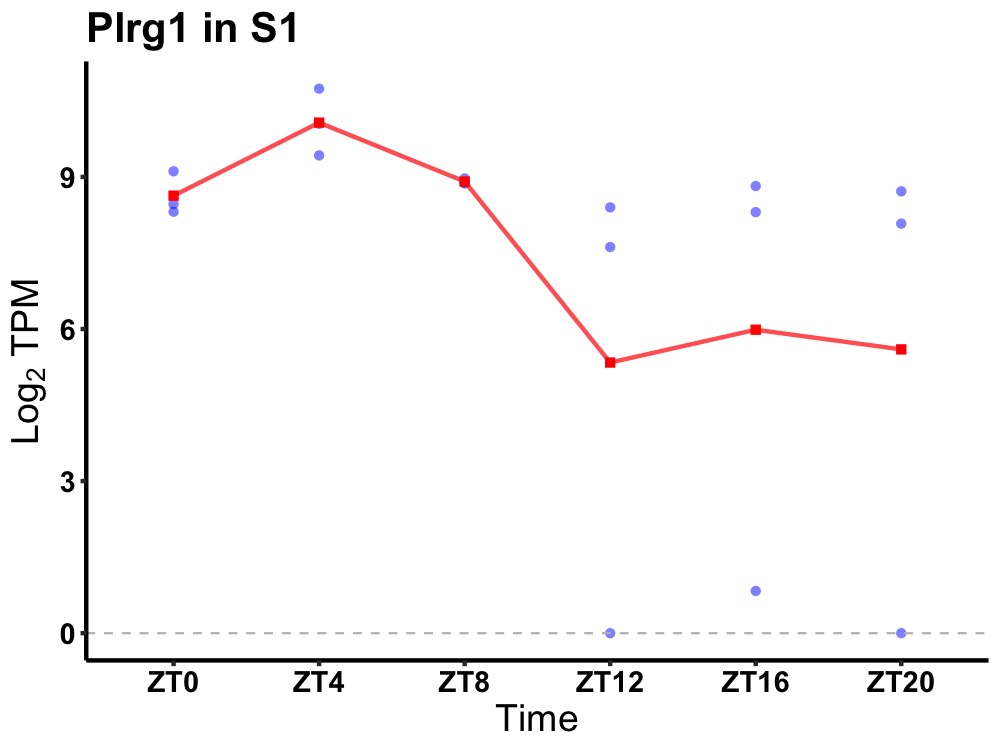 |
pleiotropic regulator 1 | 0.016 | 24 | 6 | 0.95 |
| ENSMUSG00000071866 | Flot1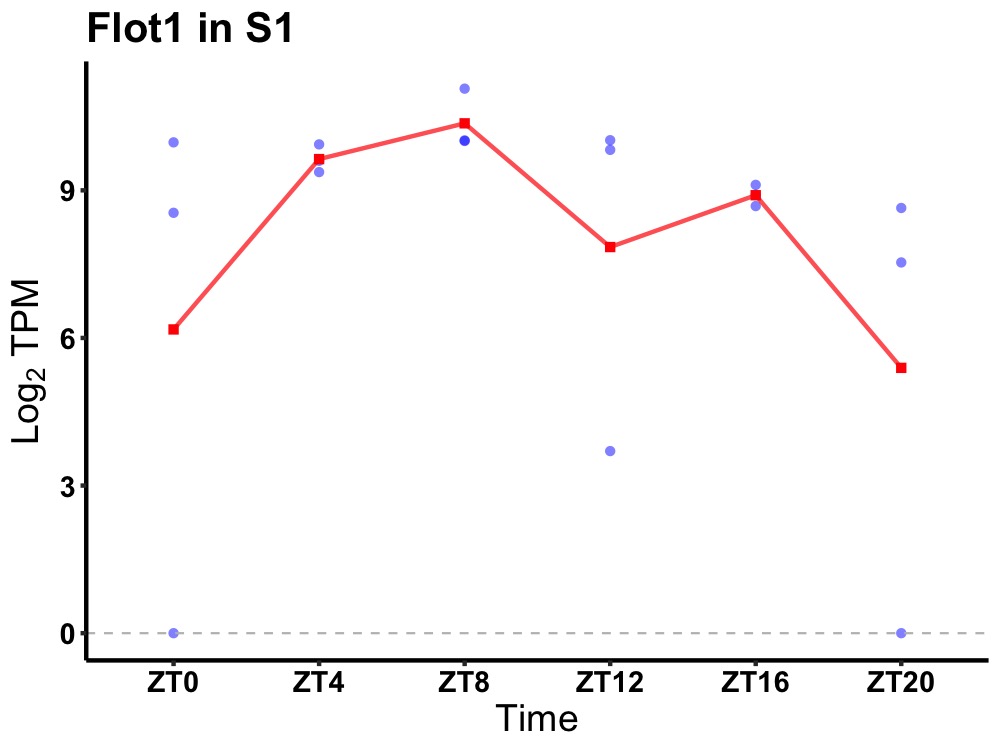 |
flotillin-1 | 0.008 | 24 | 10 | 0.95 |
| ENSMUSG00000032652 | Crebl2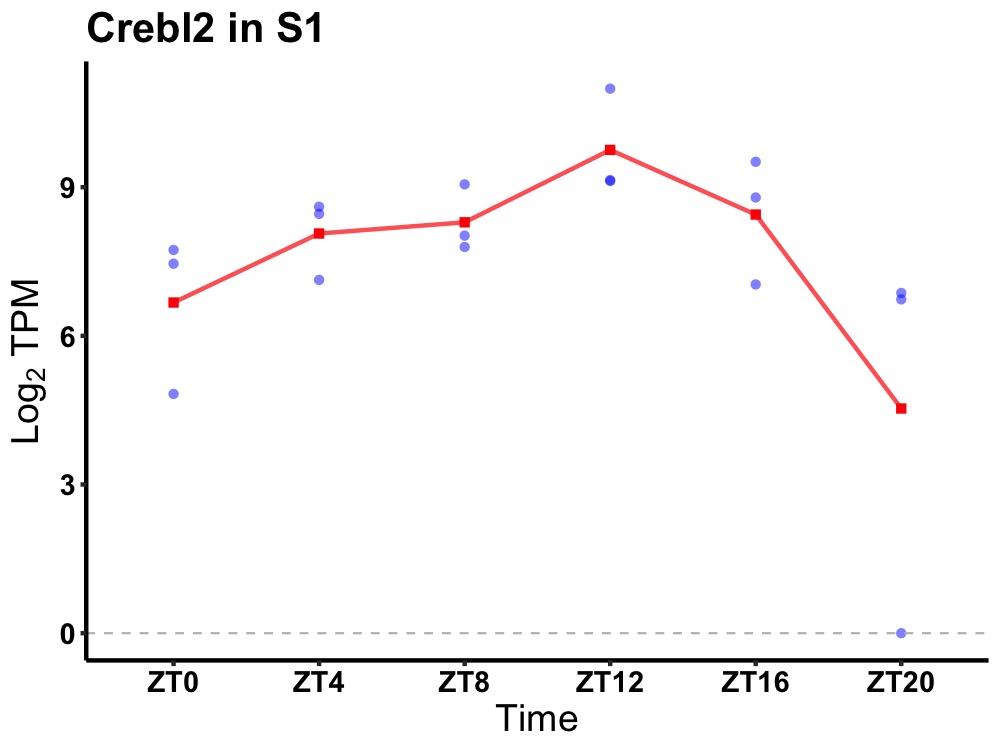 |
cAMP responsive element binding protein-like 2 | 0.002 | 20 | 12 | 0.94 |
| ENSMUSG00000029815 | Htra1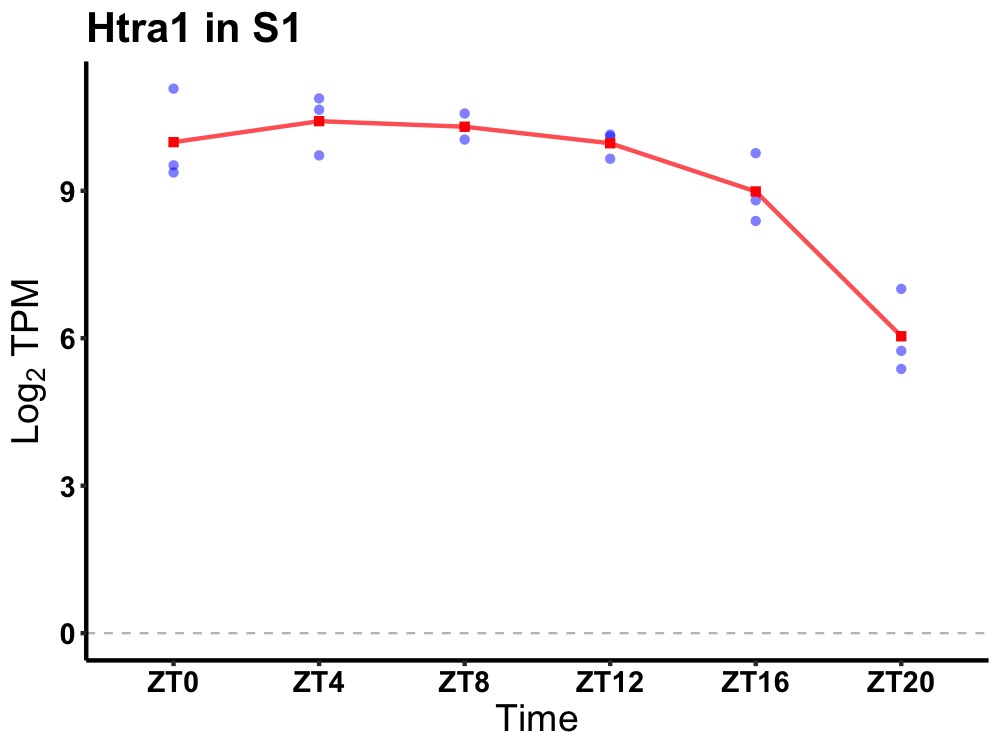 |
HtrA serine peptidase 1 | 0.007 | 24 | 8 | 0.93 |
| ENSMUSG00000028568 | Pcx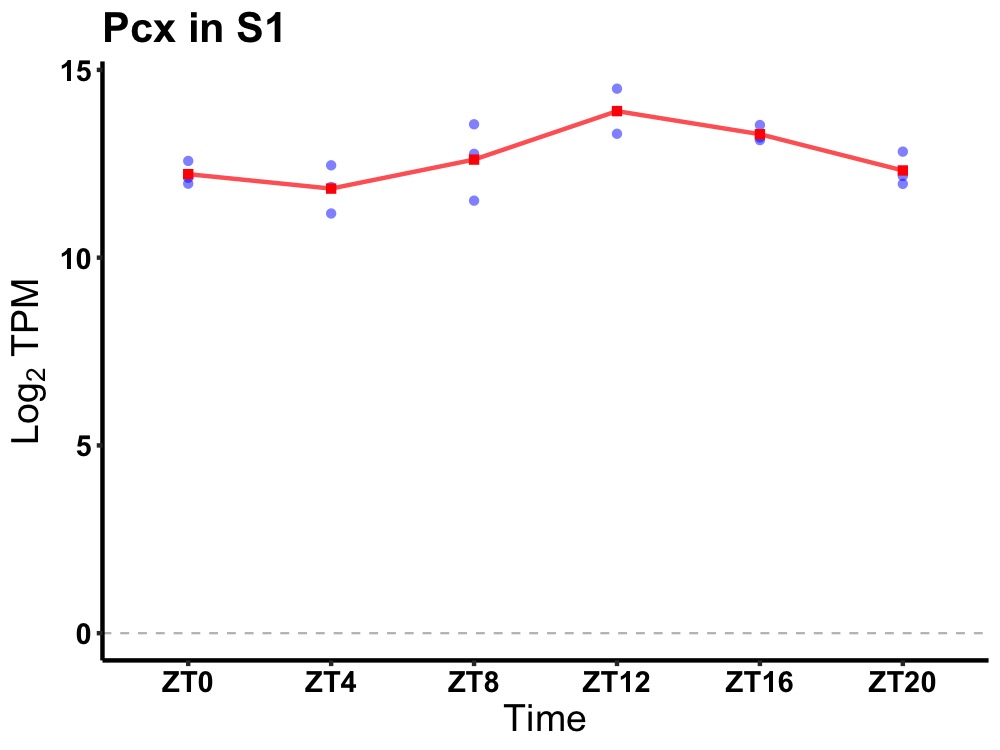 |
pyruvate carboxylase | 0.007 | 20 | 14 | 0.93 |
| ENSMUSG00000020669 | Stx16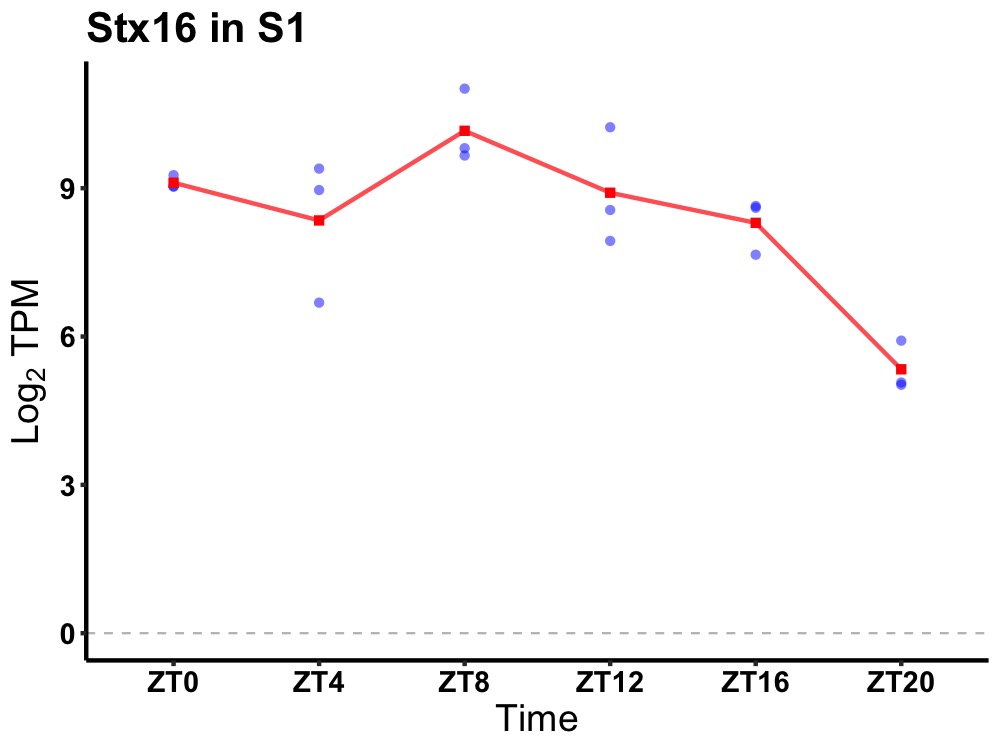 |
syntaxin-16 | 0.019 | 24 | 8 | 0.93 |
| ENSMUSG00000081111 | Sh3yl1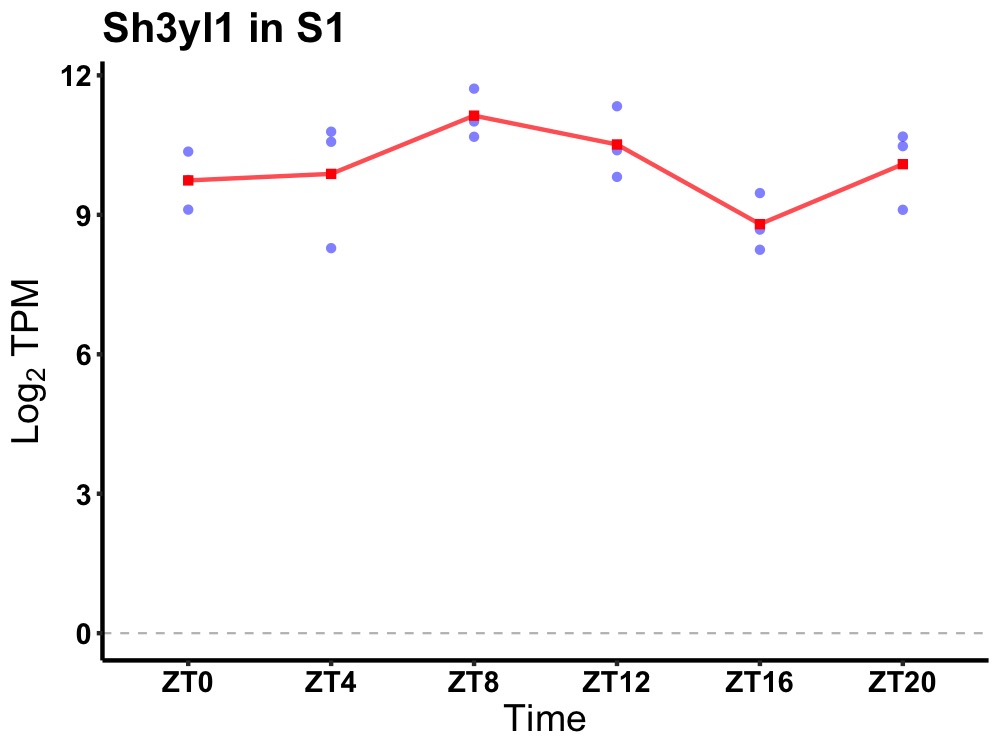 |
Sh3 domain YSC-like 1 | 0.036 | 20 | 8 | 0.92 |
| ENSMUSG00000005732 | Tbcc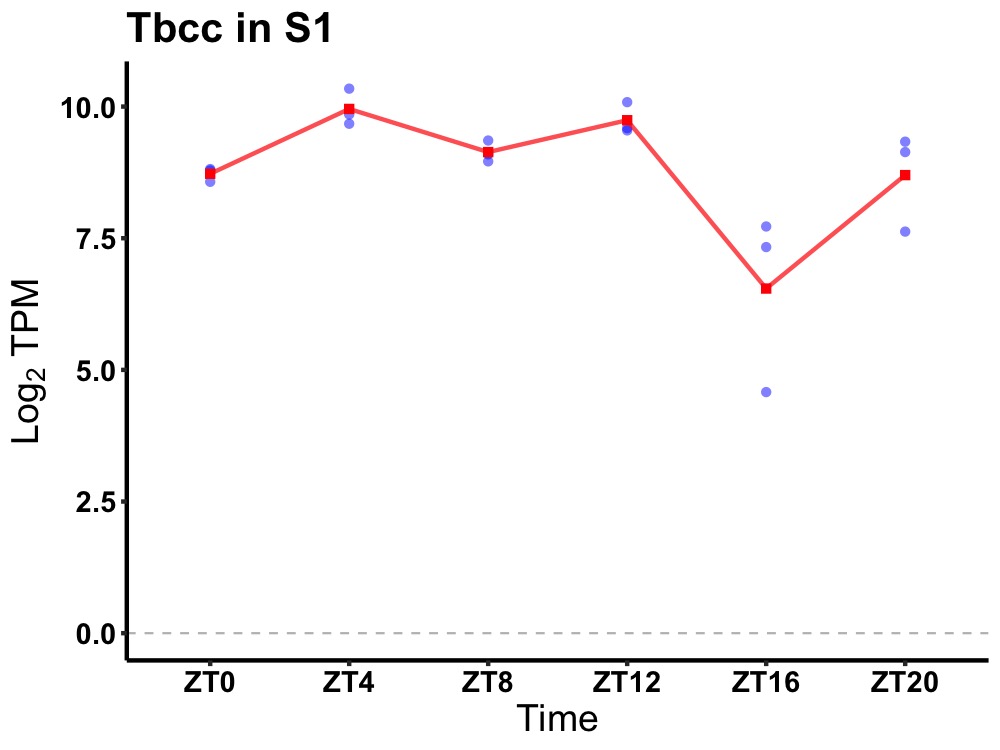 |
tubulin-specific chaperone C | 0.036 | 20 | 8 | 0.91 |
| ENSMUSG00000048731 | Clptm1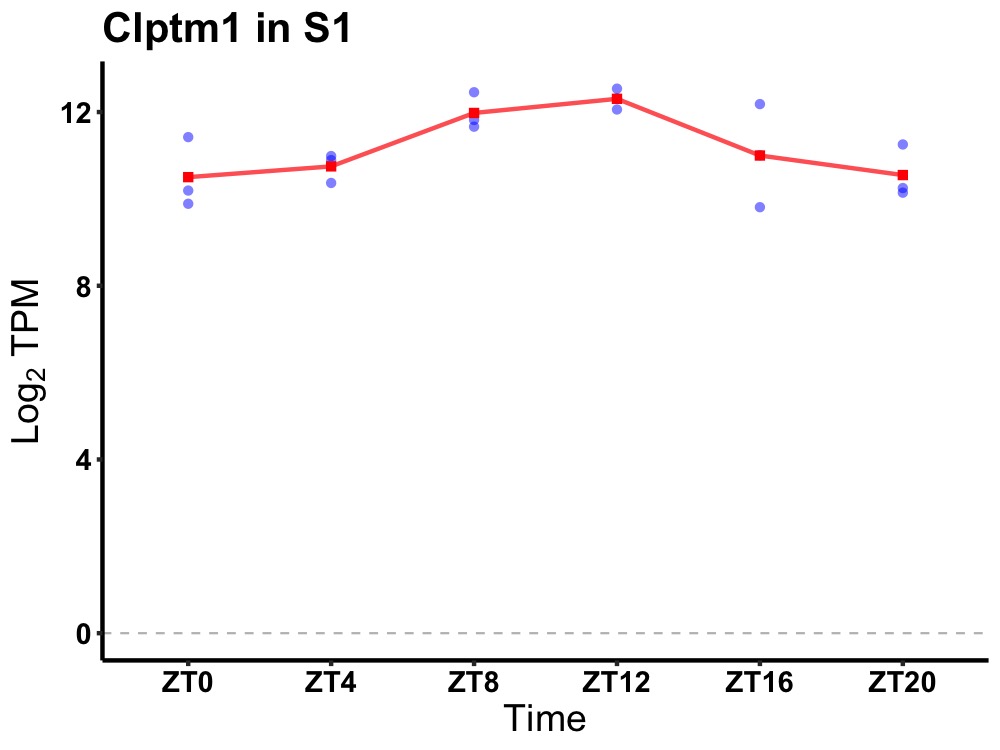 |
cleft lip and palate associated transmembrane protein 1 | 0.019 | 24 | 12 | 0.91 |
| ENSMUSG00000026753 | Pik3r2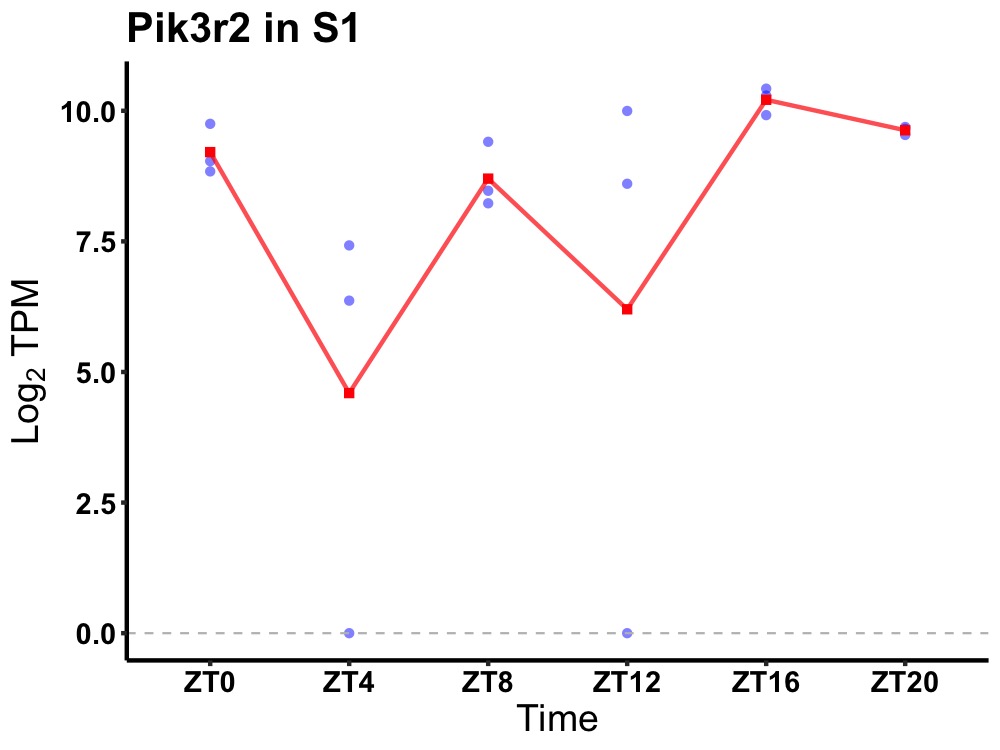 |
phosphatidylinositol 3-kinase regulatory subunit beta | 0.004 | 24 | 18 | 0.91 |
| ENSMUSG00000117249 | Gm46577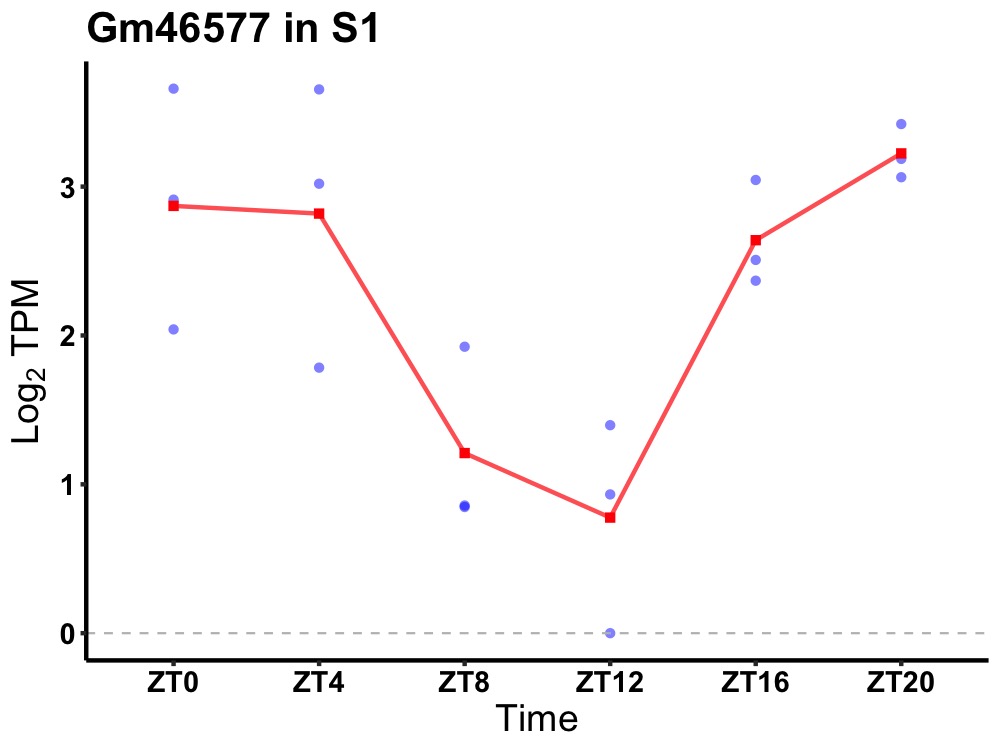 |
nuclear transport factor 2 (Nutf2) pseudogene | 0.007 | 20 | 2 | 0.90 |
| ENSMUSG00000112105 | Rps7-ps2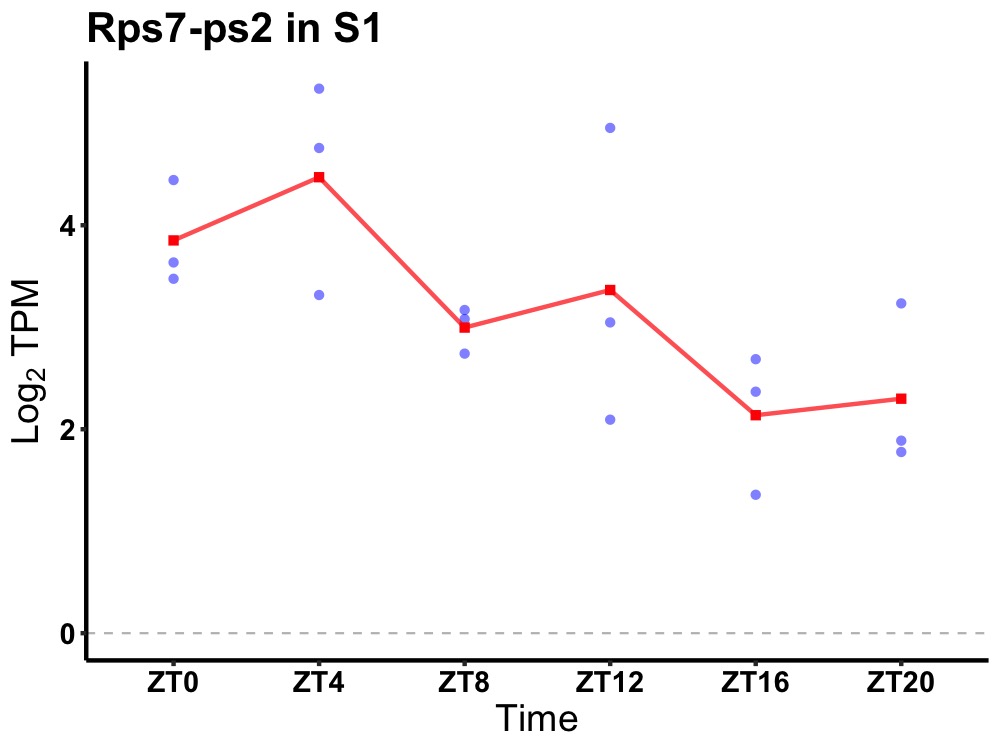 |
ribosomal protein S7, pseudogene 2 | 0.036 | 24 | 4 | 0.90 |
| ENSMUSG00000033446 | Bicc1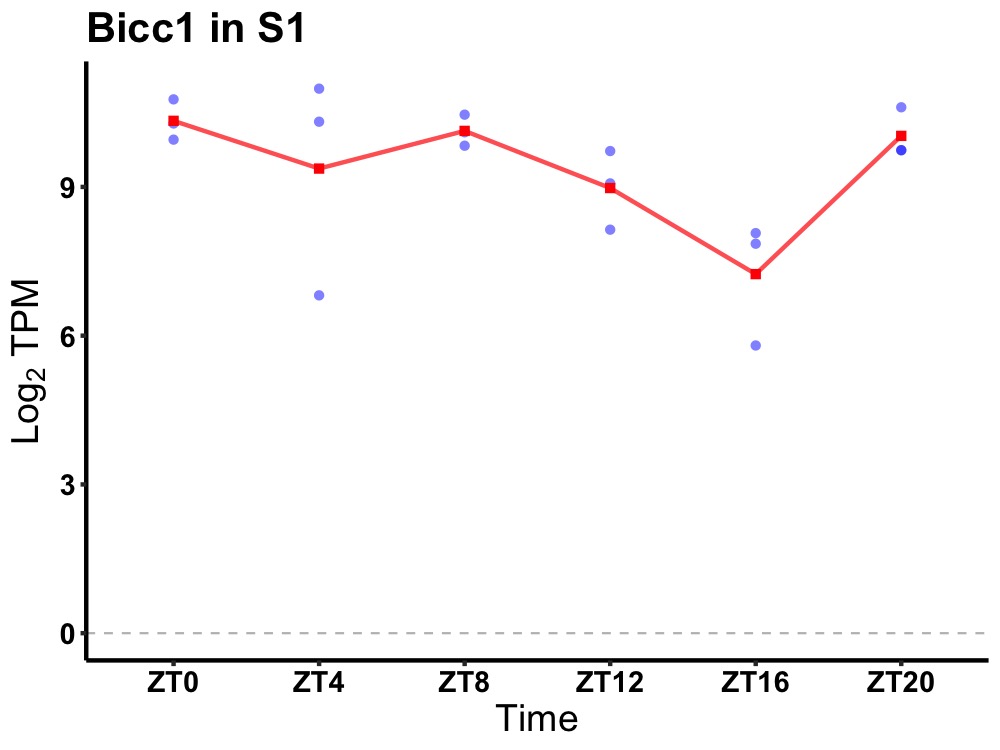 |
BicC family RNA binding protein 1 | 0.014 | 24 | 4 | 0.89 |
| ENSMUSG00000083512 | Gm12749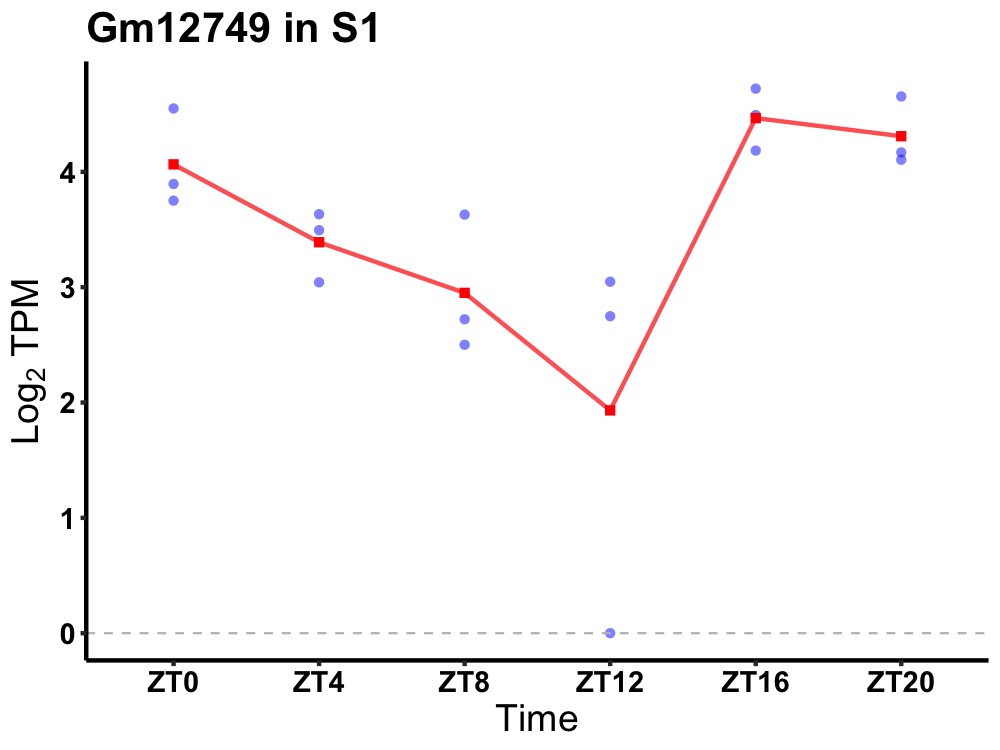 |
predicted gene 12749 | 0.010 | 24 | 22 | 0.88 |
| ENSMUSG00000069268 | Dcun1d4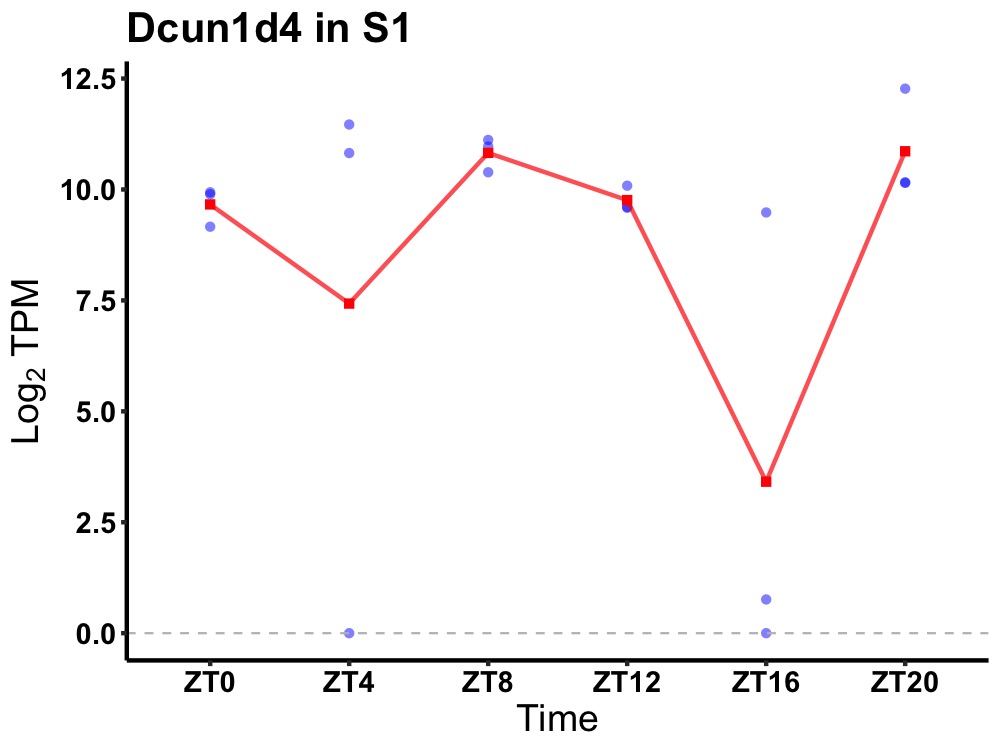 |
DCN1-like protein 4 | 0.031 | 20 | 6 | 0.87 |
| ENSMUSG00000059775 | Cpeb3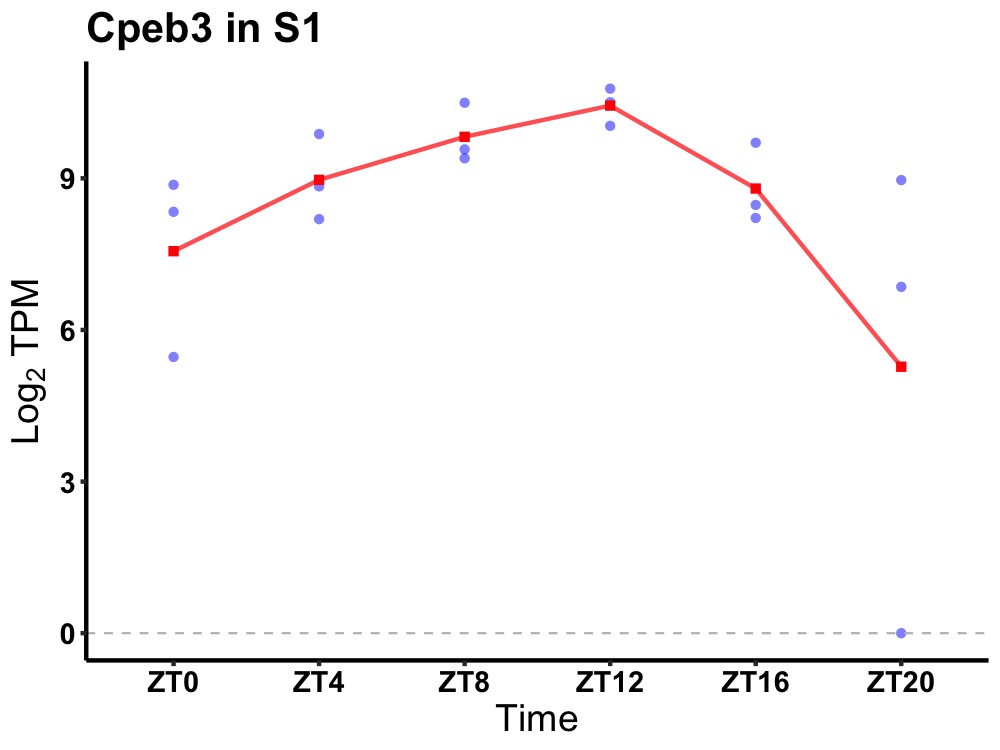 |
cytoplasmic polyadenylation element-binding protein 3 | 0.010 | 20 | 12 | 0.86 |
| ENSMUSG00000114607 | Gm8990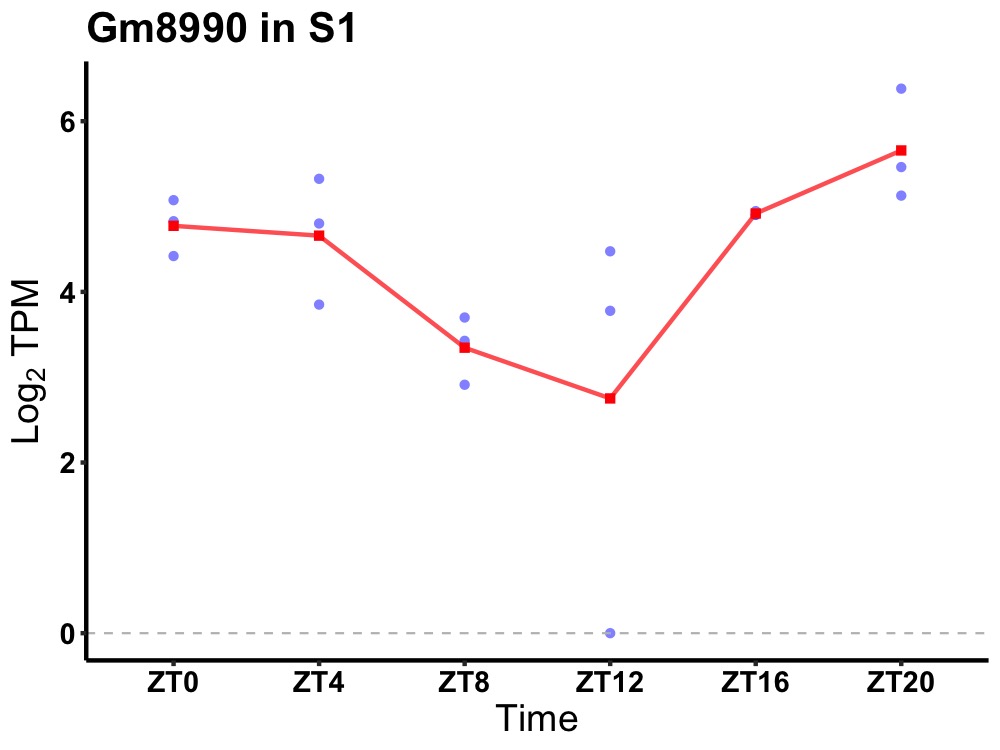 |
predicted gene 8990 | 0.001 | 24 | 22 | 0.85 |
| ENSMUSG00000108435 | Gm45051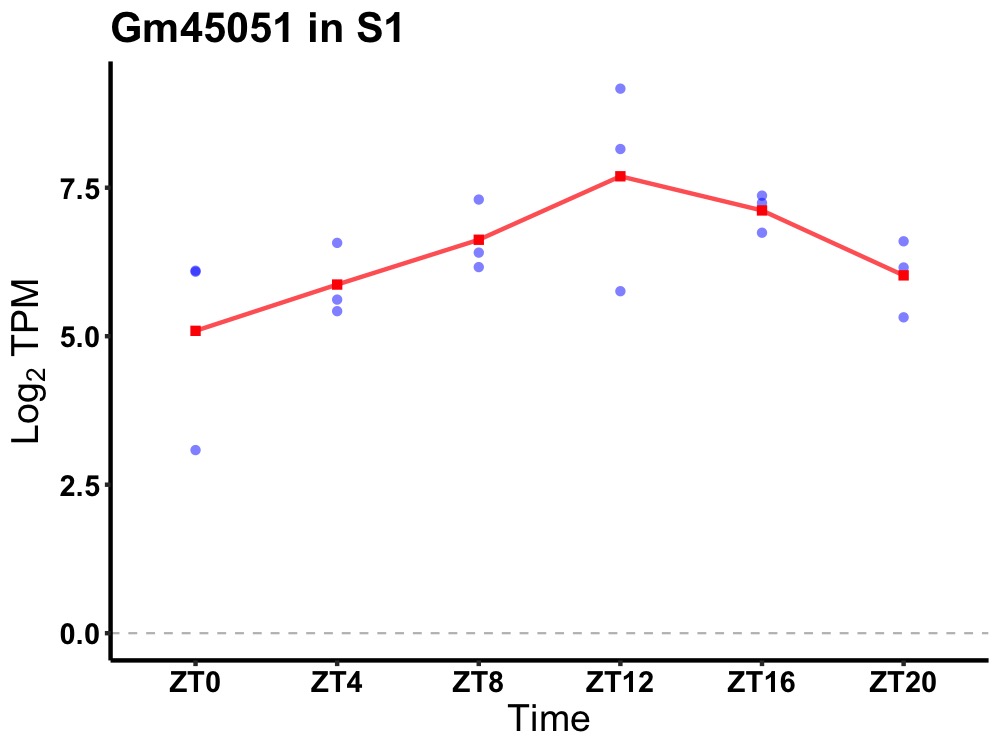 |
predicted gene 45051 | 0.027 | 24 | 14 | 0.85 |
| ENSMUSG00000110502 | Gm8804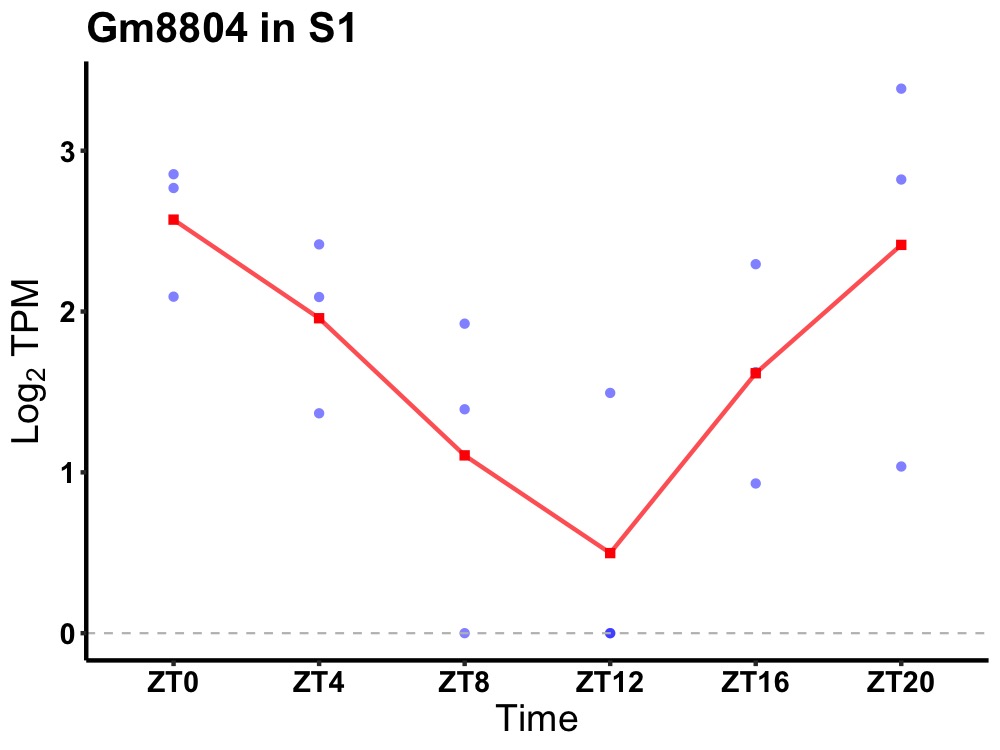 |
none | 0.027 | 20 | 2 | 0.85 |
| ENSMUSG00000027364 | Ogg1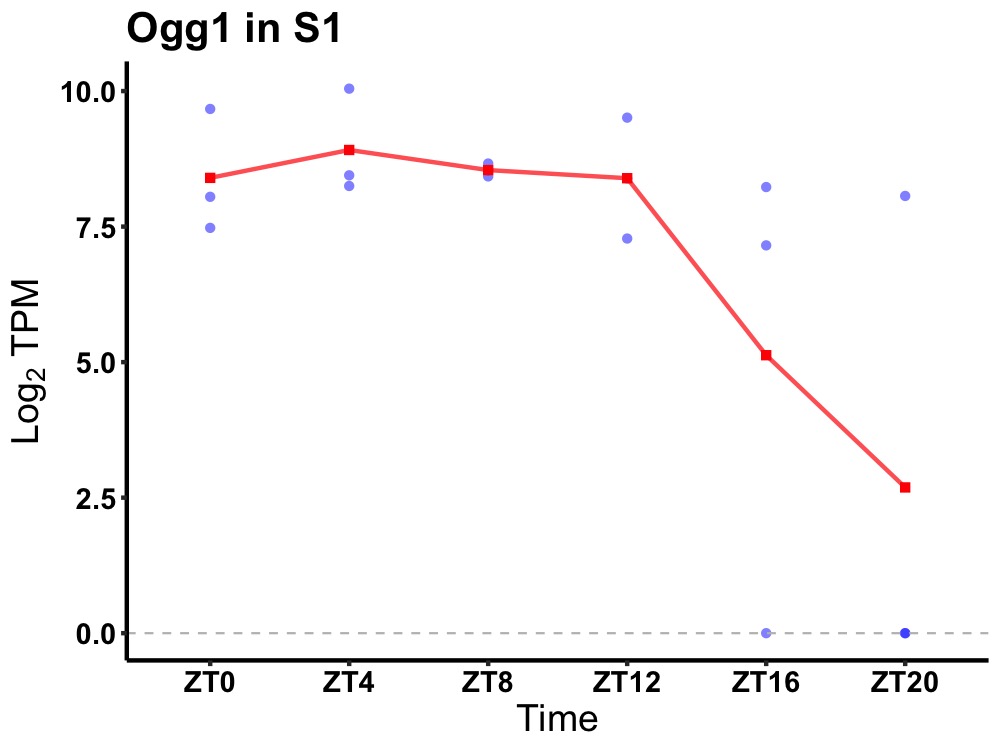 |
8-oxoguanine DNA-glycosylase 1 | 0.049 | 24 | 8 | 0.84 |
| ENSMUSG00000083562 | Gm13234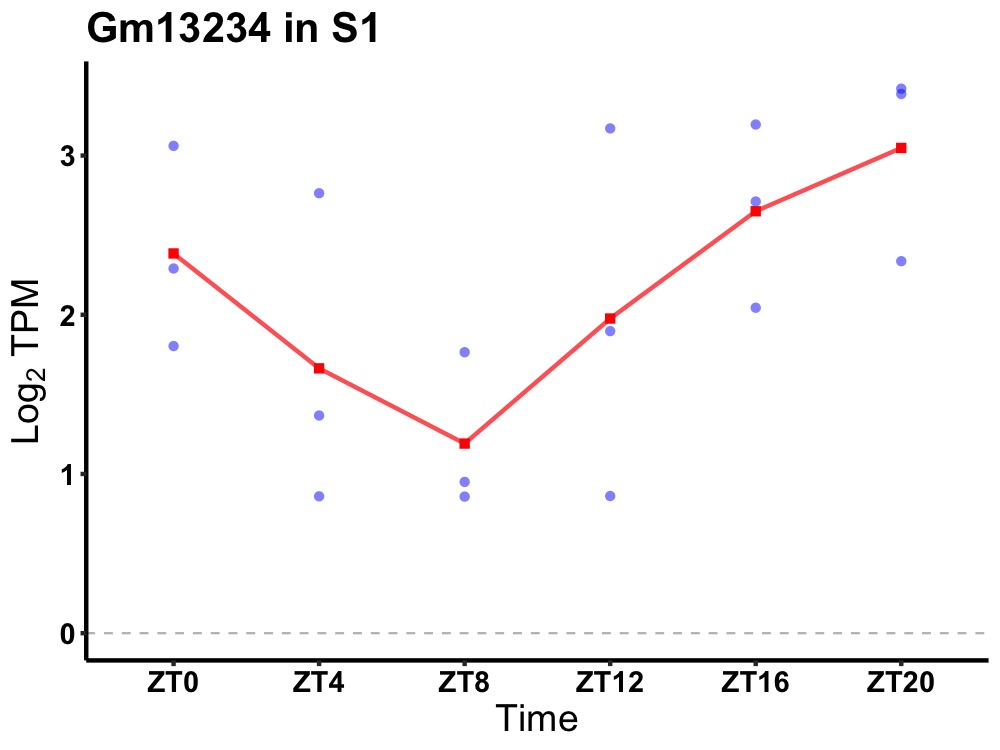 |
predicted gene 13234 | 0.019 | 24 | 20 | 0.84 |
| ENSMUSG00000095597 | Fbxo9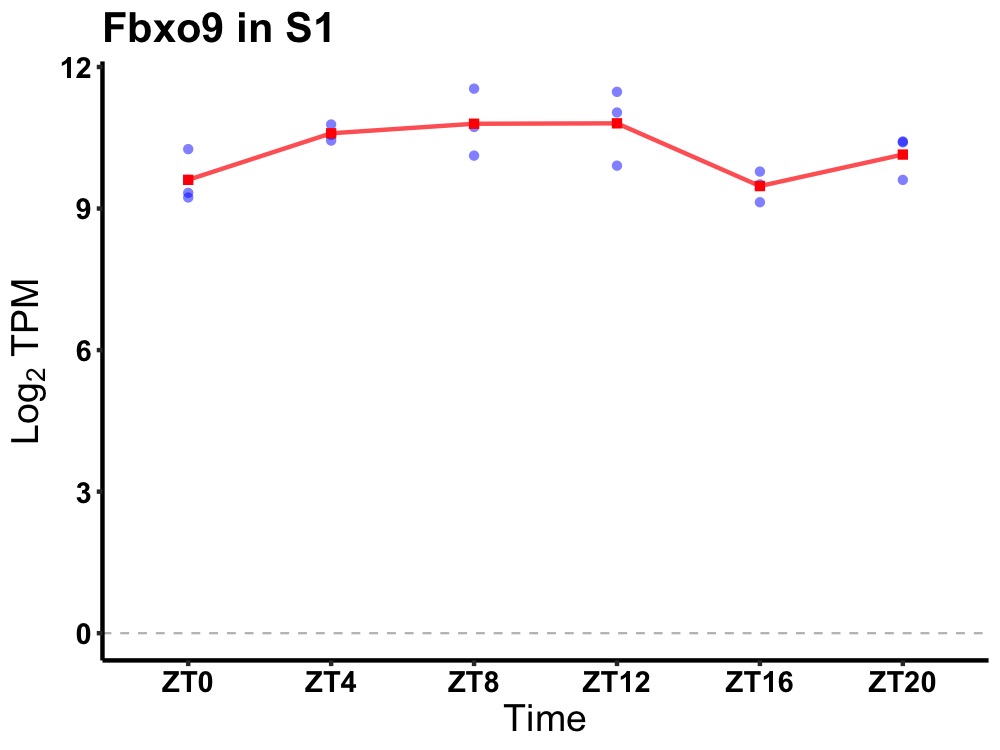 |
f-box protein 9 | 0.019 | 20 | 8 | 0.83 |
| ENSMUSG00000031168 | Klf9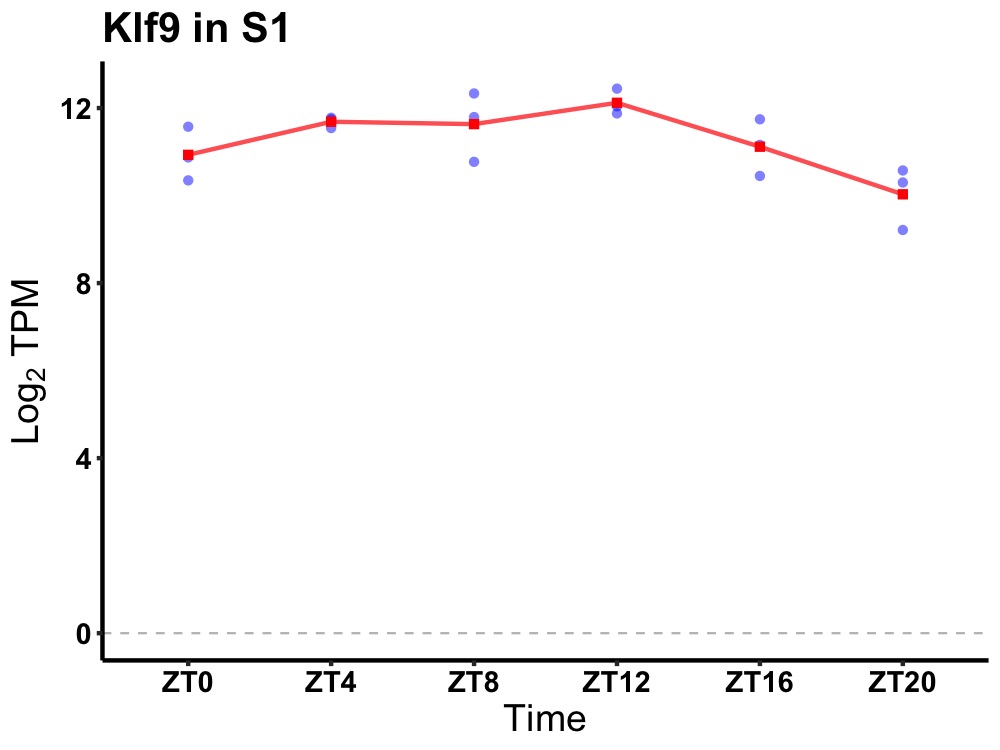 |
Kruppel-like factor 9 | 0.005 | 24 | 10 | 0.83 |
| ENSMUSG00000081128 | Gm13328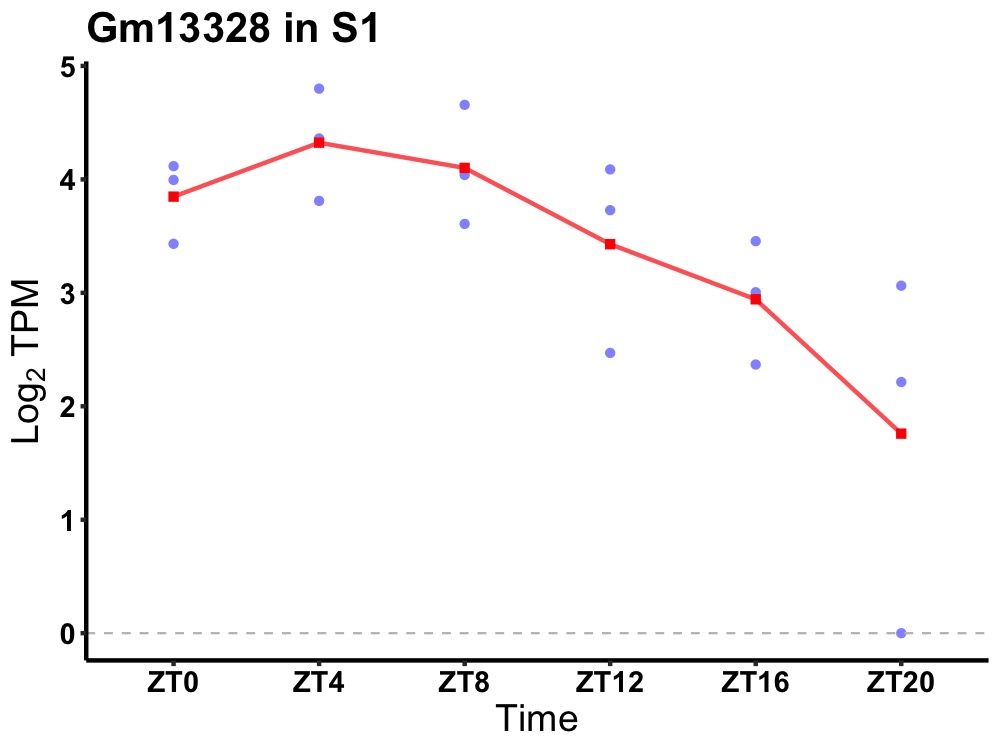 |
predicted gene 13328 | 0.019 | 24 | 6 | 0.80 |
| ENSMUSG00000028093 | Ndrg1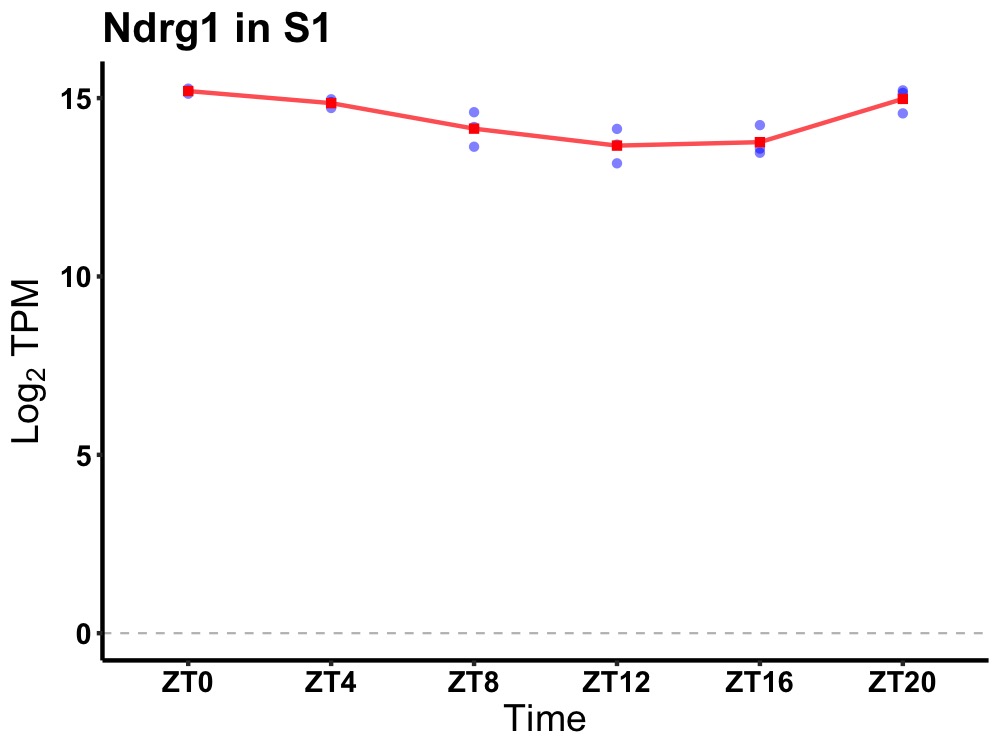 |
N-myc downstream regulated gene 1 | 0.001 | 24 | 2 | 0.80 |
| ENSMUSG00000022521 | Tnfrsf21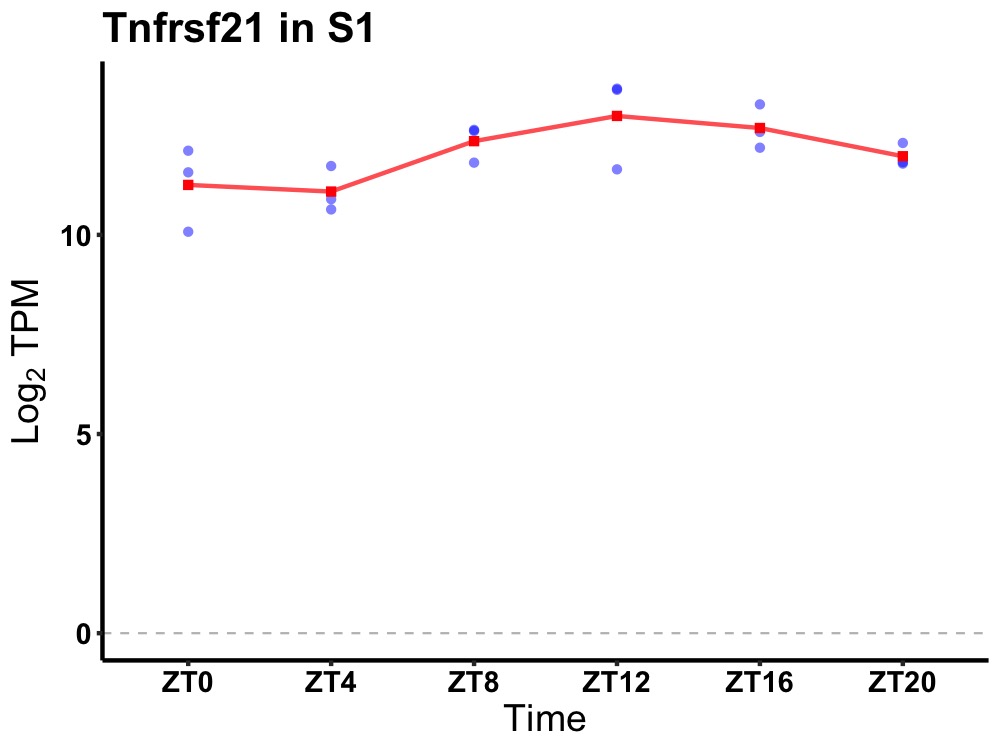 |
tumor necrosis factor receptor superfamily, member 21 | 0.019 | 24 | 14 | 0.80 |
| ENSMUSG00000106943 | Dancr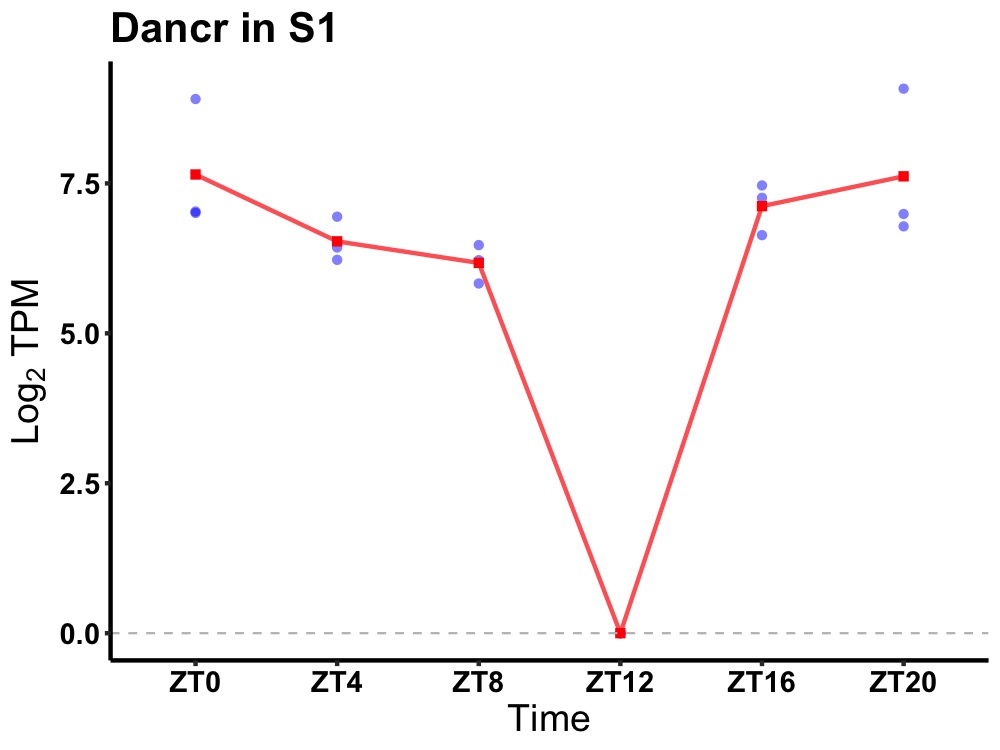 |
none | 0.005 | 24 | 0 | 0.79 |
| ENSMUSG00000082878 | Gm12583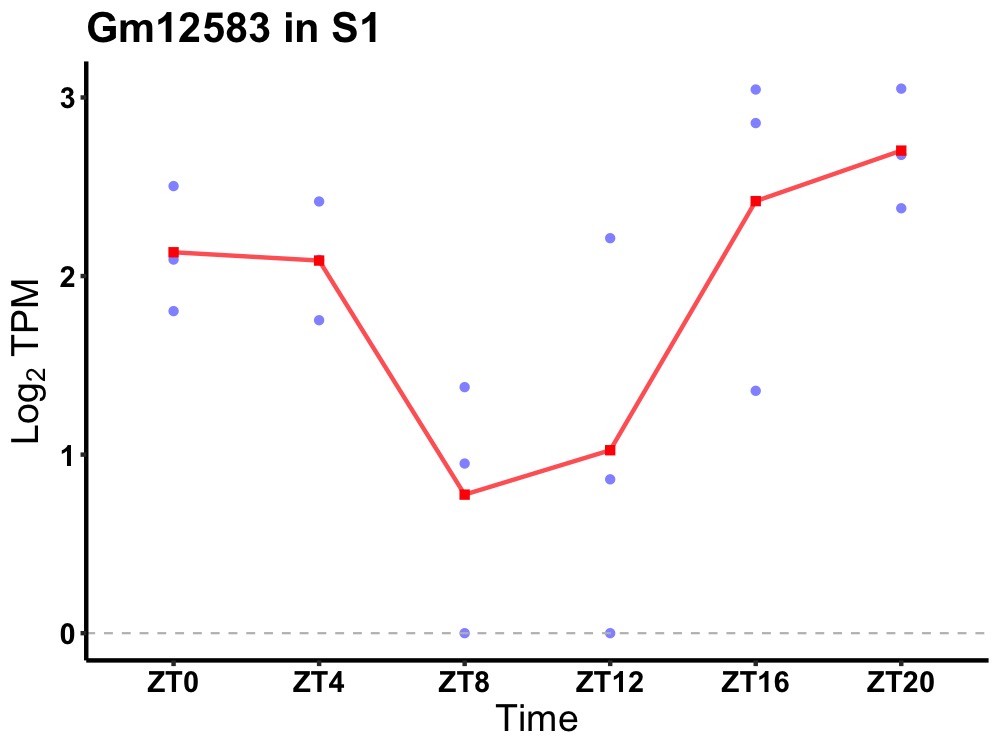 |
none | 0.043 | 24 | 22 | 0.78 |
| ENSMUSG00000111355 | Gm8911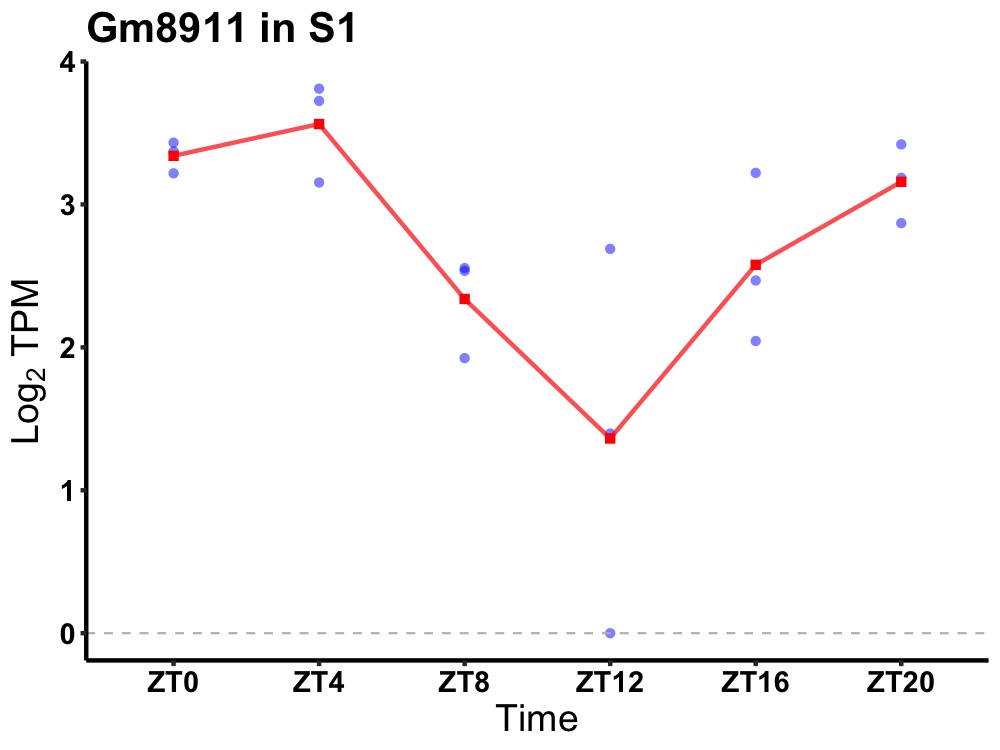 |
predicted gene 8911 | 0.010 | 24 | 2 | 0.78 |
| ENSMUSG00000096160 | Gm3436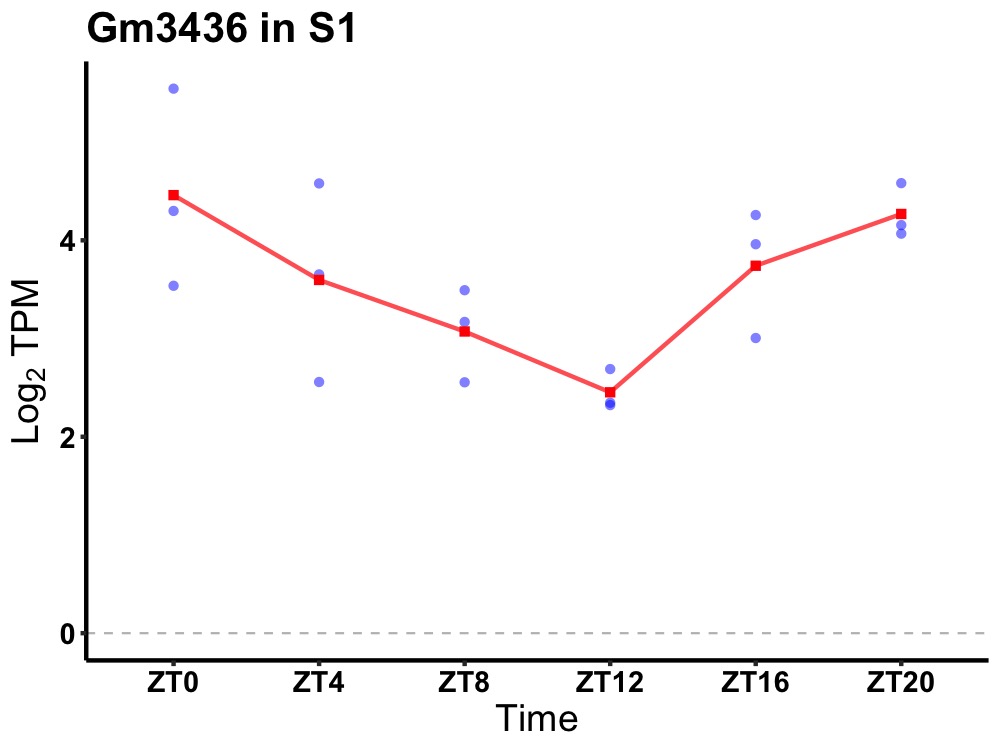 |
predicted pseudogene 3436 | 0.010 | 24 | 0 | 0.77 |
| ENSMUSG00000068205 | Stag2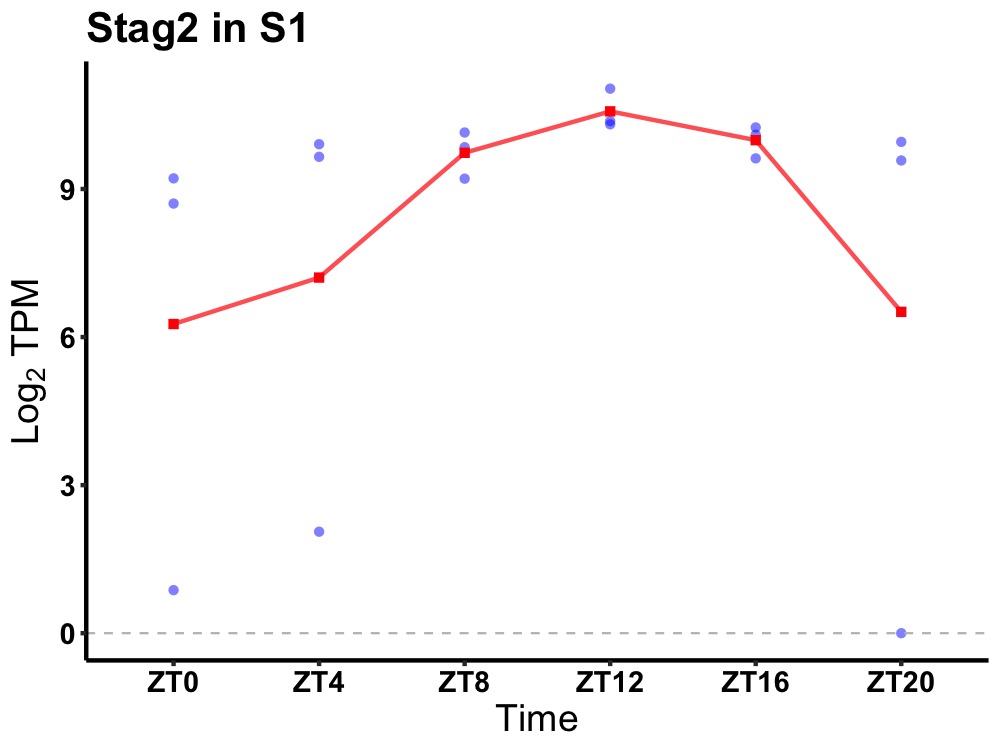 |
stromal antigen 2 | 0.007 | 24 | 14 | 0.76 |
| ENSMUSG00000087008 | Gm5530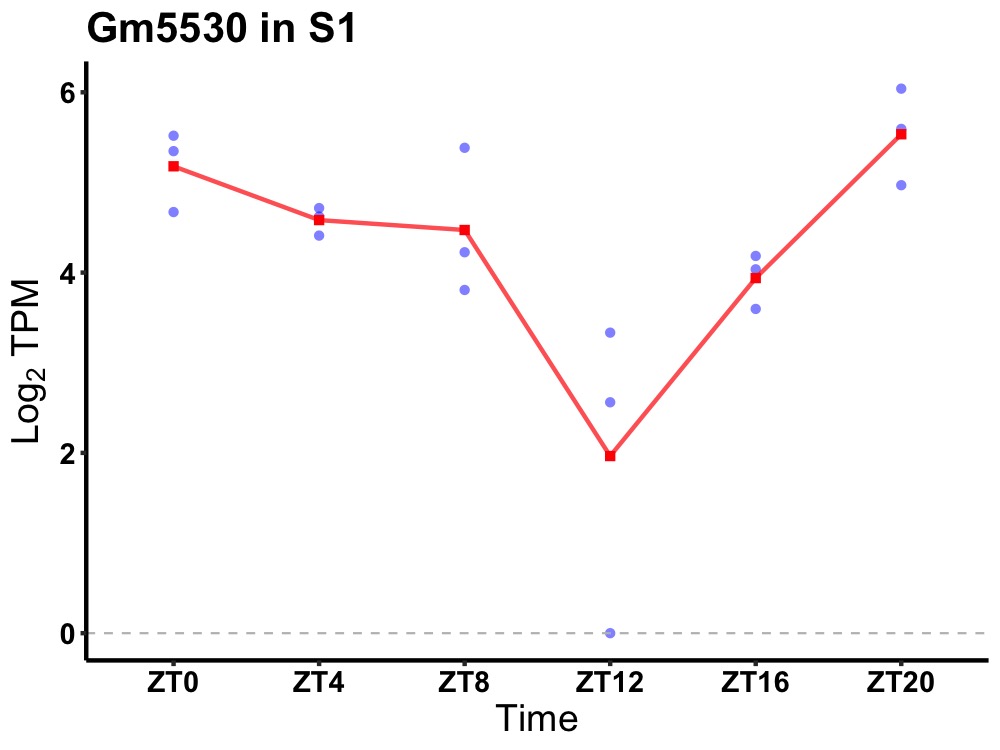 |
predicted gene 5530 | 0.000 | 20 | 2 | 0.76 |
| ENSMUSG00000005615 | Klf15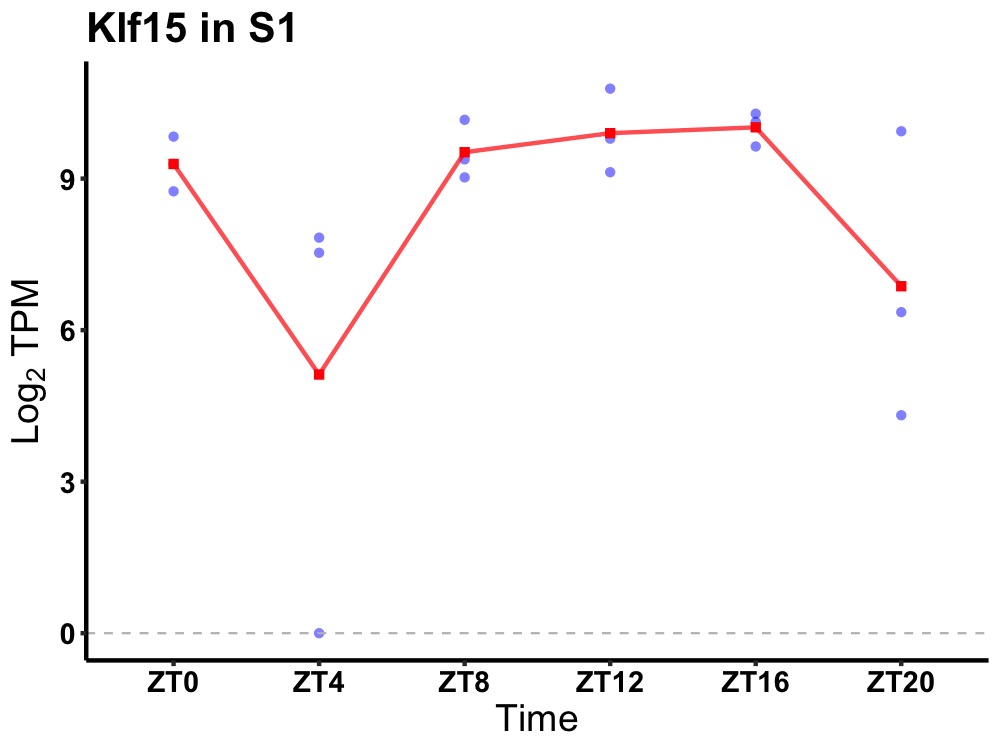 |
Kruppel-like factor 15 | 0.049 | 20 | 14 | 0.76 |
| ENSMUSG00000091697 | Arsg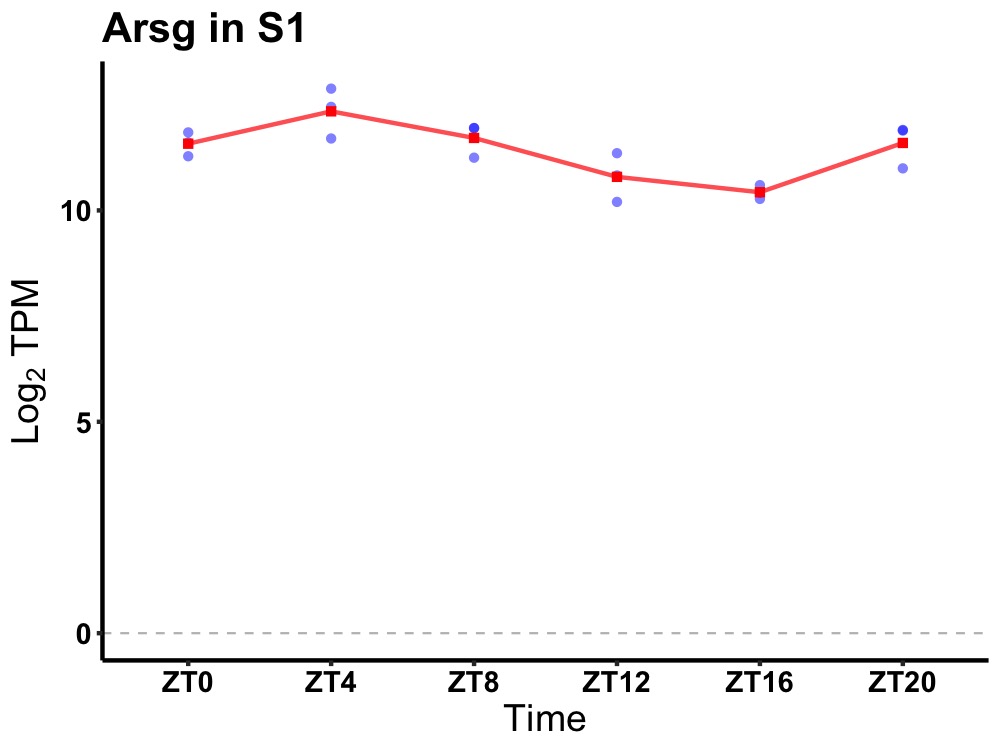 |
arylsulfatase G | 0.001 | 20 | 6 | 0.76 |
| ENSMUSG00000105243 | Mgat2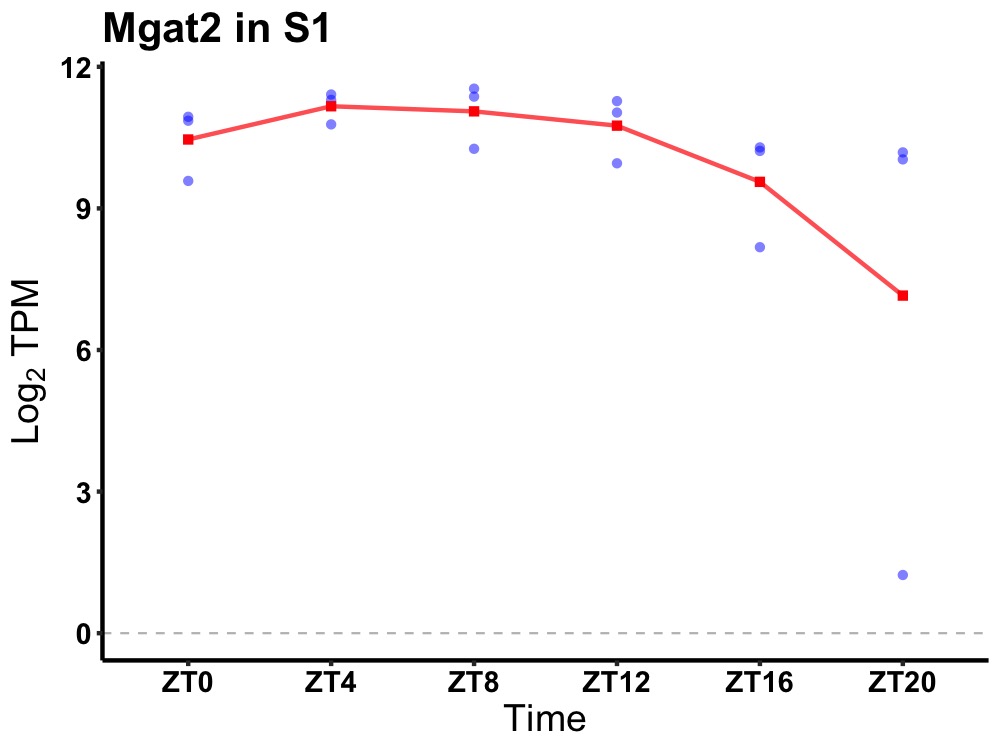 |
mannoside acetylglucosaminyltransferase 2 | 0.014 | 24 | 8 | 0.76 |
| ENSMUSG00000116835 | Gsk3b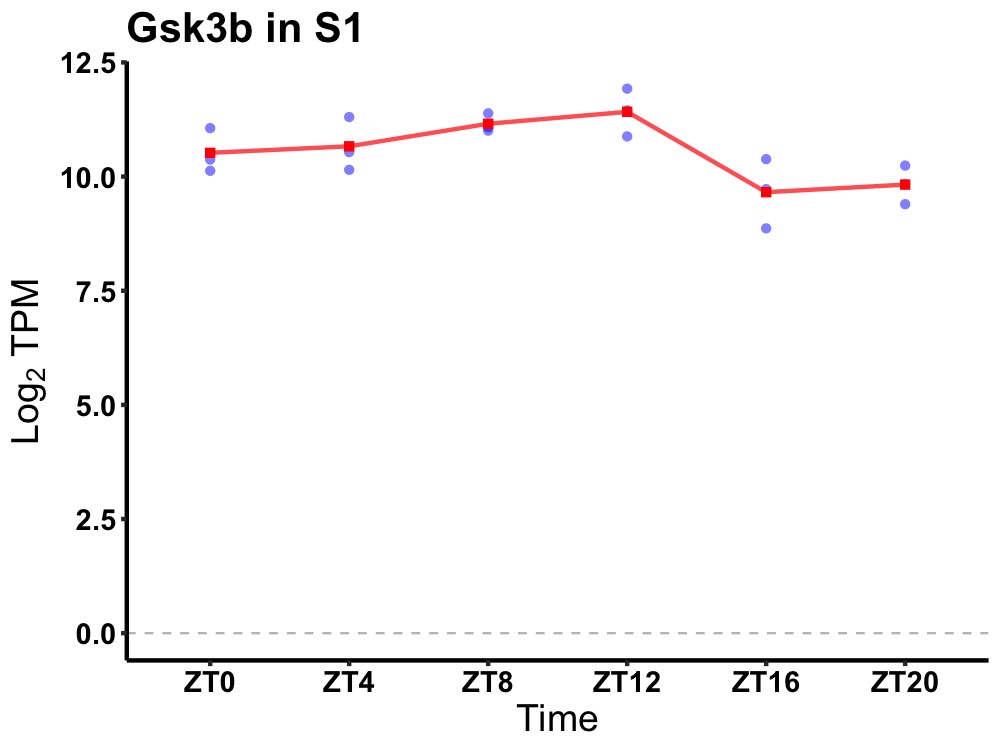 |
glycogen synthase kinase 3 beta | 0.049 | 24 | 10 | 0.75 |
| ENSMUSG00000035337 | Uchl4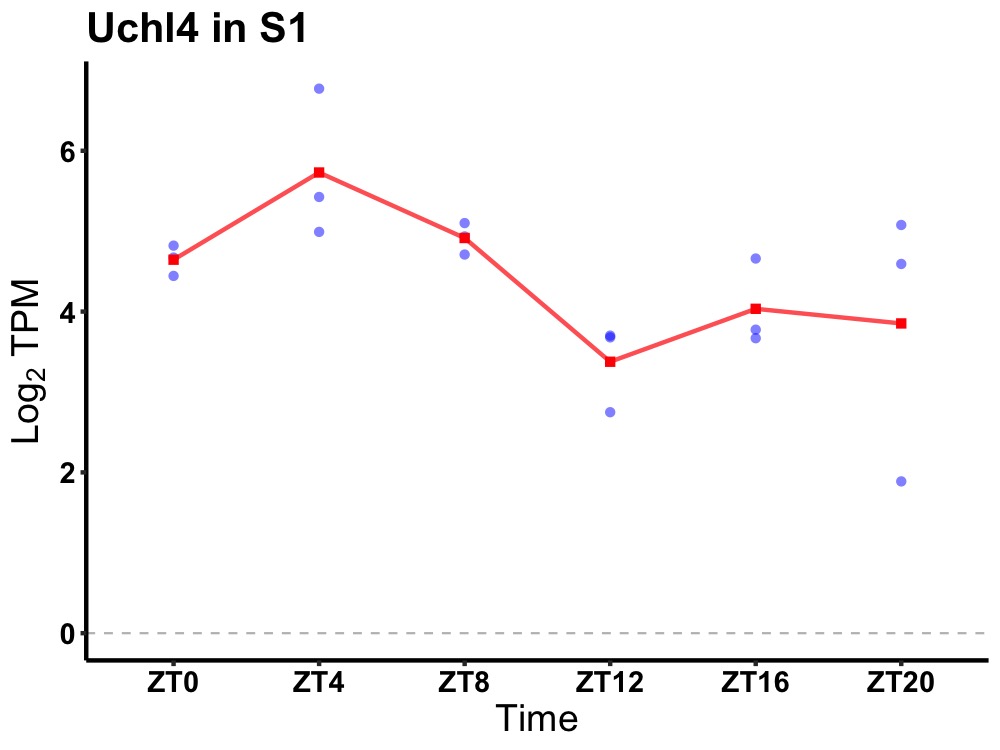 |
ubiquitin carboxyl-terminal esterase L4 | 0.014 | 20 | 6 | 0.75 |
| ENSMUSG00000042251 | Eif3d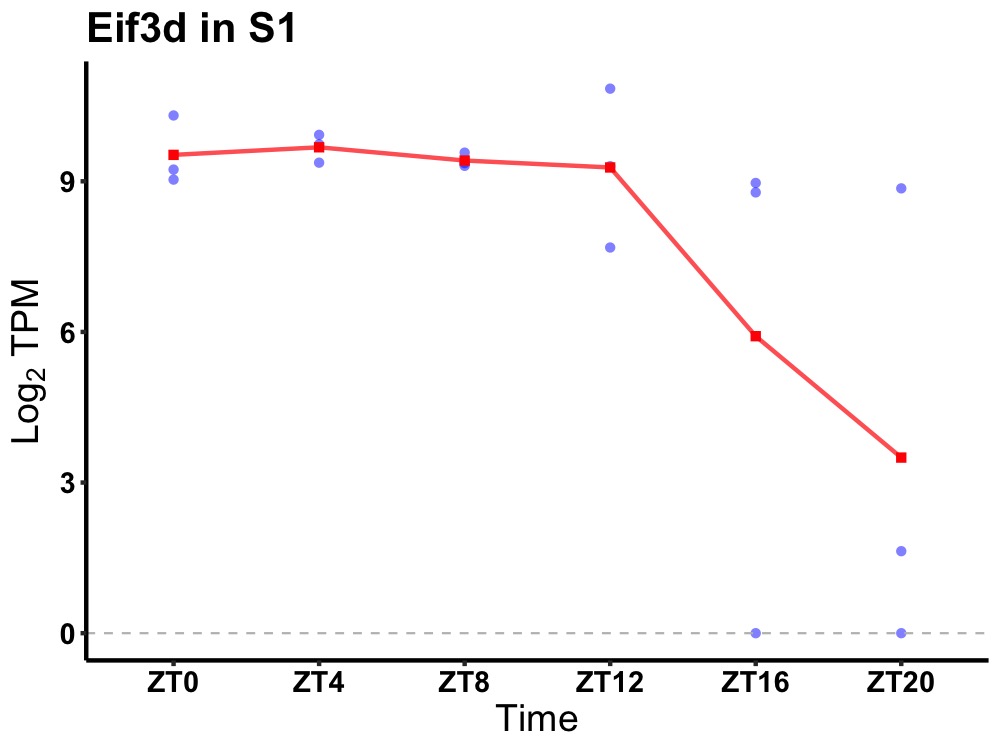 |
eukaryotic translation initiation factor 3, subunit D | 0.023 | 24 | 6 | 0.75 |
| ENSMUSG00000005413 | Reep3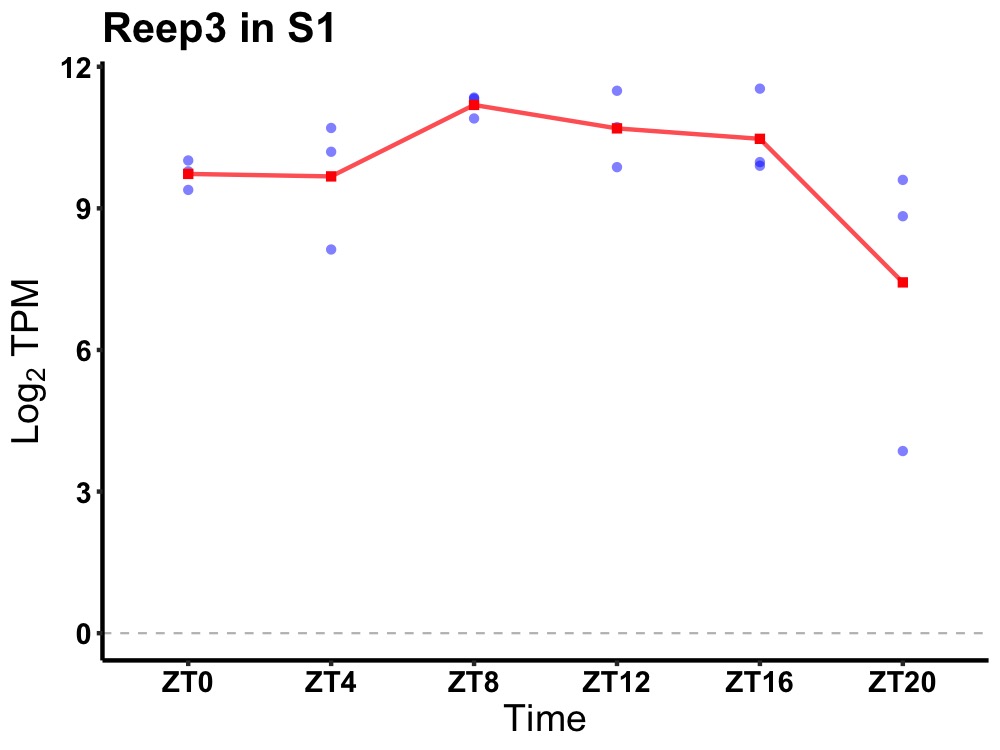 |
receptor accessory protein 3 | 0.027 | 24 | 10 | 0.73 |
| ENSMUSG00000061353 | Oplah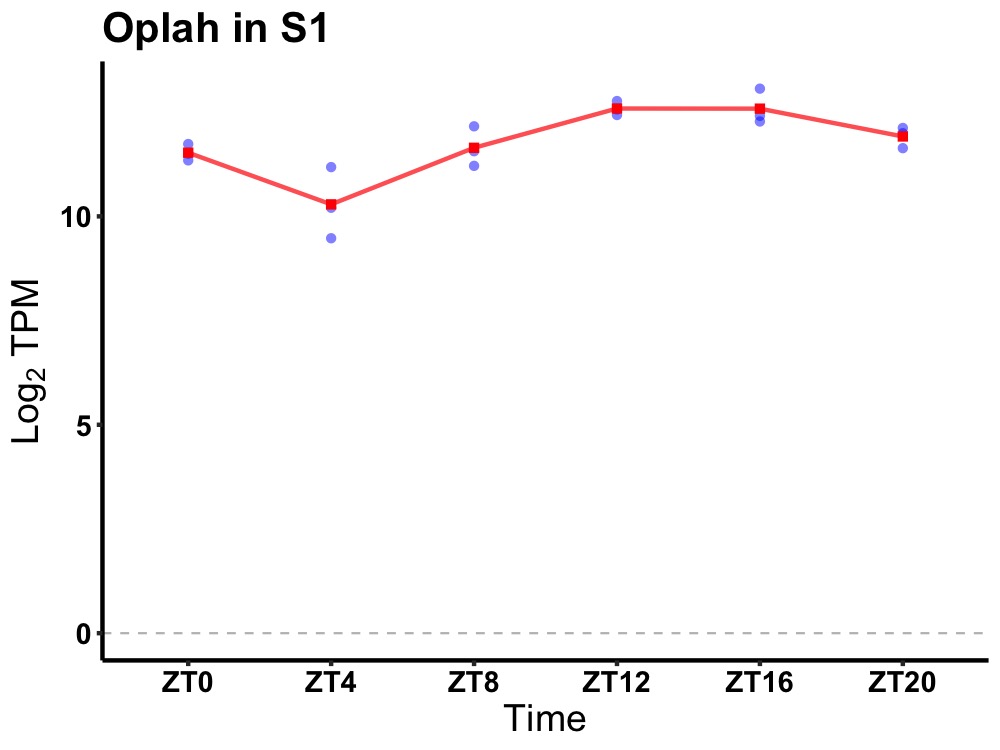 |
5-oxoprolinase (ATP-hydrolysing) | 0.000 | 24 | 16 | 0.73 |
| ENSMUSG00000068226 | Tmed2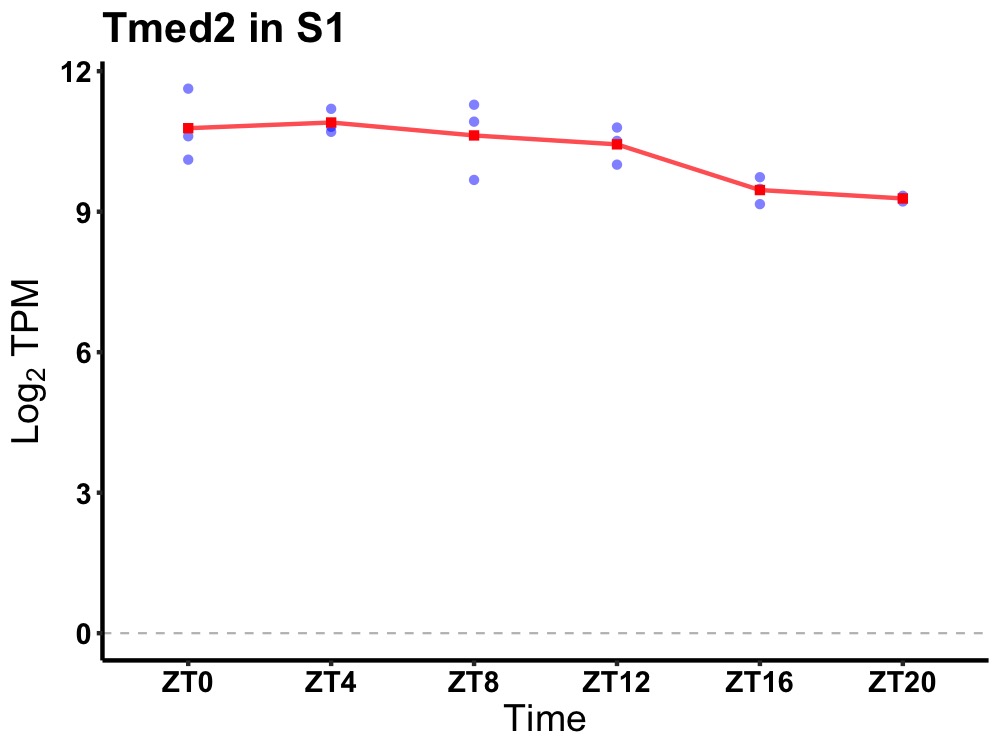 |
transmembrane p24 trafficking protein 2 | 0.019 | 24 | 8 | 0.73 |
| ENSMUSG00000022722 | Nt5e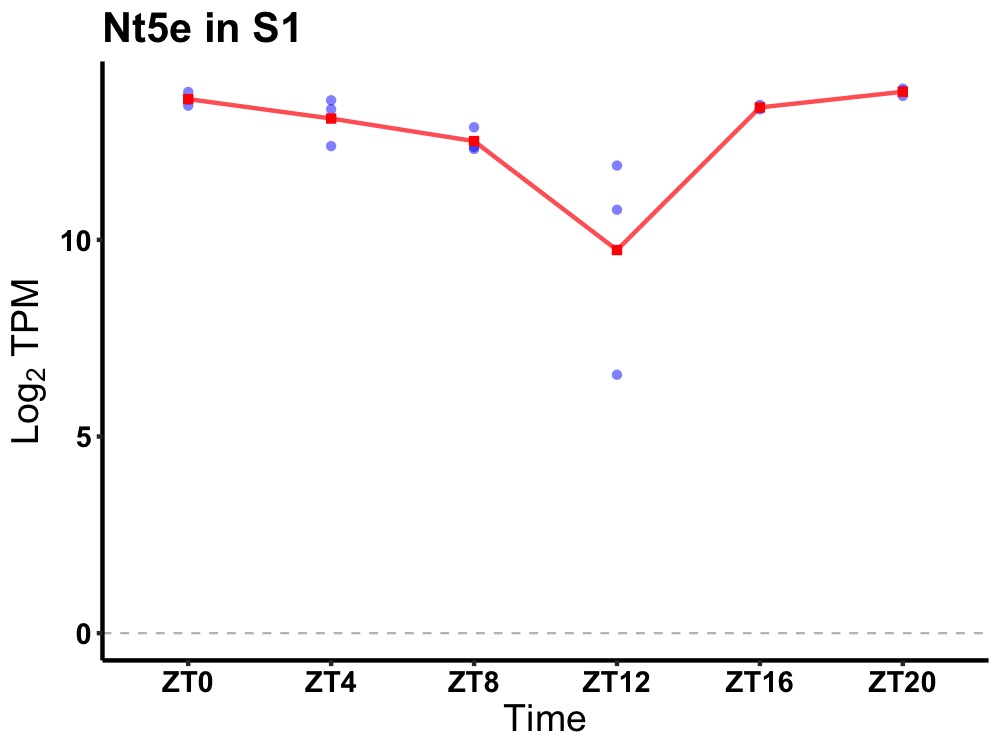 |
5' nucleotidase, ecto | 0.000 | 20 | 2 | 0.73 |
| ENSMUSG00000117935 | AC111023.1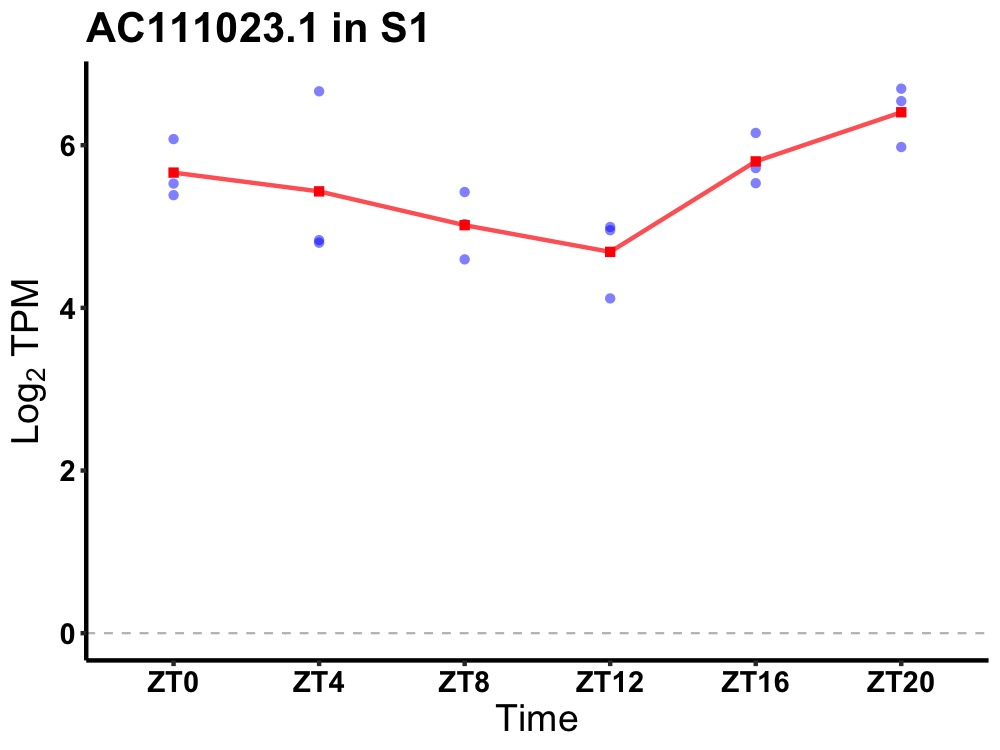 |
none | 0.049 | 24 | 20 | 0.72 |
| ENSMUSG00000025917 | Odc1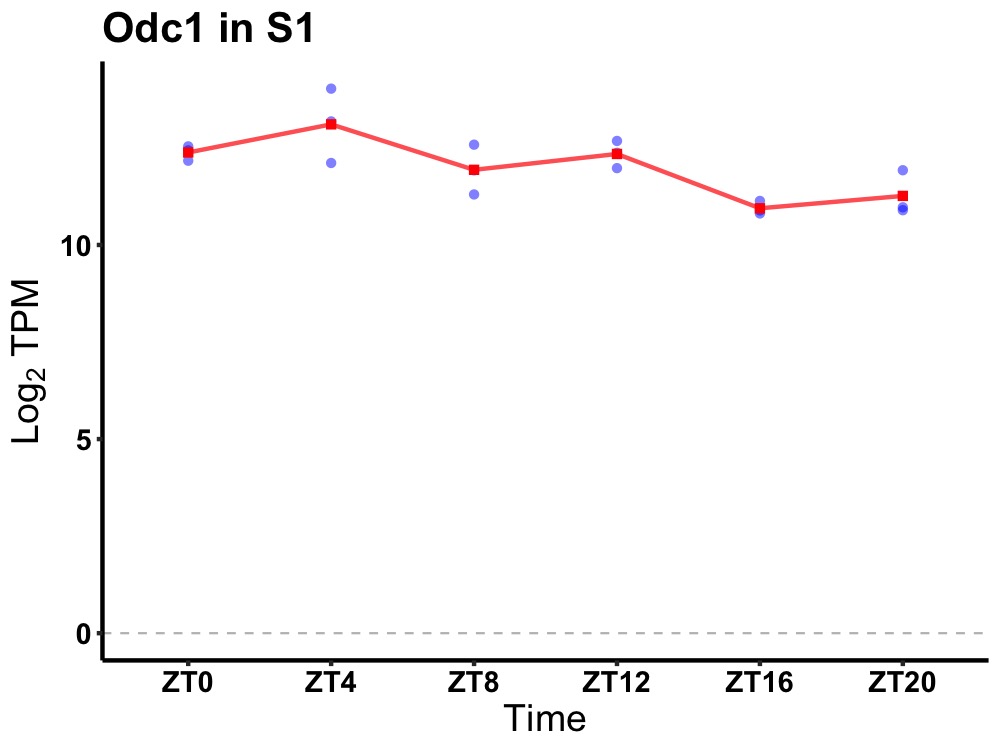 |
ornithine decarboxylase, structural 1 | 0.027 | 24 | 6 | 0.72 |
| ENSMUSG00000050628 | Ubald2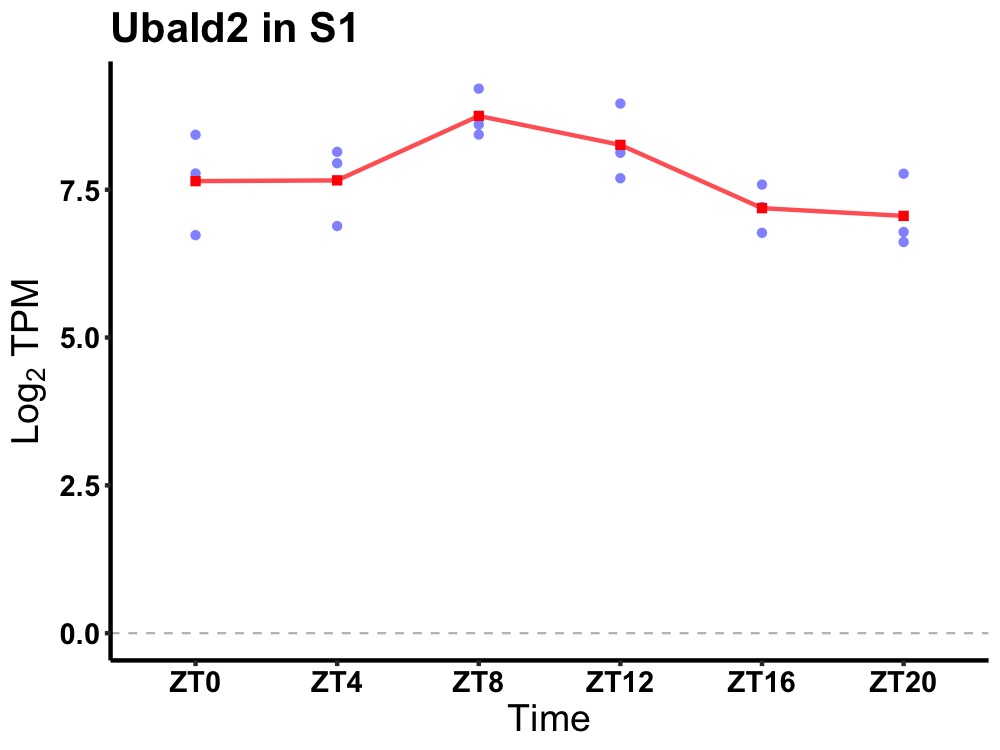 |
UBA-like domain containing 2 | 0.036 | 24 | 10 | 0.72 |
| ENSMUSG00000108562 | Mrpl10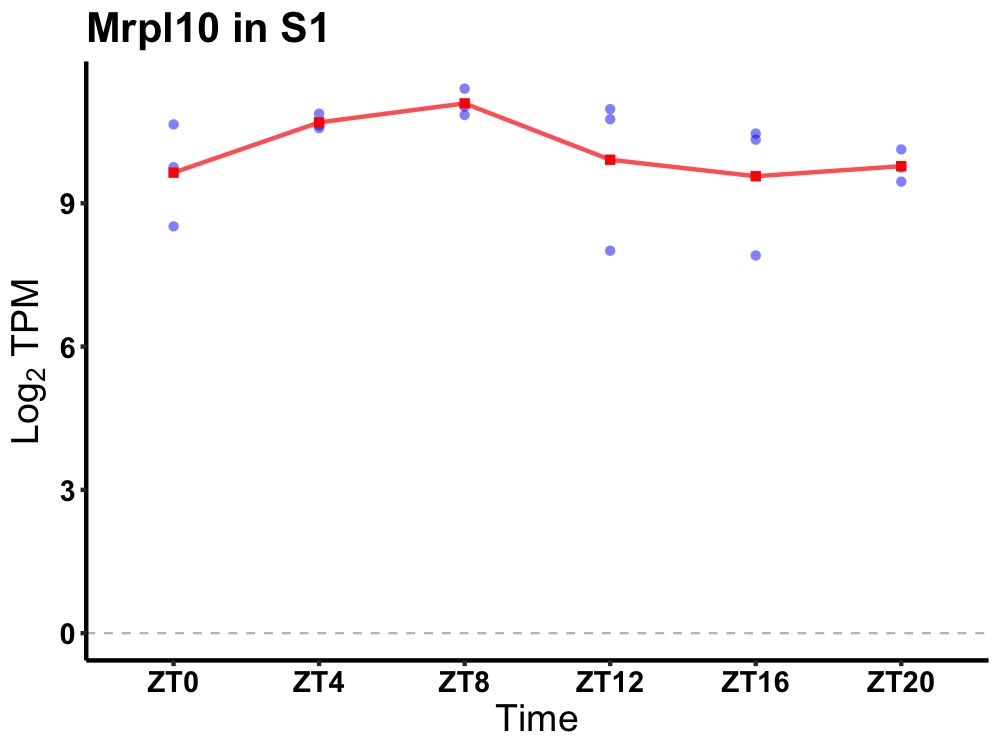 |
mitochondrial ribosomal protein L10 | 0.036 | 24 | 10 | 0.71 |
| ENSMUSG00000038970 | Acad12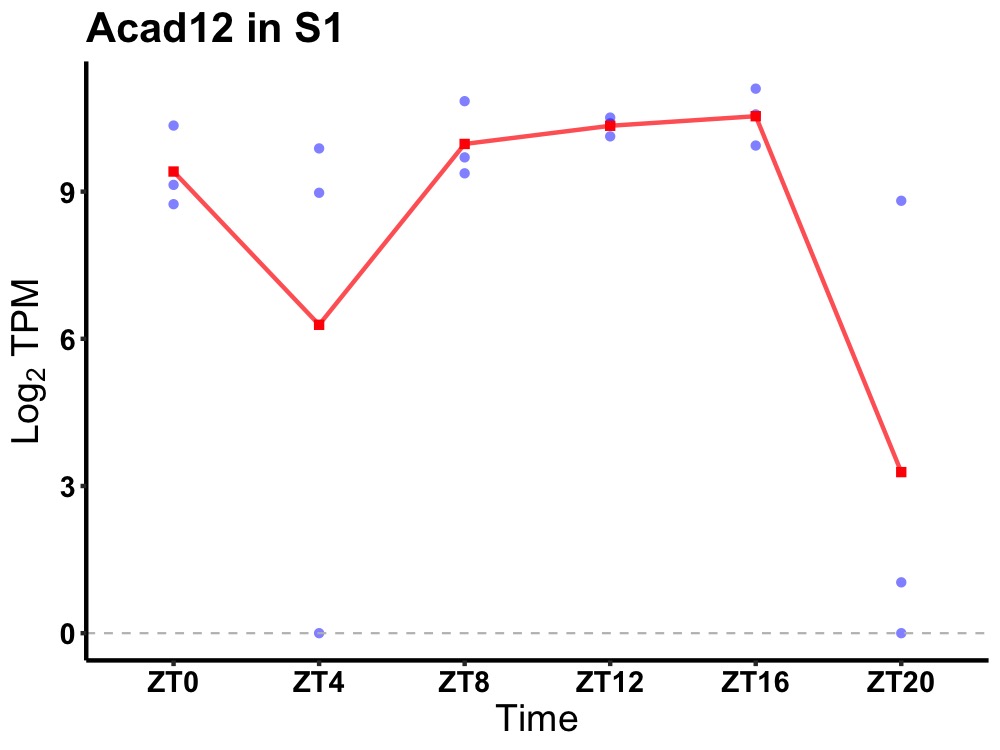 |
acyl-Coenzyme A dehydrogenase family, member 12 | 0.031 | 20 | 14 | 0.71 |
| ENSMUSG00000100441 | Atg101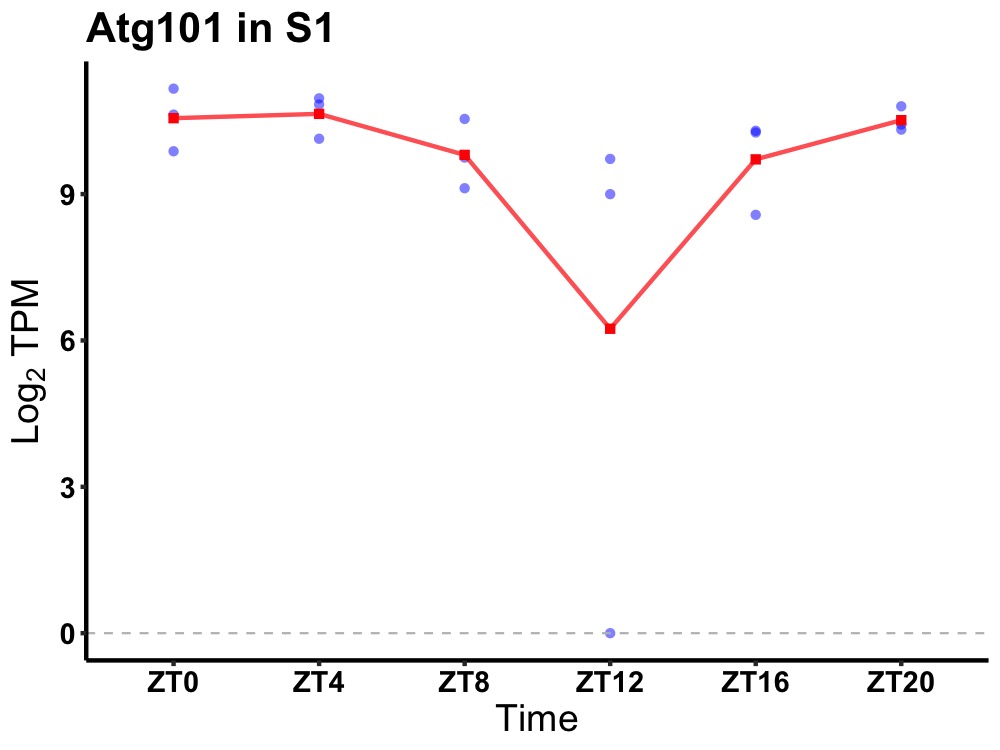 |
autophagy related 101 | 0.027 | 24 | 2 | 0.71 |
| ENSMUSG00000115938 | Tvp23b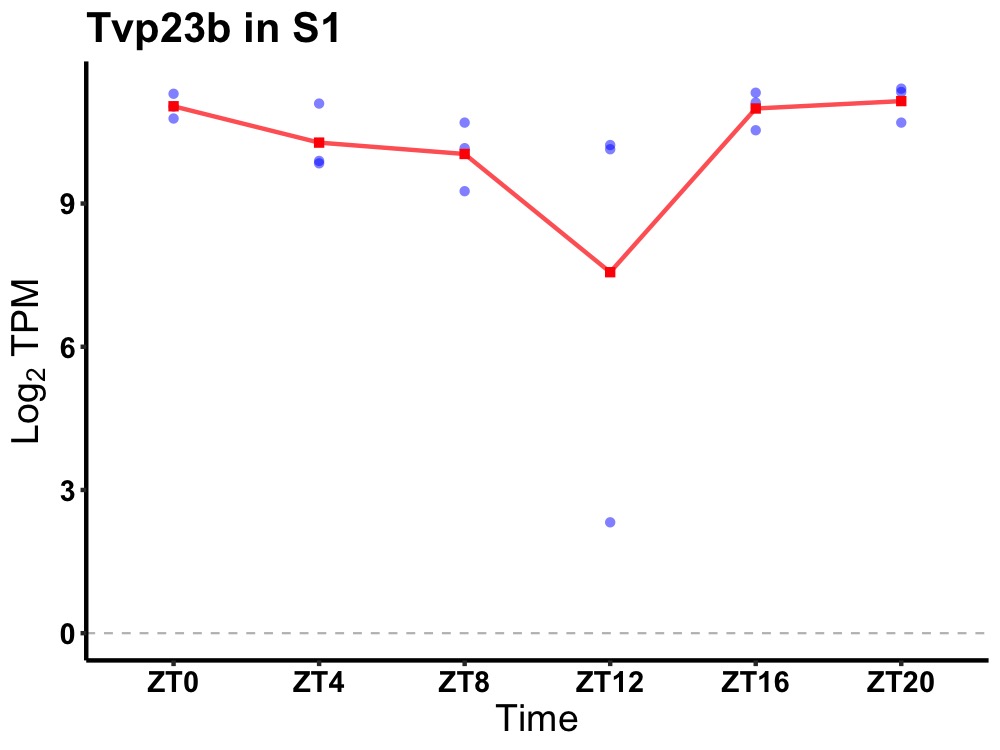 |
trans-golgi network vesicle protein 23B | 0.049 | 24 | 22 | 0.71 |
| ENSMUSG00000075415 | Acp6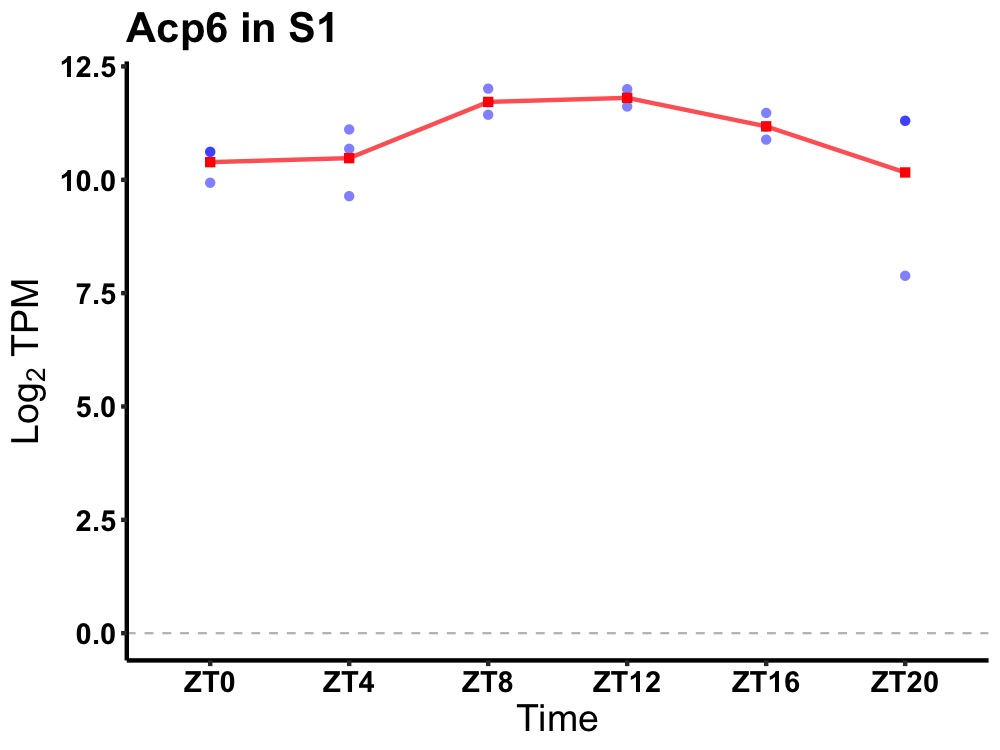 |
acid phosphatase 6, lysophosphatidic | 0.005 | 24 | 12 | 0.70 |
| ENSMUSG00000078348 | Sys1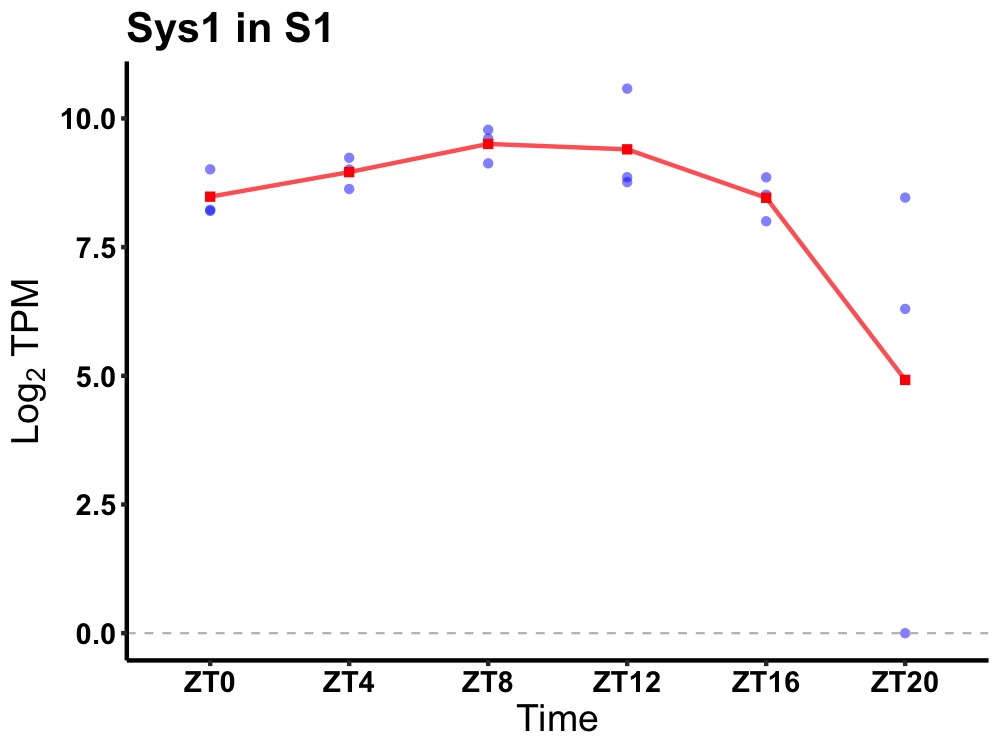 |
SYS1 Golgi-localized integral membrane protein homolog (S. cerevisiae) | 0.005 | 24 | 10 | 0.70 |
| ENSMUSG00000115538 | Hspd1-ps5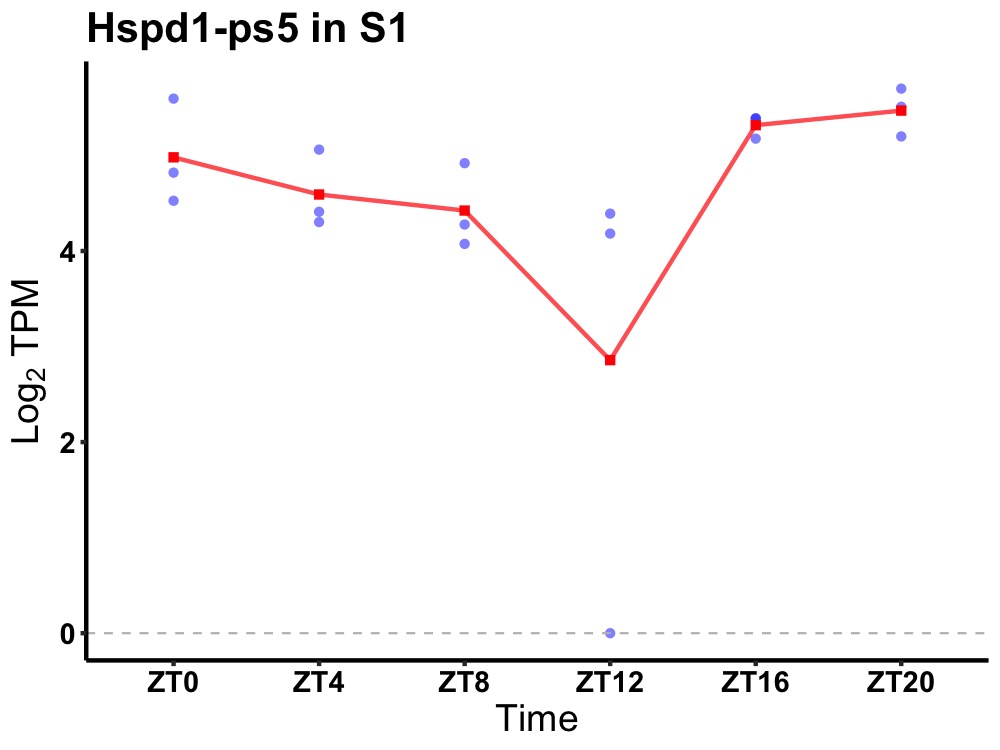 |
heat shock protein 1 (chaperonin), pseudogene 5 | 0.007 | 24 | 22 | 0.70 |
| ENSMUSG00000081731 | Calr-ps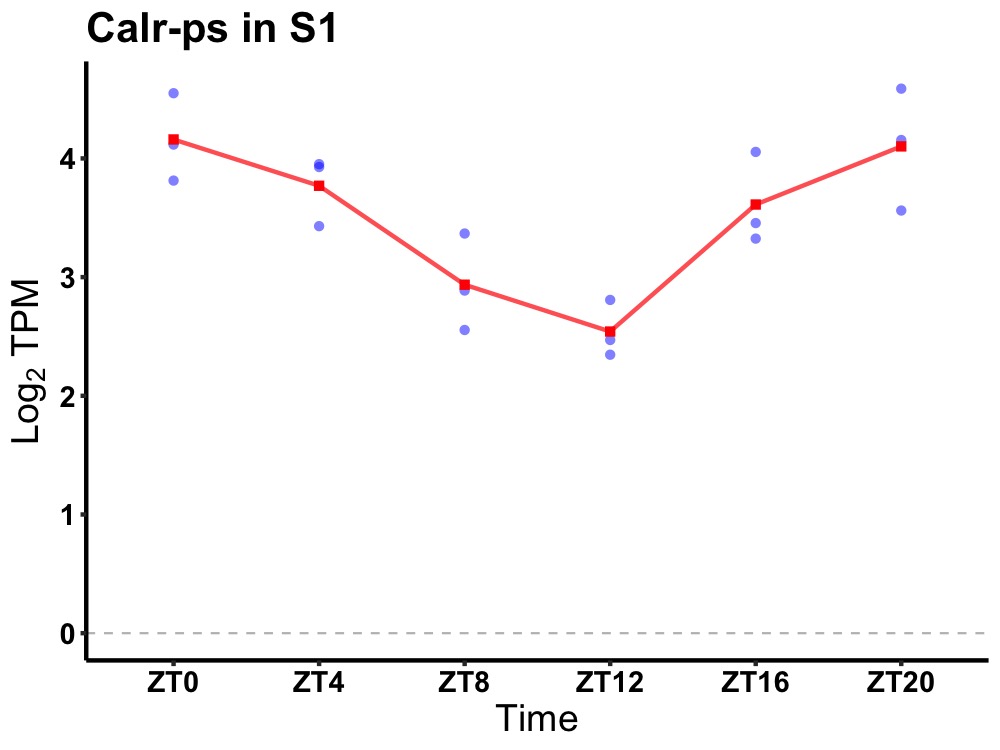 |
calreticulin, pseudogene | 0.000 | 20 | 2 | 0.70 |
| ENSMUSG00000060657 | Sat2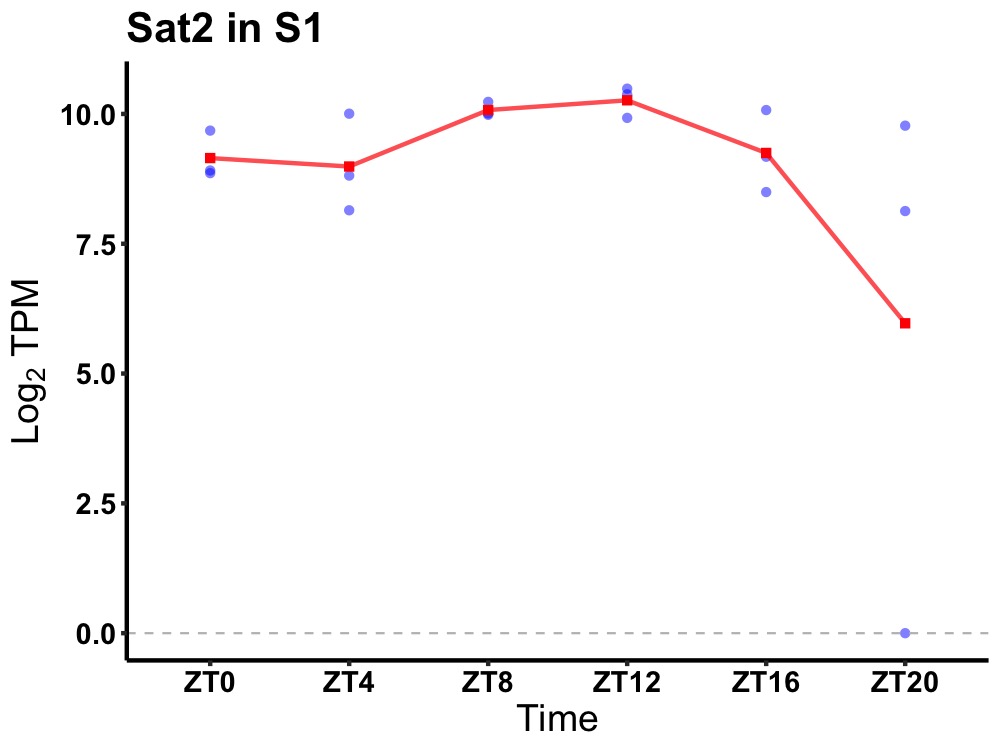 |
spermidine/spermine N1-acetyl transferase 2 | 0.036 | 20 | 12 | 0.69 |
| ENSMUSG00000084874 | Gm12906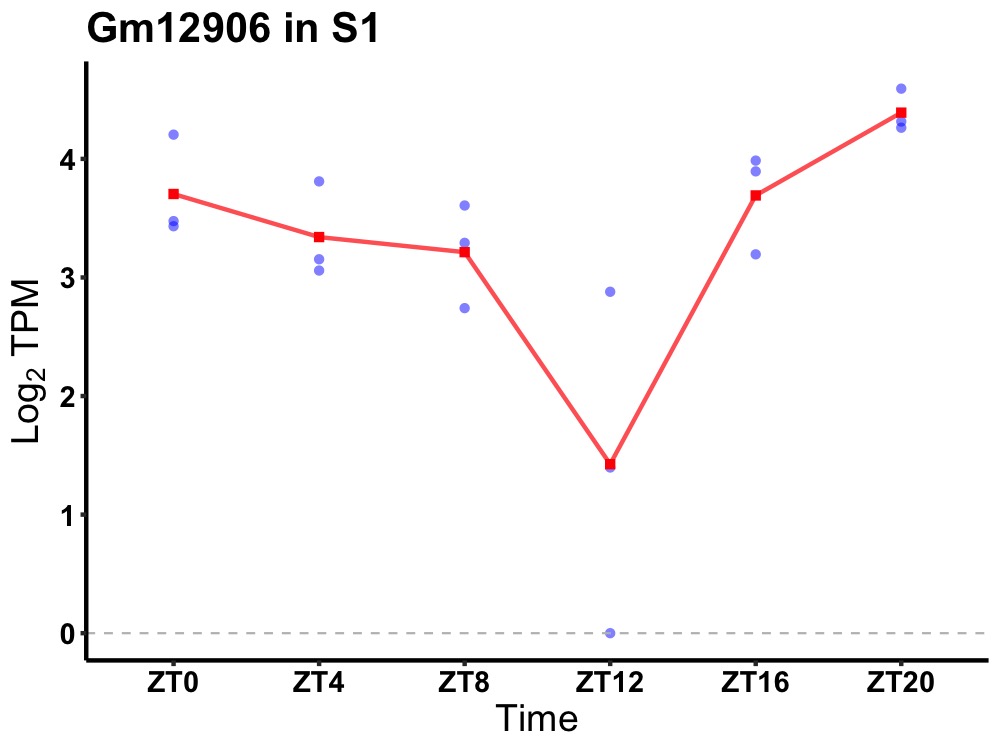 |
predicted gene 12906 | 0.007 | 24 | 22 | 0.69 |
| ENSMUSG00000104664 | Gm35570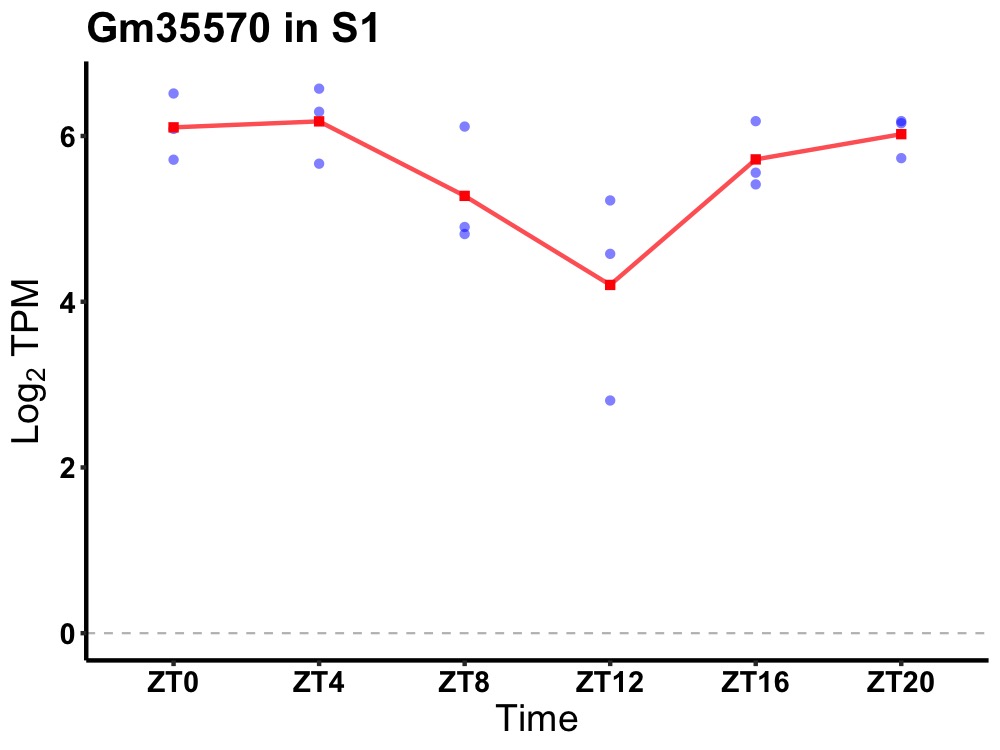 |
predicted gene, 35570 | 0.036 | 20 | 2 | 0.68 |
| ENSMUSG00000021025 | Pmm1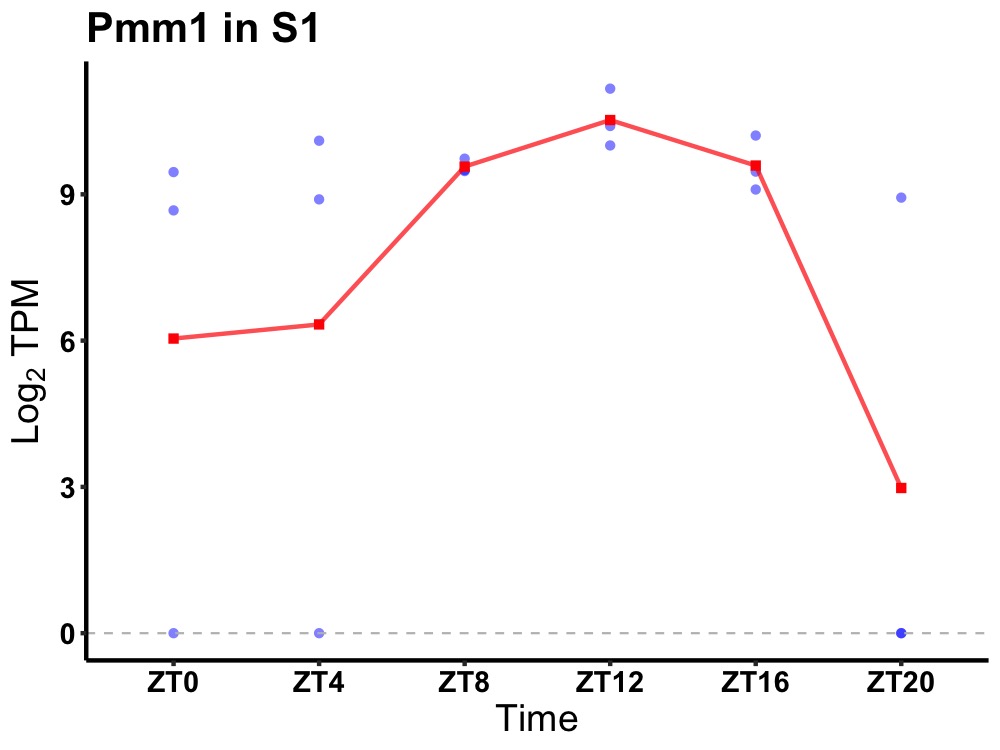 |
phosphomannomutase 1 | 0.003 | 20 | 12 | 0.68 |
| ENSMUSG00000059796 | Tardbp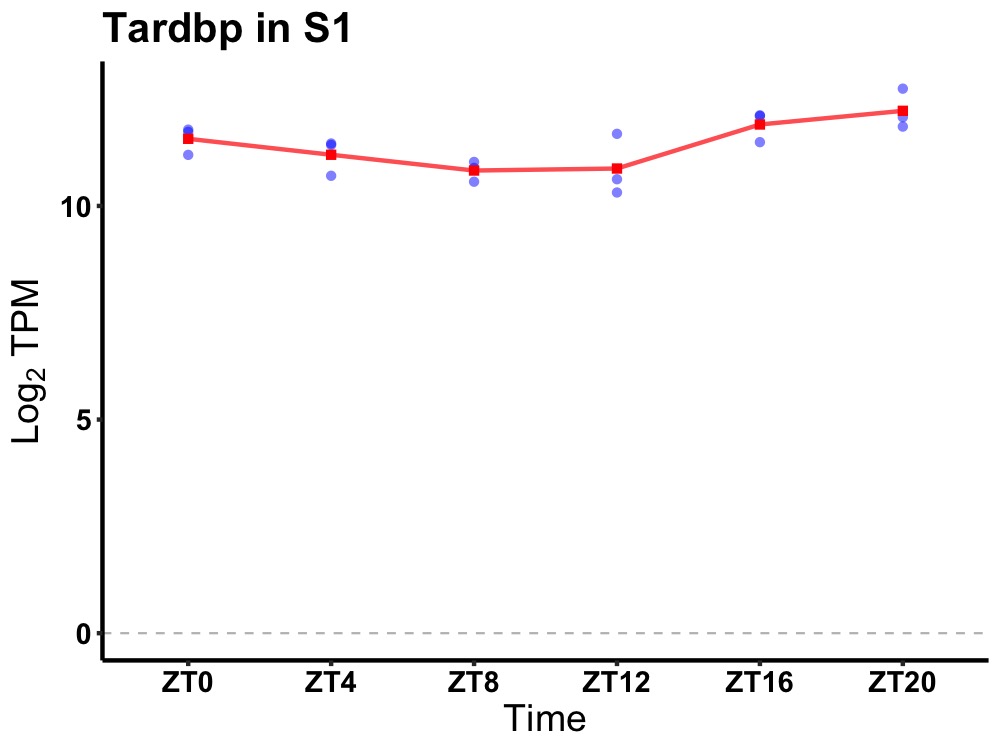 |
TAR DNA binding protein | 0.007 | 24 | 20 | 0.68 |
| ENSMUSG00000024604 | Rwdd1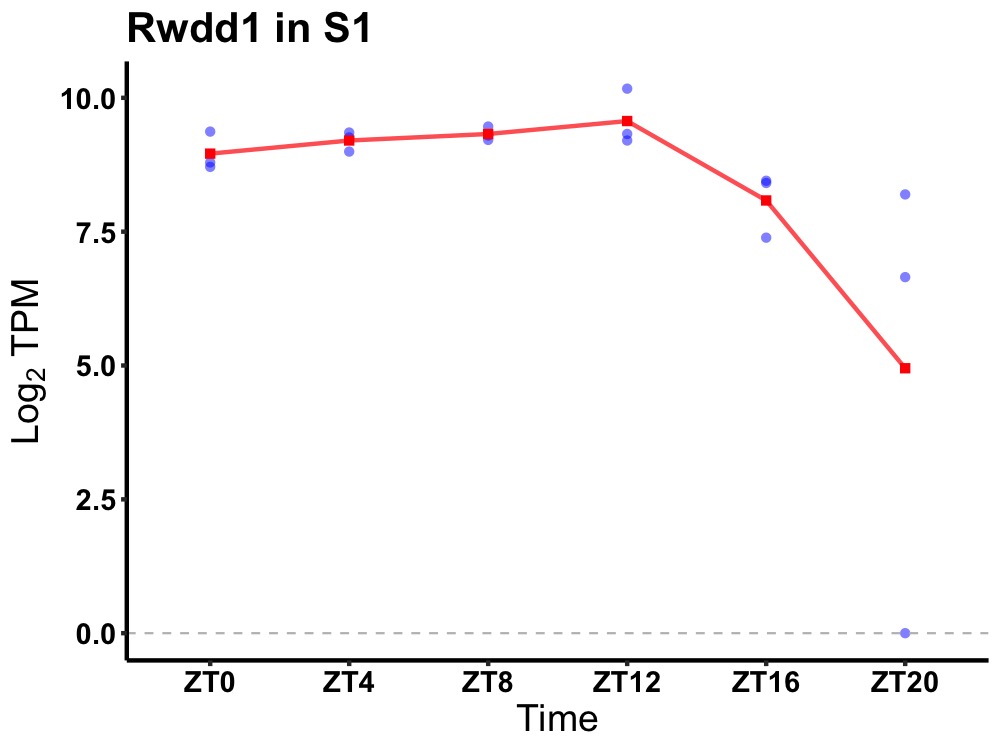 |
RWD domain containing 1 | 0.005 | 24 | 8 | 0.67 |
| ENSMUSG00000080847 | Atp1b3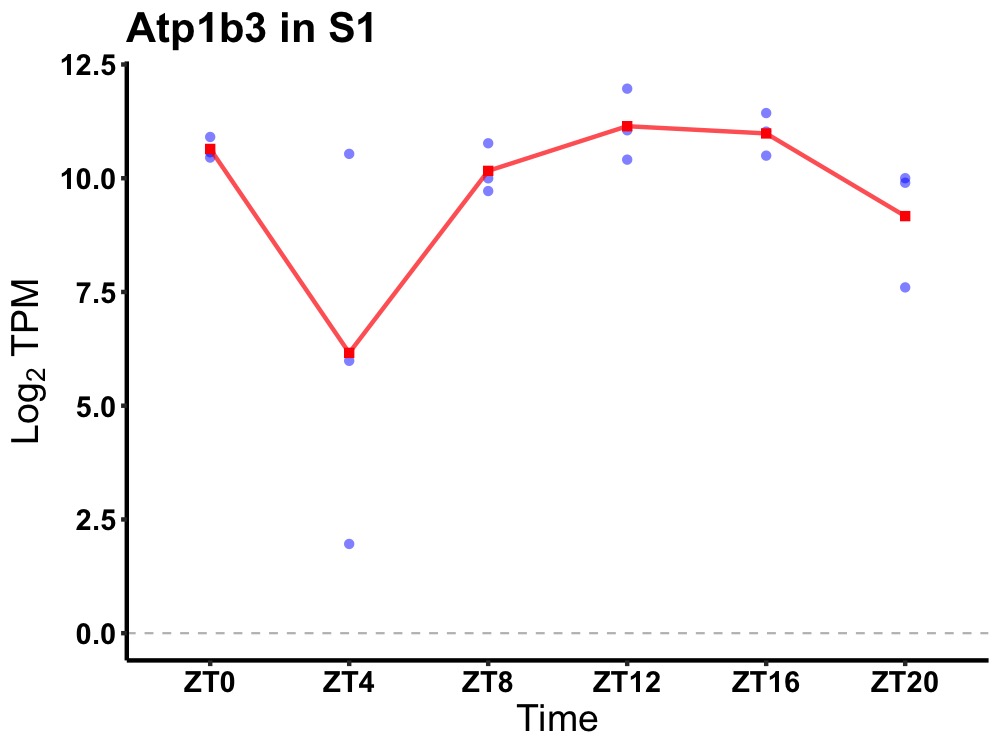 |
ATPase, Na+/K+ transporting, beta 3 polypeptide | 0.036 | 20 | 16 | 0.66 |
| ENSMUSG00000007850 | Nr1d2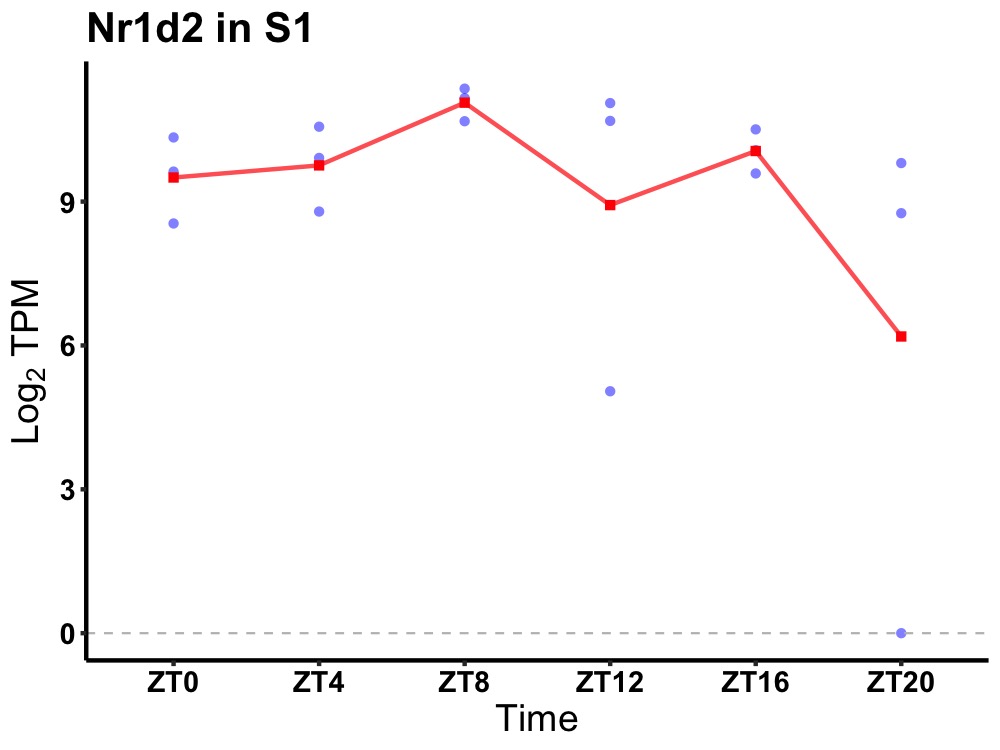 |
nuclear receptor subfamily 1 group D member 2 | 0.049 | 24 | 10 | 0.66 |
| ENSMUSG00000037709 | Psme4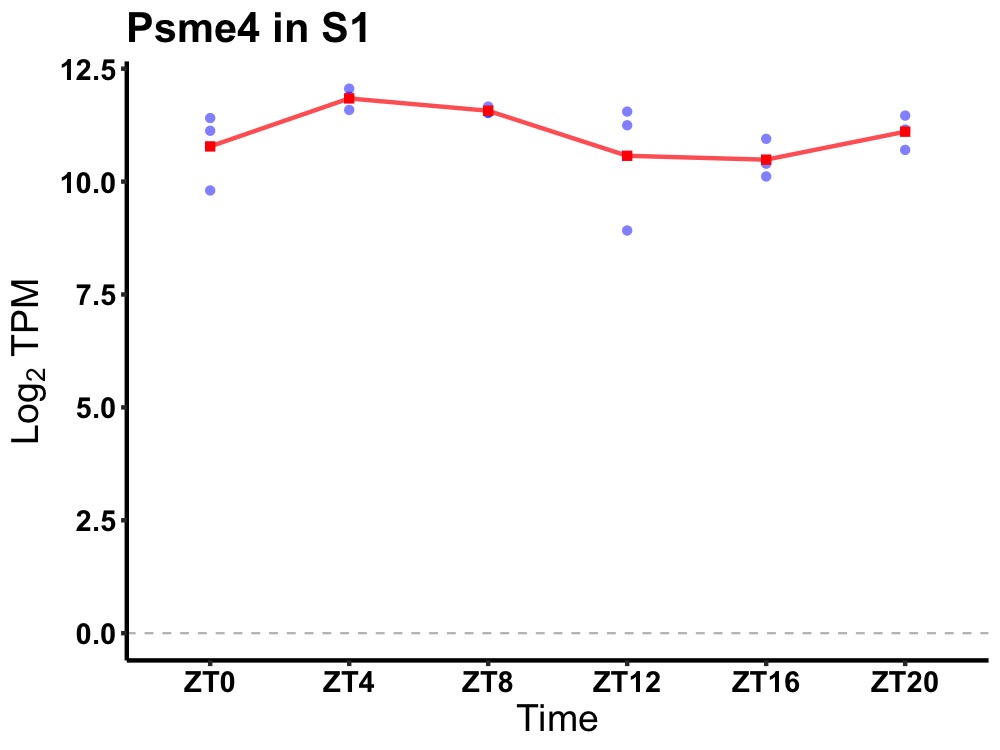 |
proteasome (prosome, macropain) activator subunit 4 | 0.007 | 20 | 6 | 0.66 |
| ENSMUSG00000085711 | Gm15163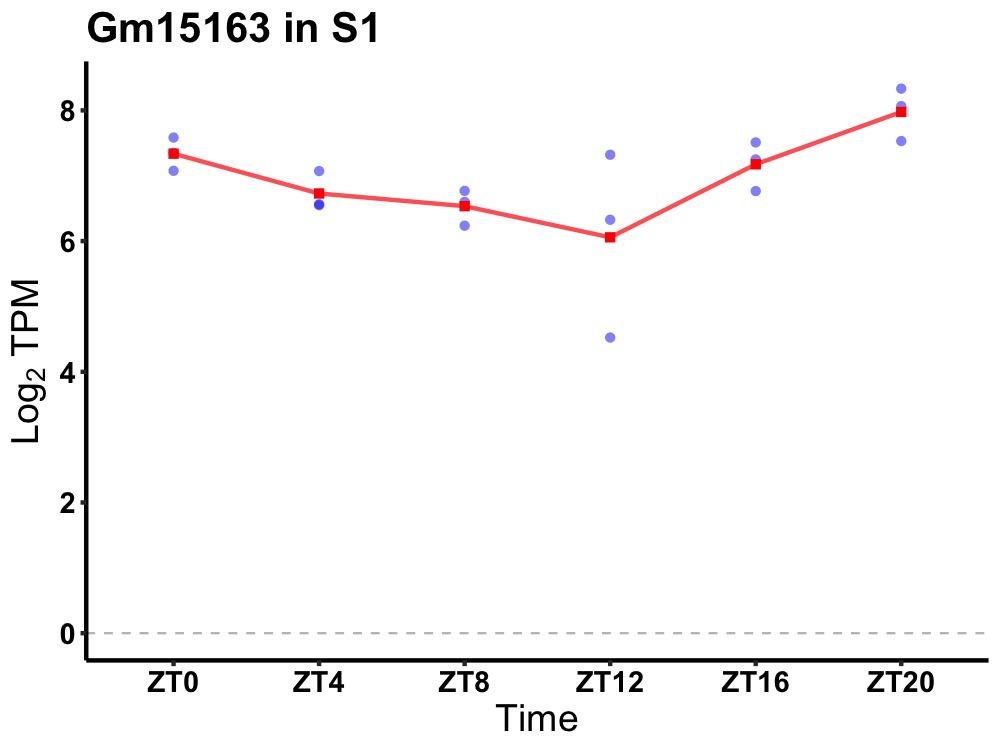 |
predicted gene 15163 | 0.003 | 24 | 22 | 0.66 |
| ENSMUSG00000061848 | Rab34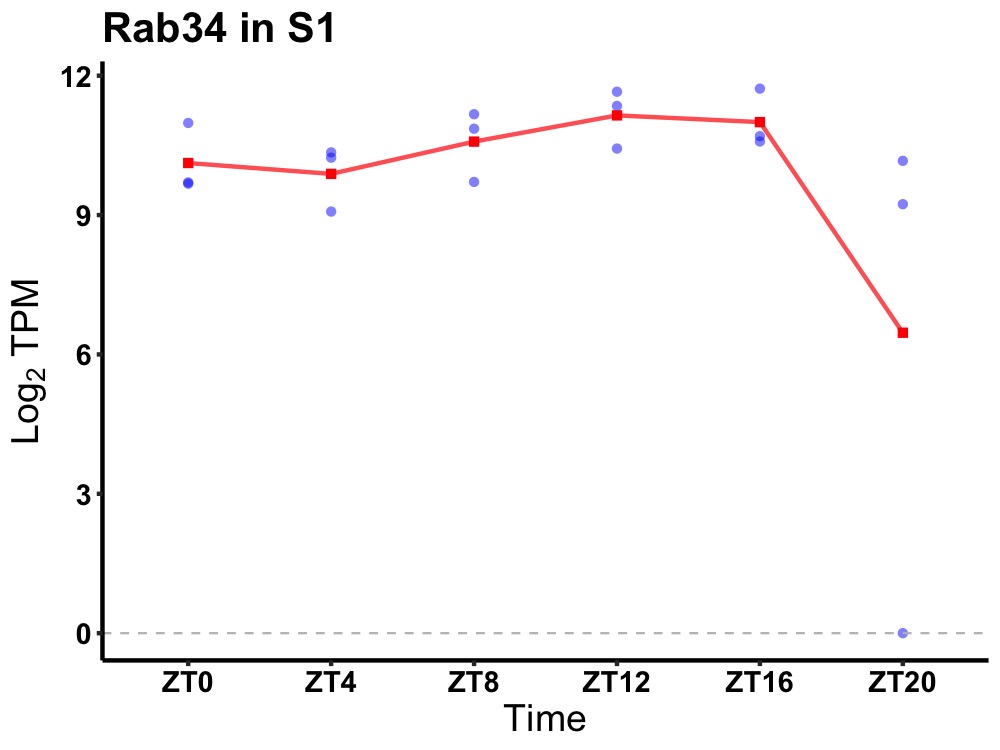 |
RAB34, member RAS oncogene family | 0.049 | 20 | 12 | 0.65 |
| ENSMUSG00000081957 | Ak3l2-ps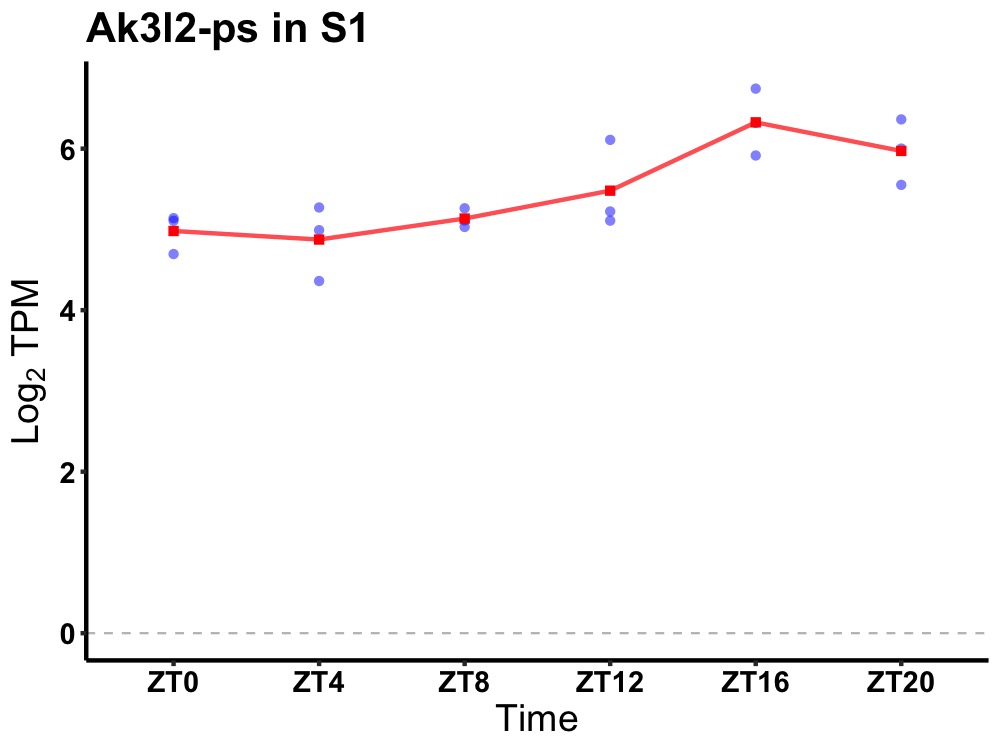 |
adenylate kinase 3-like 2, pseudogene | 0.014 | 24 | 18 | 0.65 |
| ENSMUSG00000069682 | Sec31a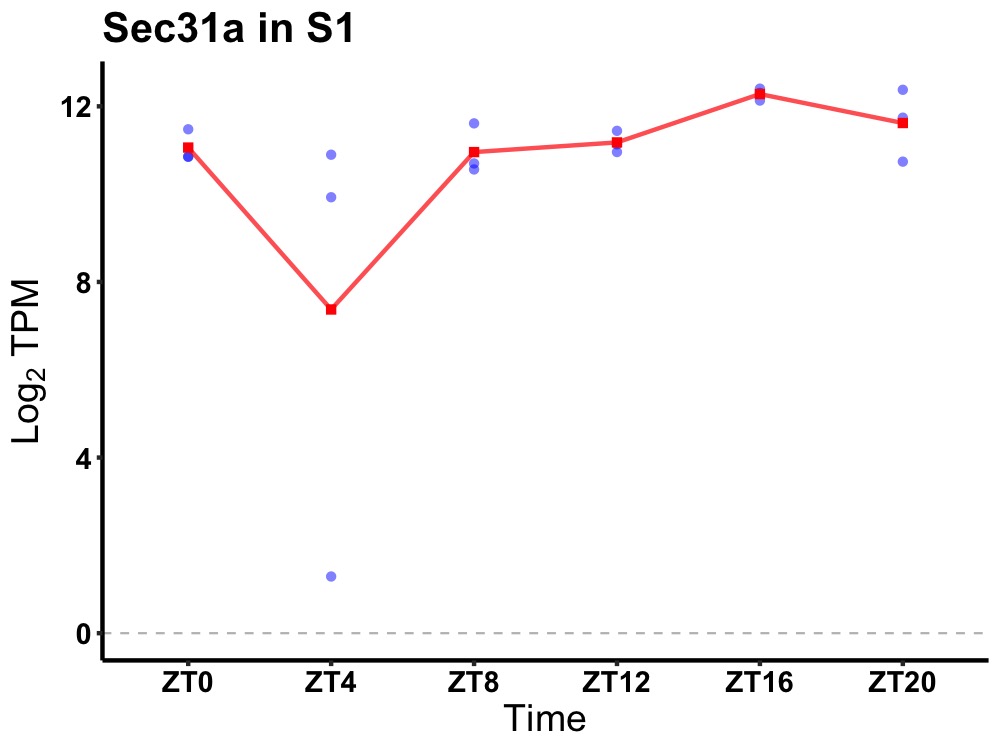 |
Sec31 homolog A (S. cerevisiae) | 0.007 | 24 | 18 | 0.65 |
| ENSMUSG00000056116 | Ostc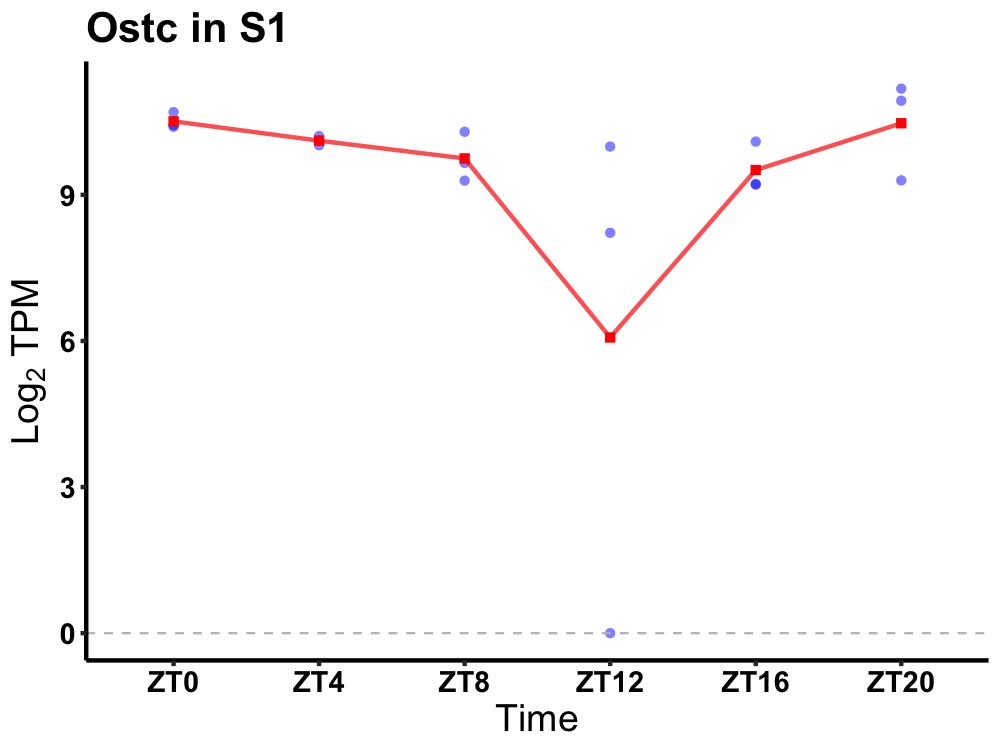 |
oligosaccharyltransferase complex subunit (non-catalytic) | 0.014 | 20 | 2 | 0.64 |
| ENSMUSG00000118345 | Pck1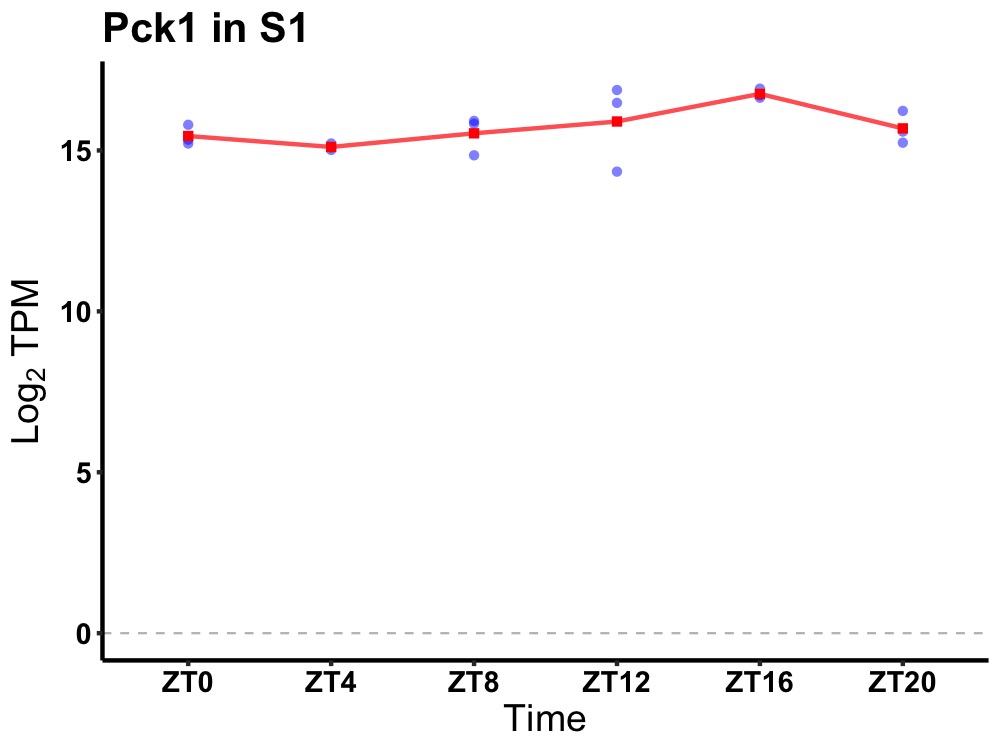 |
phosphoenolpyruvate carboxykinase 1, cytosolic | 0.036 | 24 | 16 | 0.63 |
| ENSMUSG00000034647 | Alas1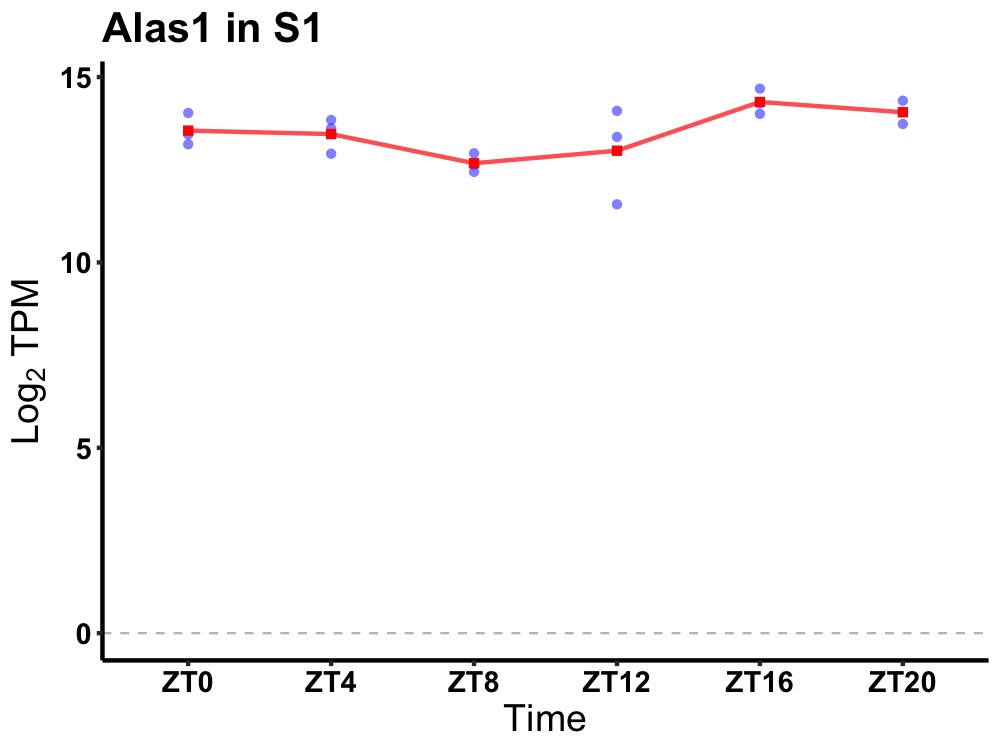 |
aminolevulinic acid synthase 1 | 0.049 | 24 | 20 | 0.62 |
| ENSMUSG00000032478 | Ppp2r5a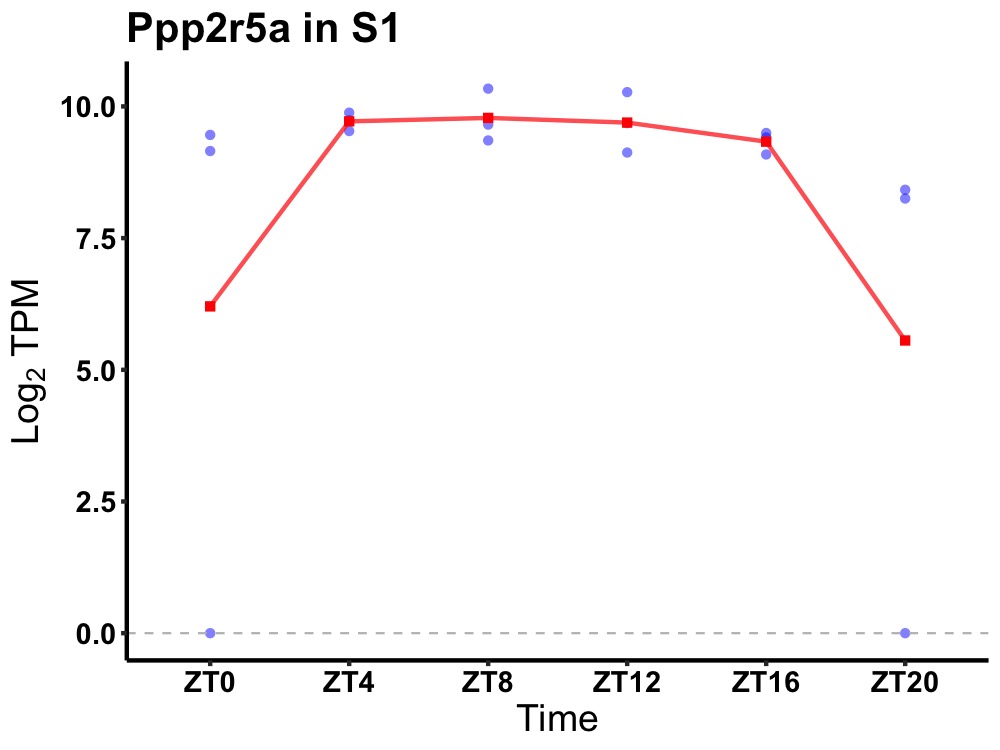 |
protein phosphatase 2, regulatory subunit B', alpha | 0.023 | 24 | 10 | 0.62 |
| ENSMUSG00000078291 | Gm3940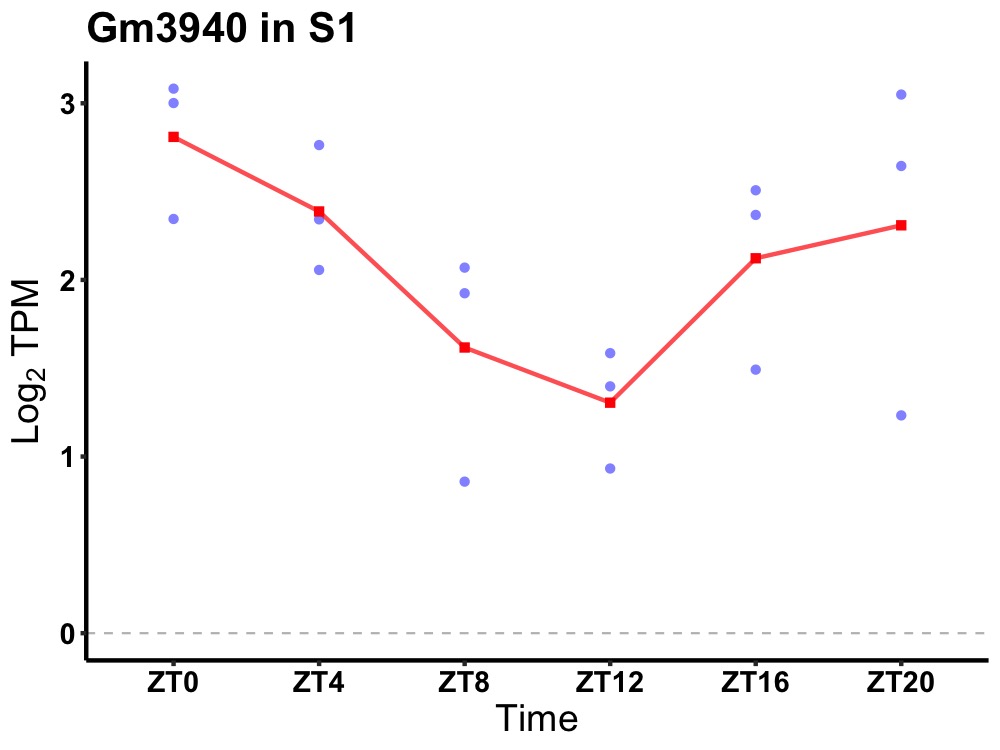 |
predicted gene 3940 | 0.036 | 24 | 0 | 0.62 |
| ENSMUSG00000113843 | Gns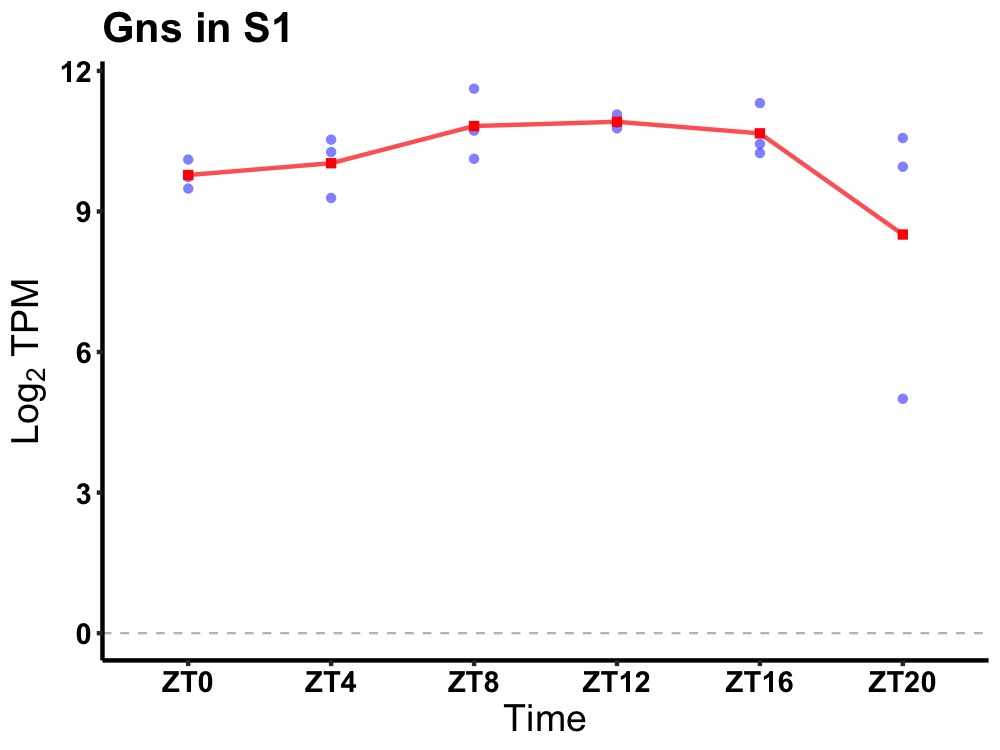 |
glucosamine (N-acetyl)-6-sulfatase | 0.049 | 24 | 12 | 0.62 |
| ENSMUSG00000082896 | Gm5844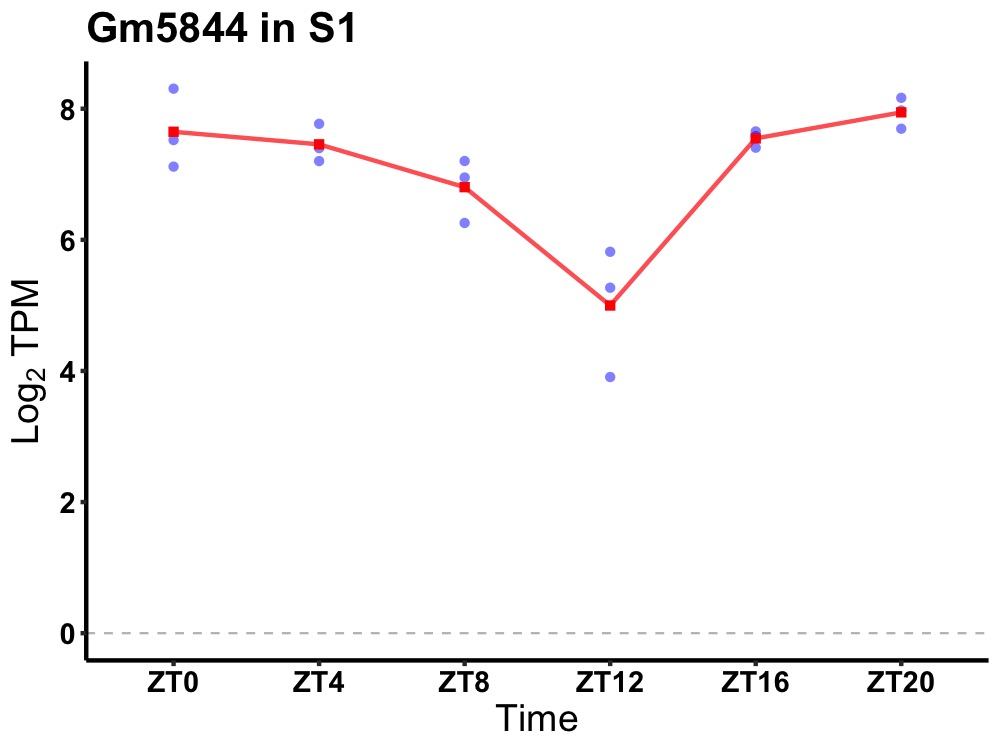 |
predicted gene 5844 | 0.003 | 20 | 2 | 0.61 |
| ENSMUSG00000024665 | Tram1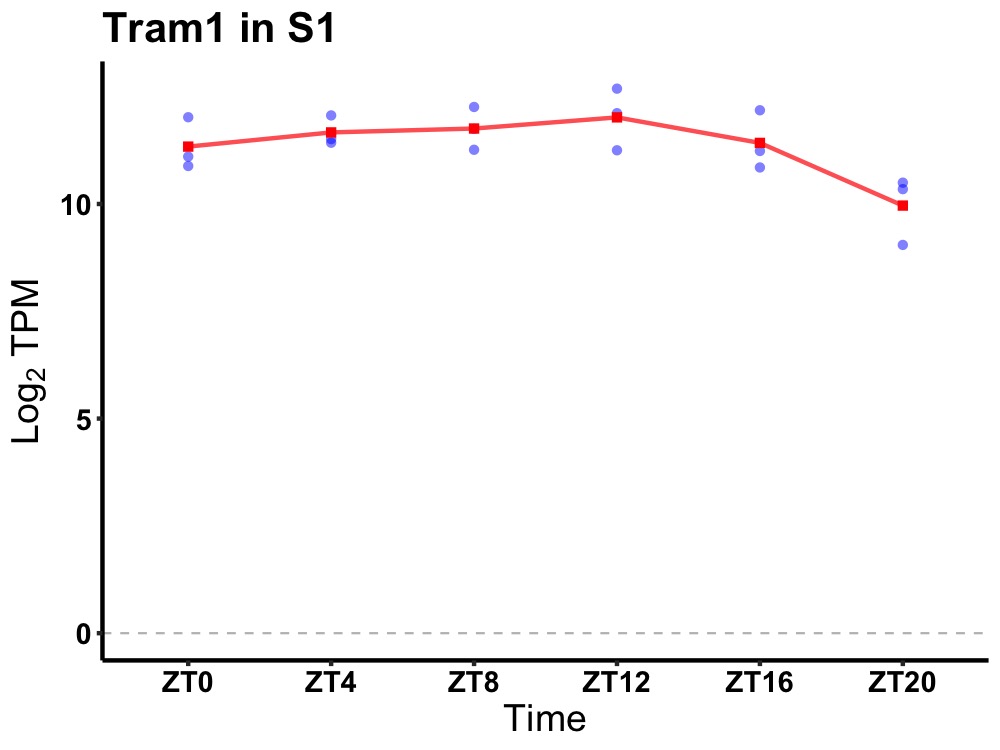 |
translocating chain-associating membrane protein 1 | 0.036 | 24 | 10 | 0.61 |
| ENSMUSG00000007458 | Stmn1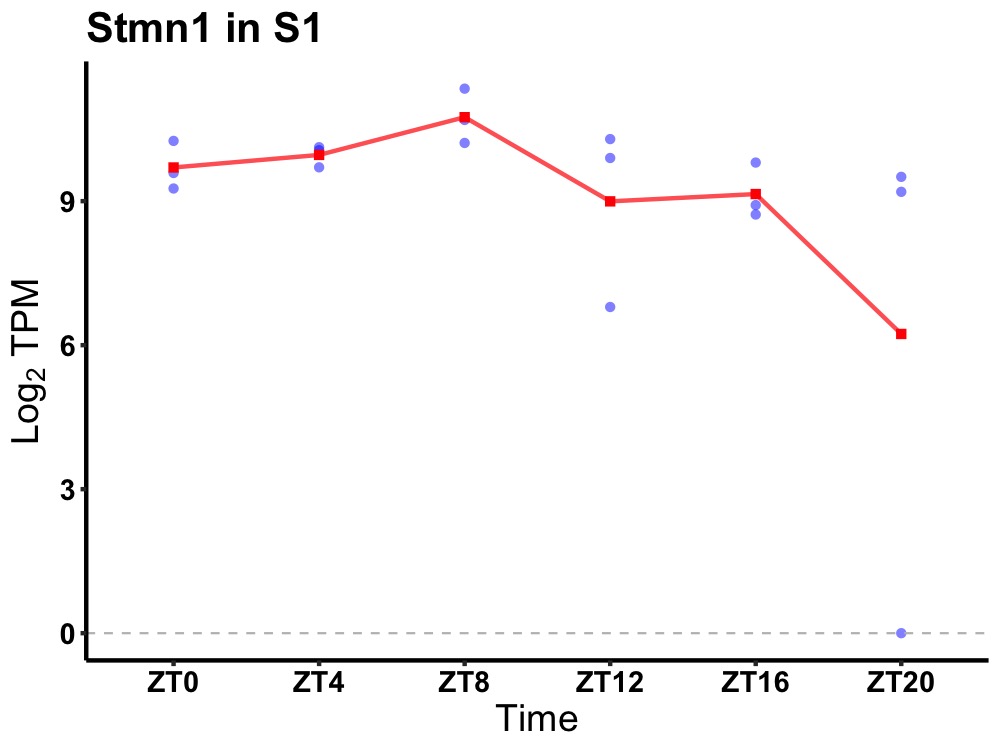 |
stathmin 1 | 0.019 | 24 | 8 | 0.61 |
| ENSMUSG00000021850 | Pcyt2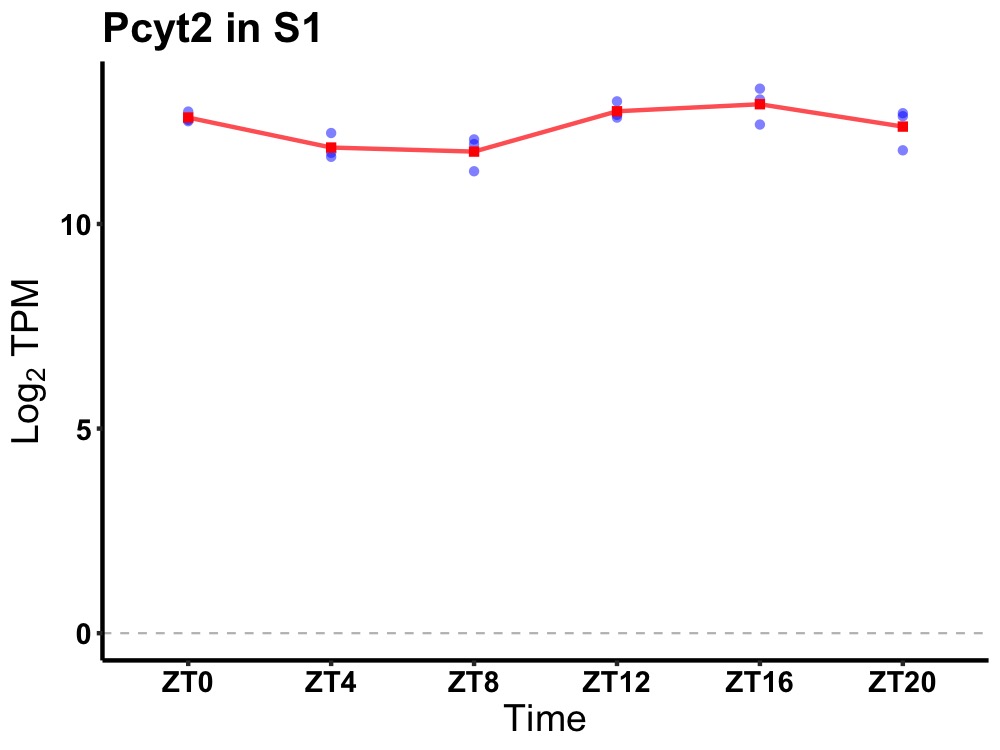 |
phosphate cytidylyltransferase 2, ethanolamine | 0.014 | 20 | 16 | 0.61 |
| ENSMUSG00000057160 | Gm16372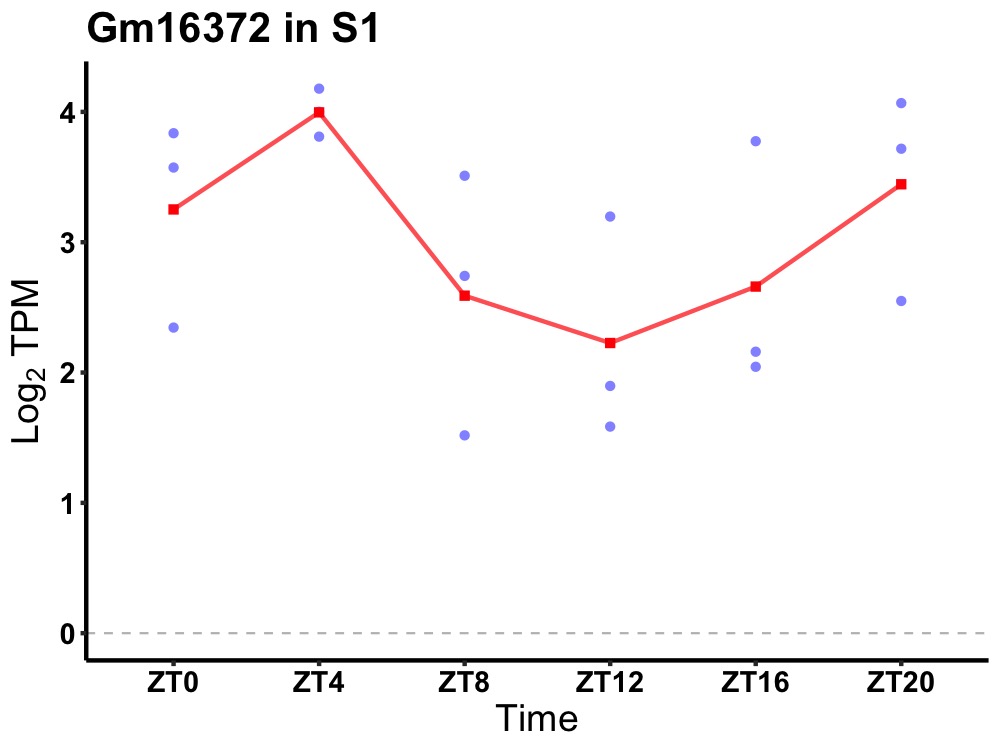 |
predicted pseudogene 16372 | 0.027 | 20 | 4 | 0.61 |
| ENSMUSG00000080848 | Dnajc14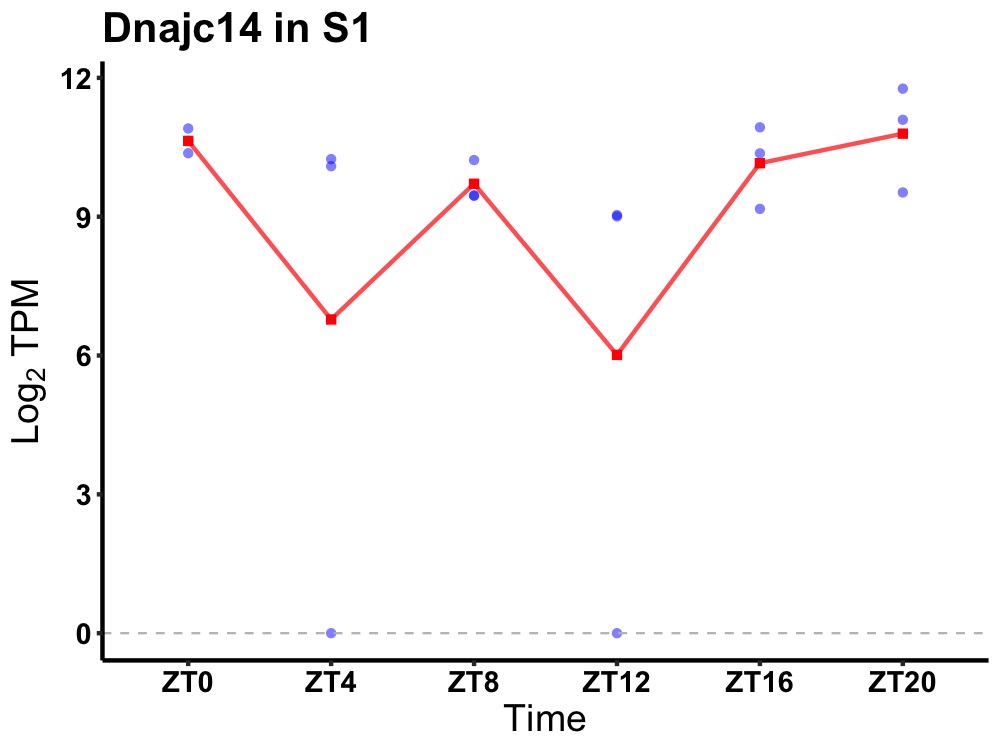 |
DnaJ heat shock protein family (Hsp40) member C14 | 0.031 | 20 | 2 | 0.60 |
| ENSMUSG00000105866 | Gm6745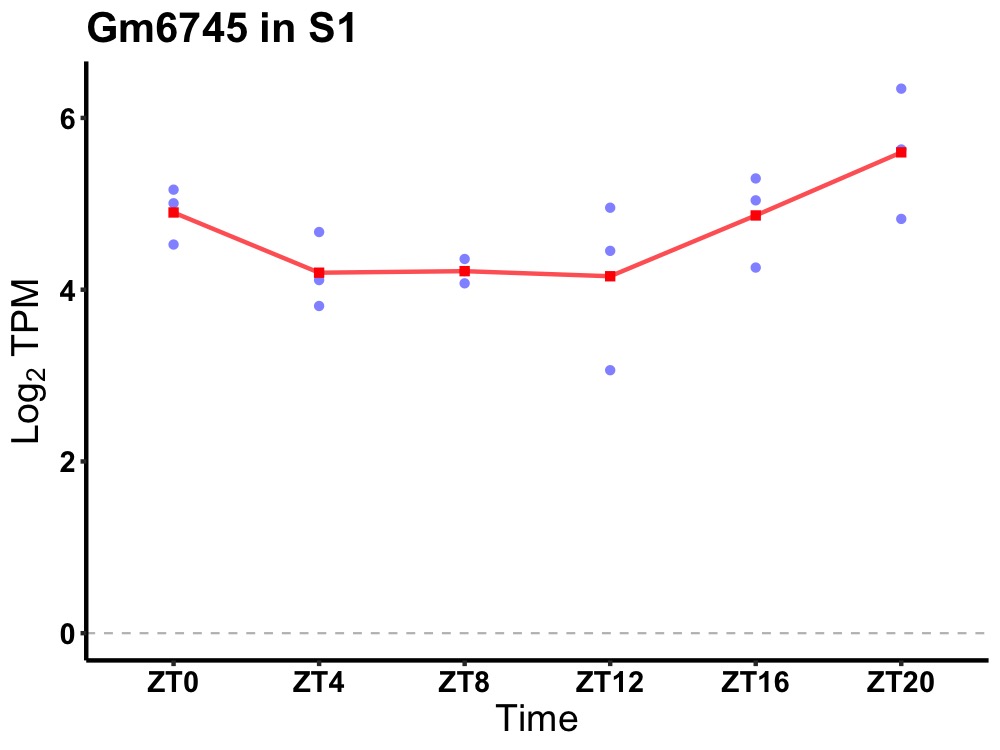 |
predicted gene 6745 | 0.027 | 24 | 20 | 0.60 |
| ENSMUSG00000078134 | Ergic2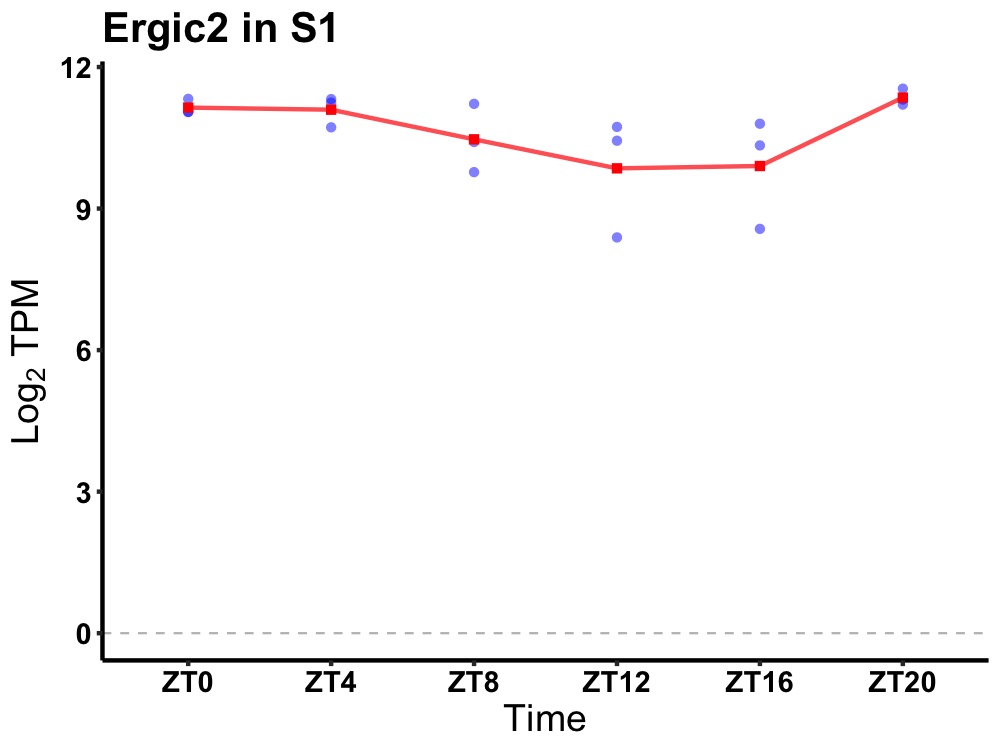 |
ERGIC and golgi 2 | 0.049 | 20 | 2 | 0.59 |
| ENSMUSG00000082585 | Gm15387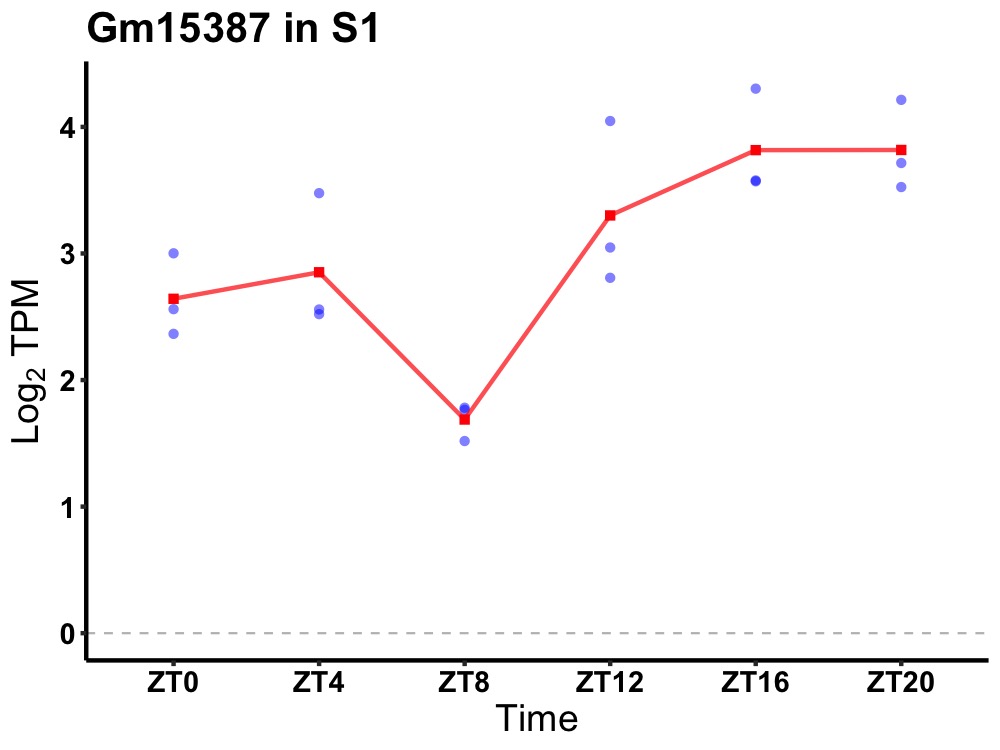 |
predicted gene 15387 | 0.010 | 24 | 20 | 0.59 |
| ENSMUSG00000050553 | Hectd1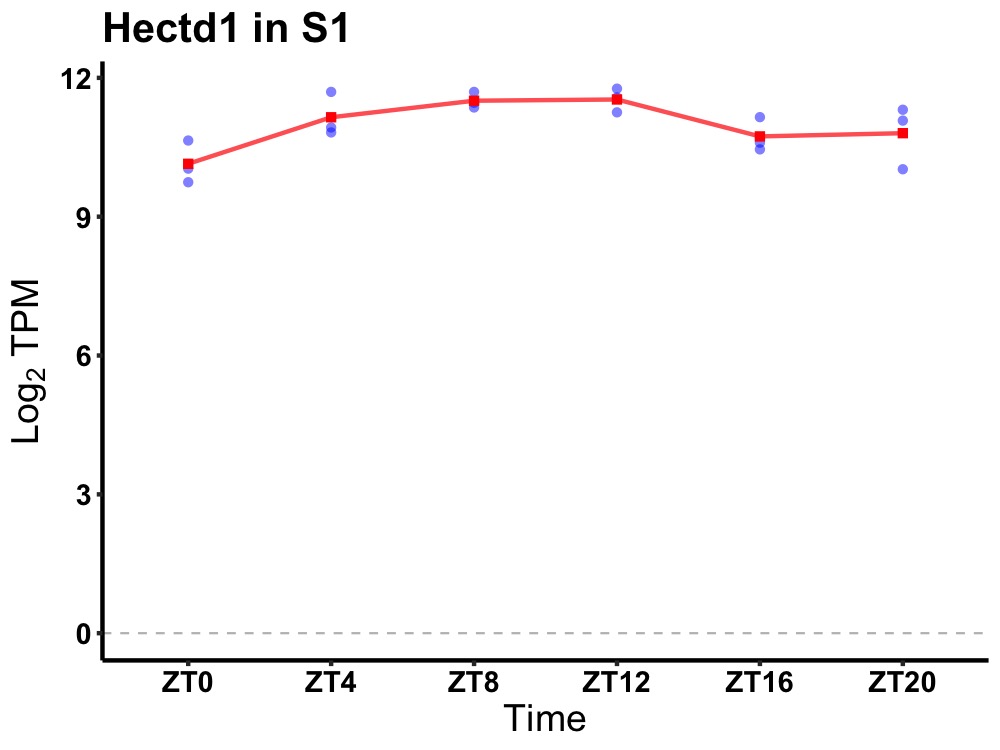 |
HECT domain E3 ubiquitin protein ligase 1 | 0.019 | 20 | 10 | 0.59 |
| ENSMUSG00000083899 | Gm12346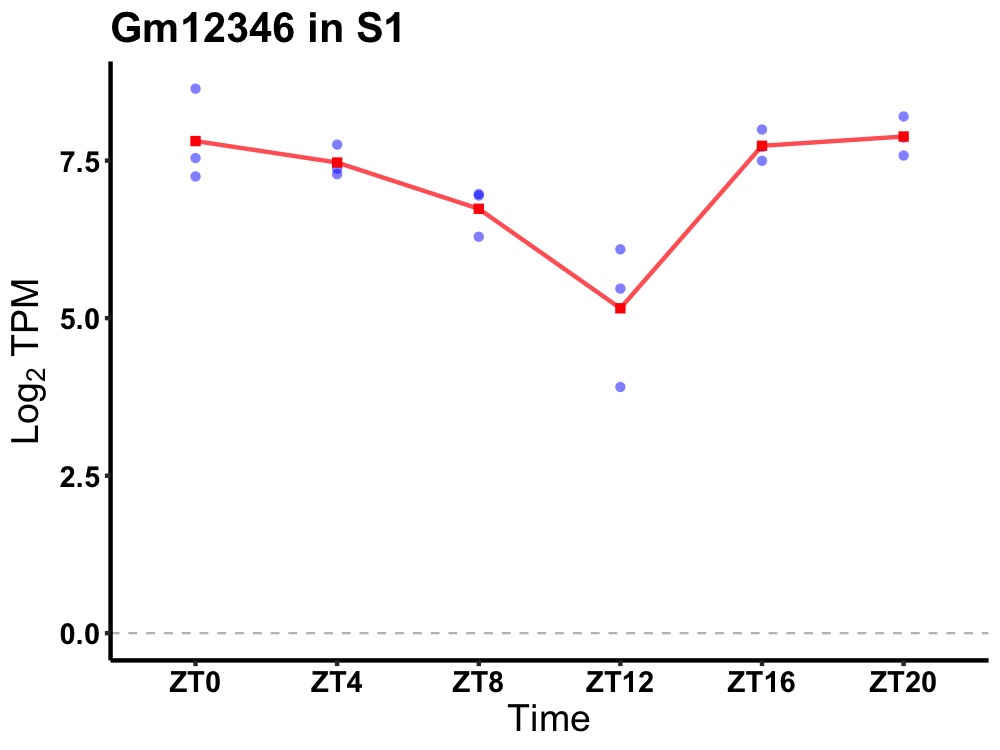 |
predicted gene 12346 | 0.007 | 20 | 2 | 0.59 |
| ENSMUSG00000022788 | Tm9sf3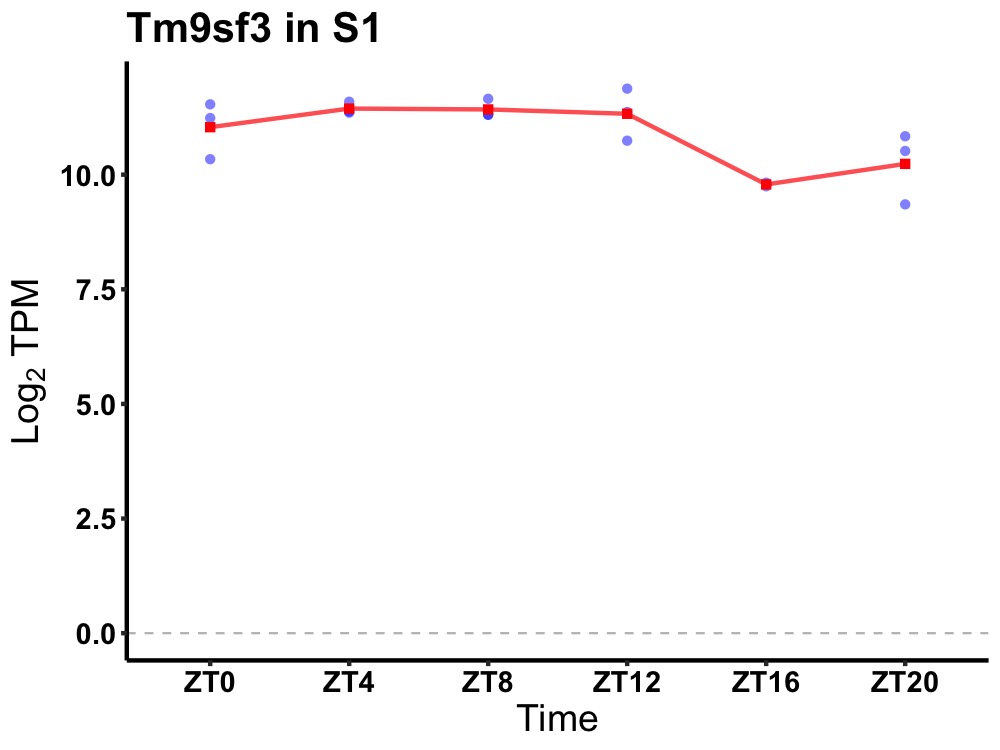 |
transmembrane 9 superfamily member 3 | 0.036 | 24 | 6 | 0.59 |
| ENSMUSG00000034947 | Nmi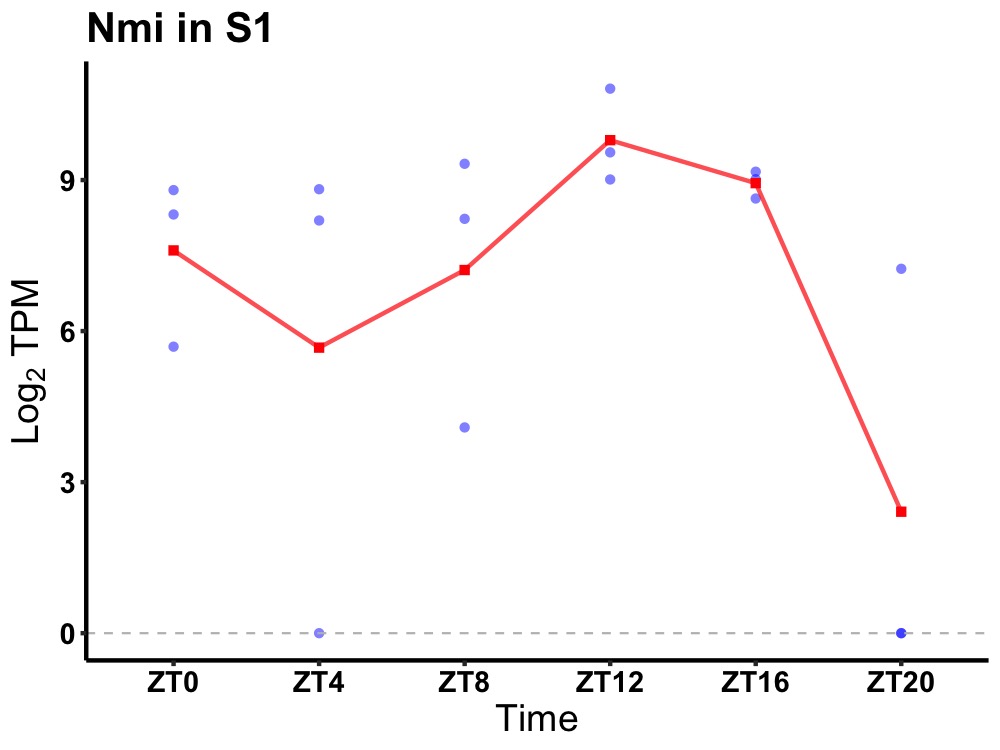 |
N-myc-interactor | 0.036 | 20 | 12 | 0.59 |
| ENSMUSG00000107092 | Tpgs1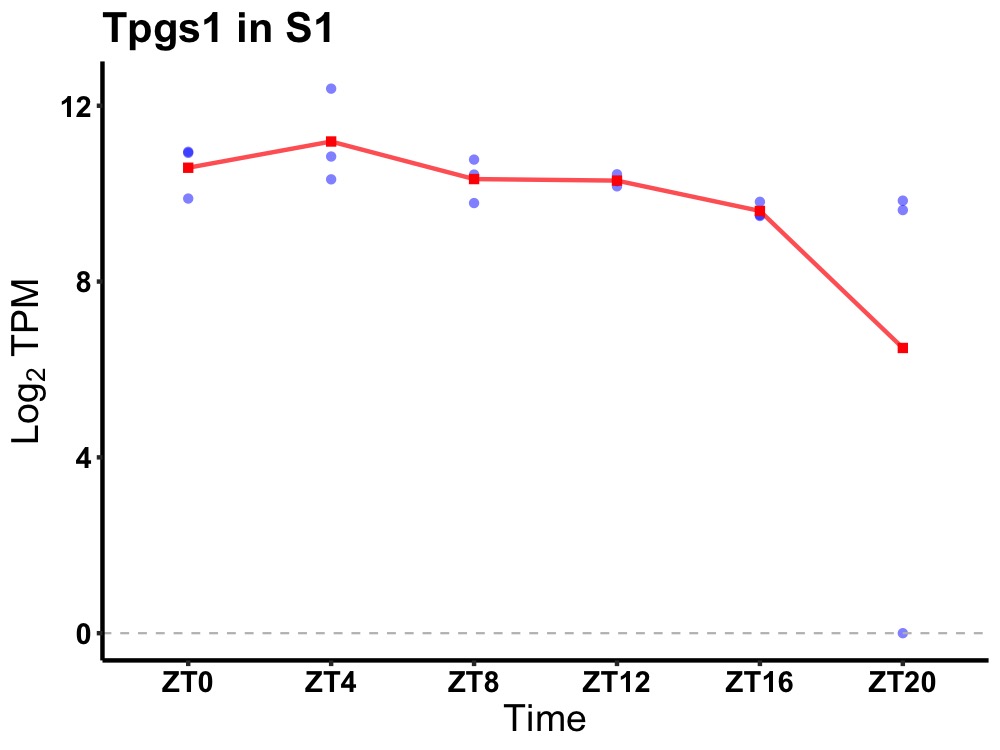 |
tubulin polyglutamylase complex subunit 1 | 0.027 | 24 | 6 | 0.59 |
| ENSMUSG00000081067 | Gm12912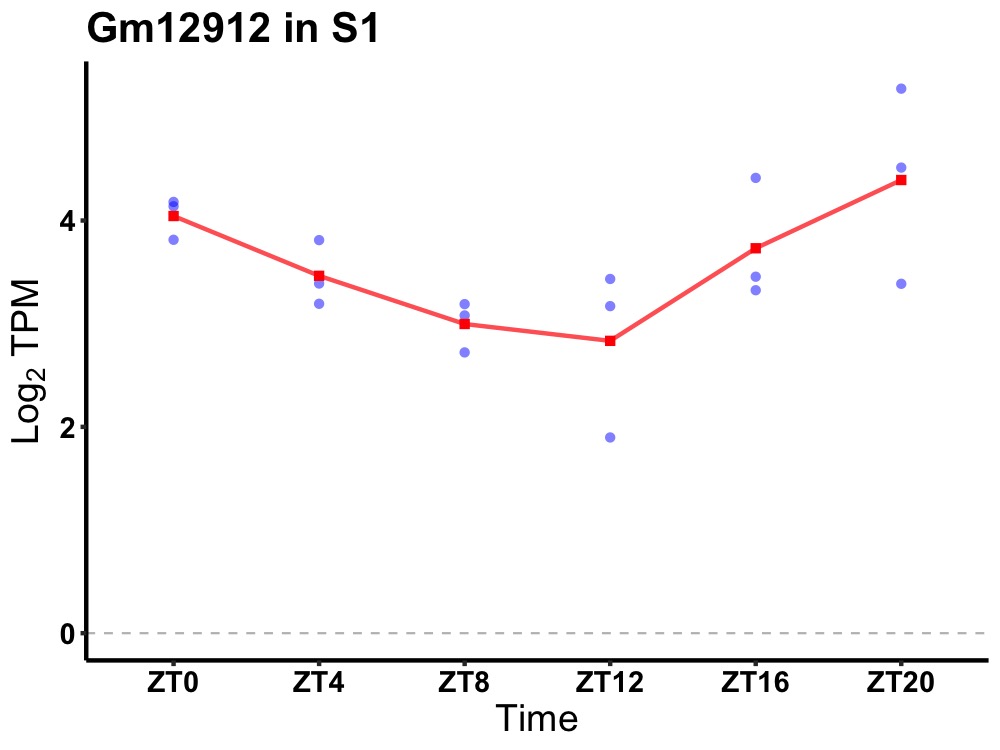 |
predicted gene 12912 | 0.003 | 24 | 22 | 0.58 |
| ENSMUSG00000080902 | Ywhaq-ps3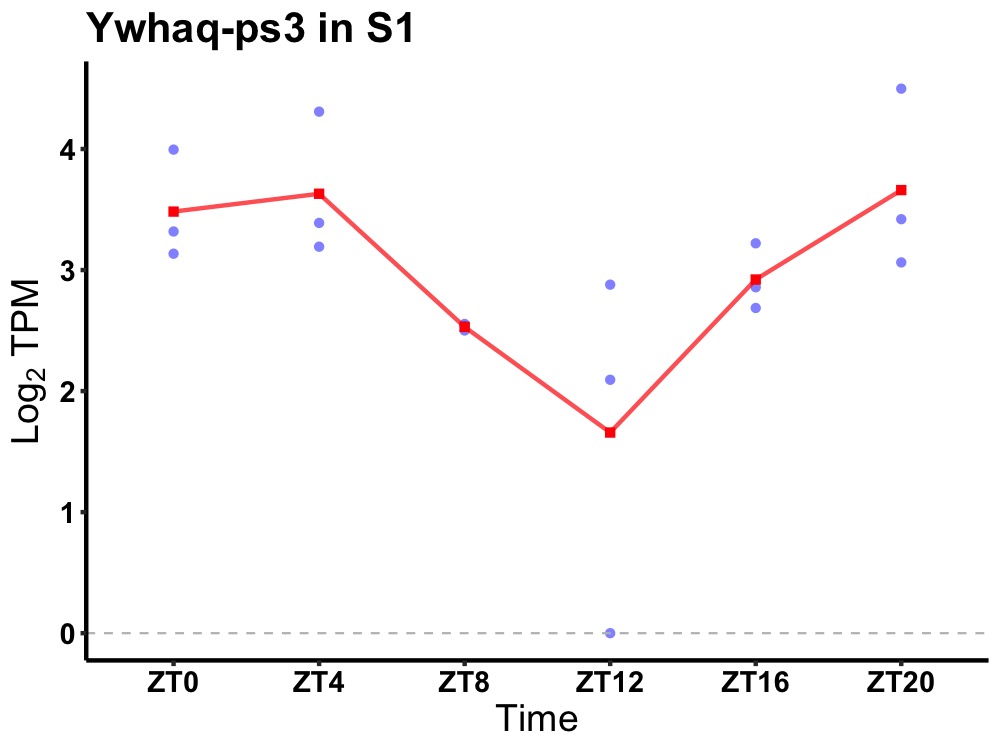 |
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein theta, pseudogene 3 | 0.005 | 20 | 2 | 0.57 |
| ENSMUSG00000056772 | Rps6-ps2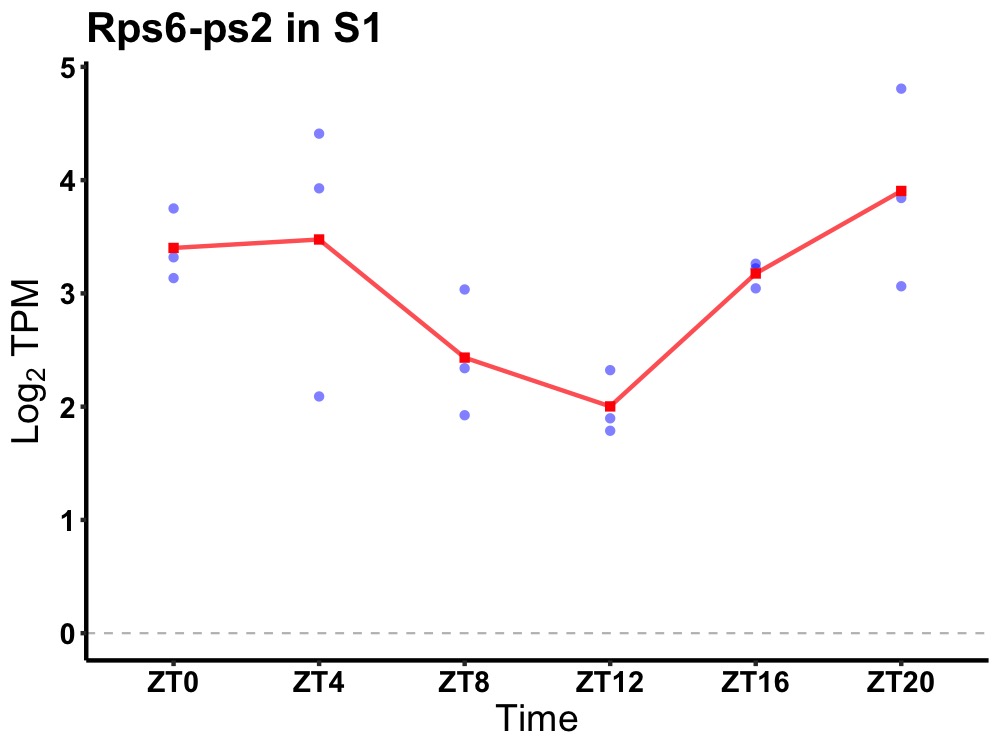 |
ribosomal protein S6, pseudogene 2 | 0.010 | 20 | 2 | 0.57 |
| ENSMUSG00000058622 | Gm5265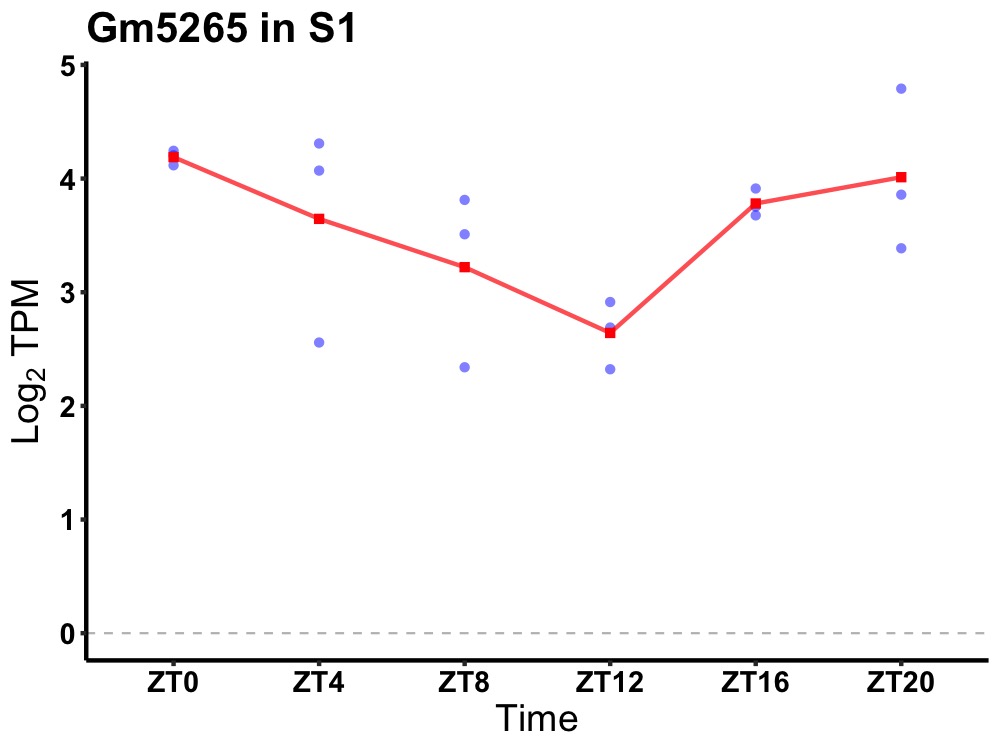 |
predicted pseudogene 5265 | 0.027 | 24 | 0 | 0.56 |
| ENSMUSG00000031483 | Lamp1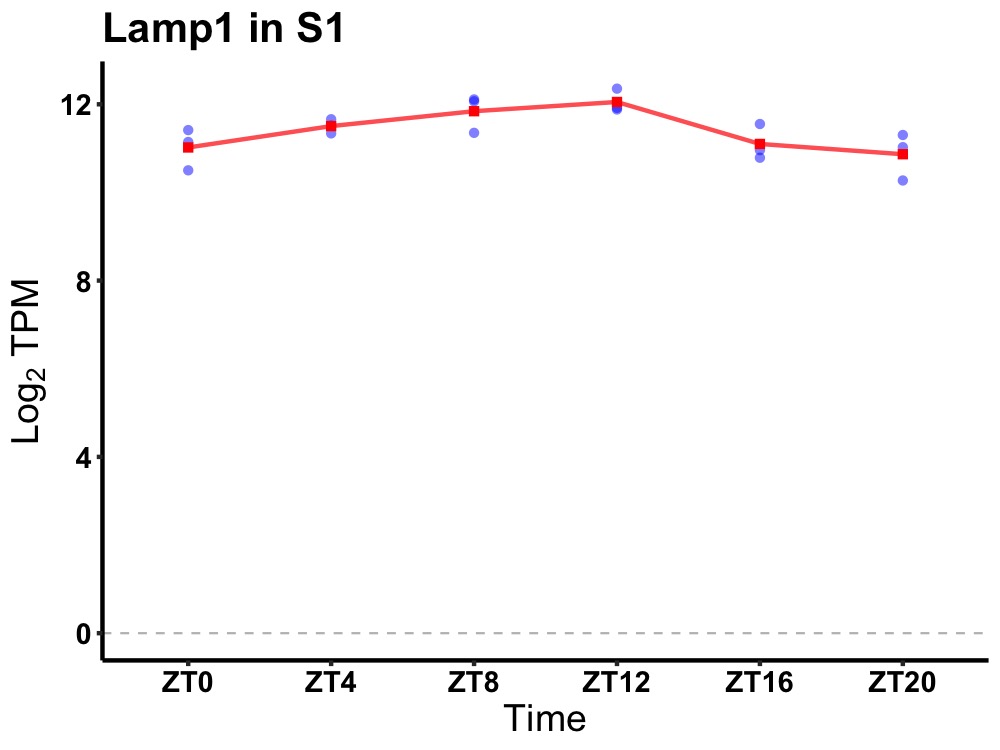 |
lysosomal-associated membrane protein 1 | 0.007 | 24 | 10 | 0.56 |
| ENSMUSG00000110537 | Gm4316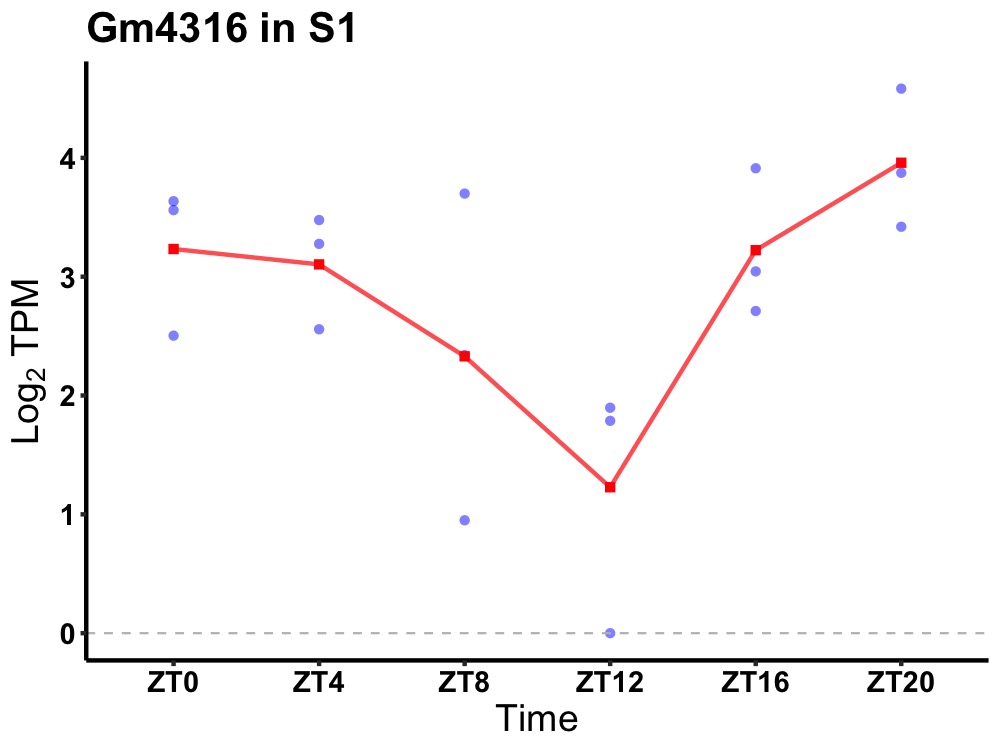 |
predicted gene 4316 | 0.049 | 20 | 2 | 0.55 |
| ENSMUSG00000036932 | mt-Nd6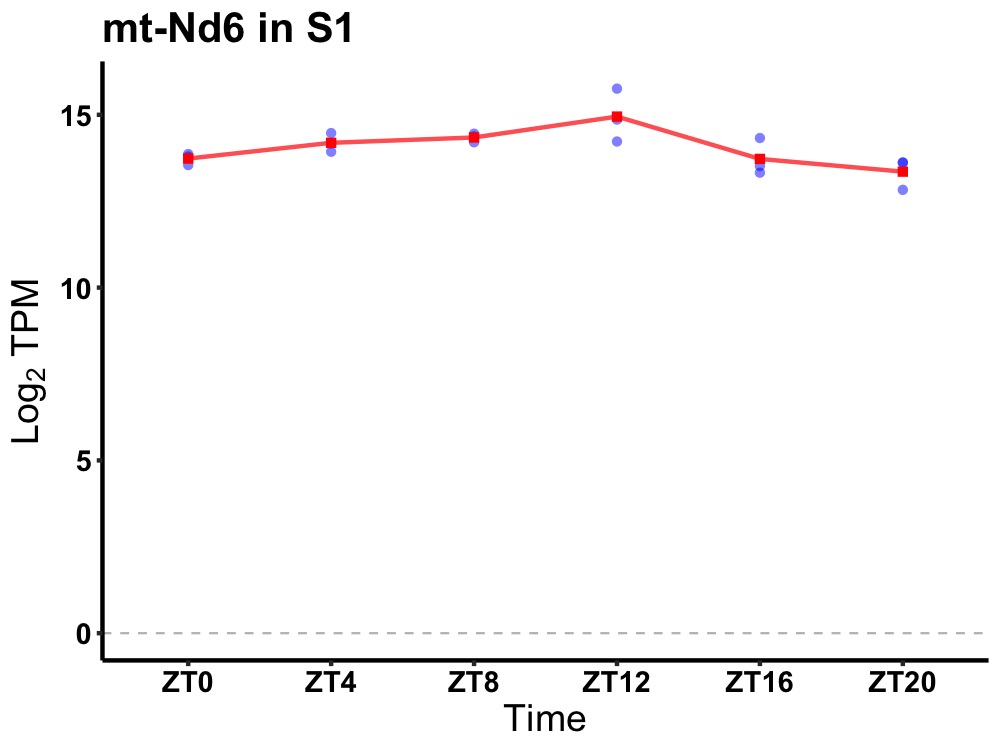 |
mitochondrially encoded NADH dehydrogenase 6 | 0.027 | 24 | 10 | 0.55 |
| ENSMUSG00000096647 | Gm6223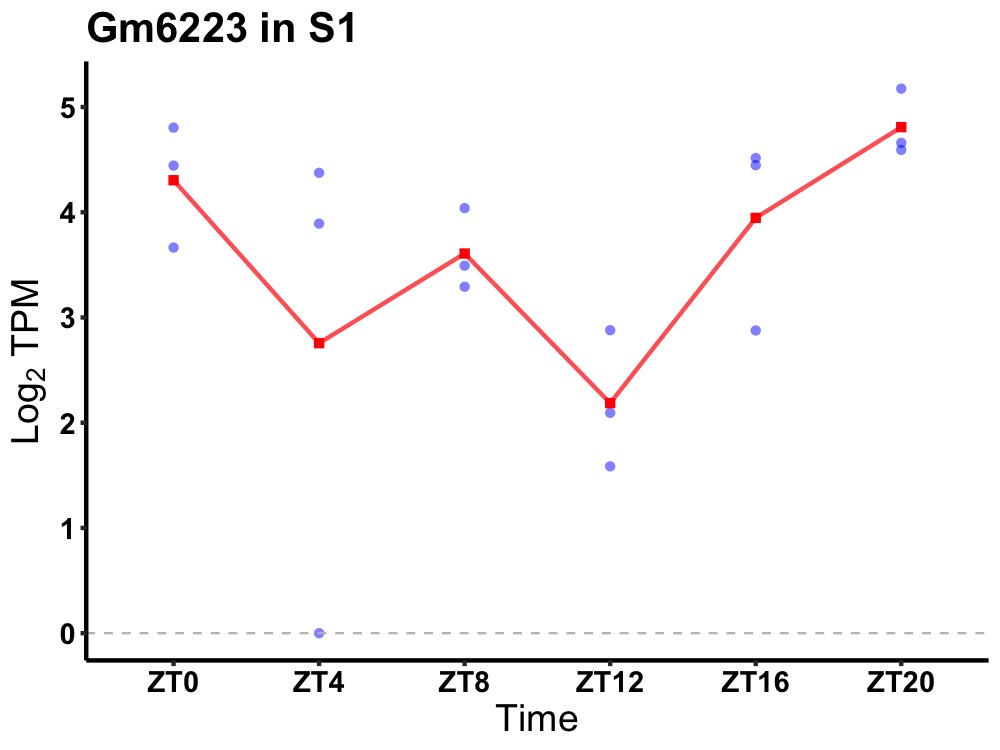 |
predicted gene 6223 | 0.027 | 20 | 2 | 0.55 |
| ENSMUSG00000043295 | Tnpo1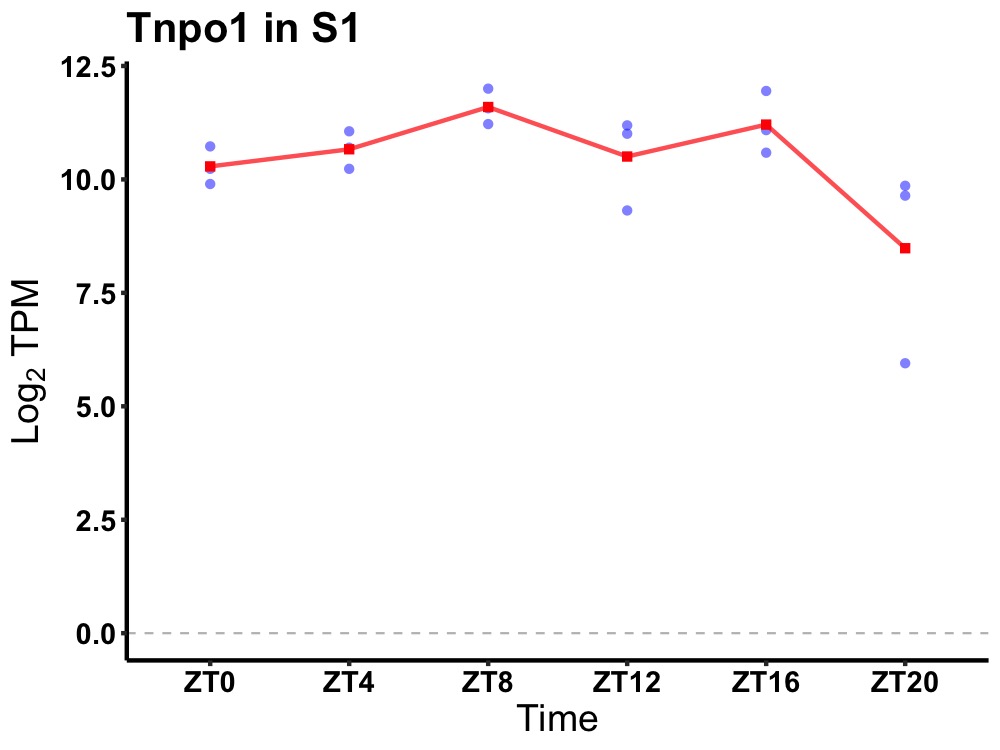 |
transportin 1 | 0.036 | 24 | 10 | 0.55 |
| ENSMUSG00000097245 | Rab17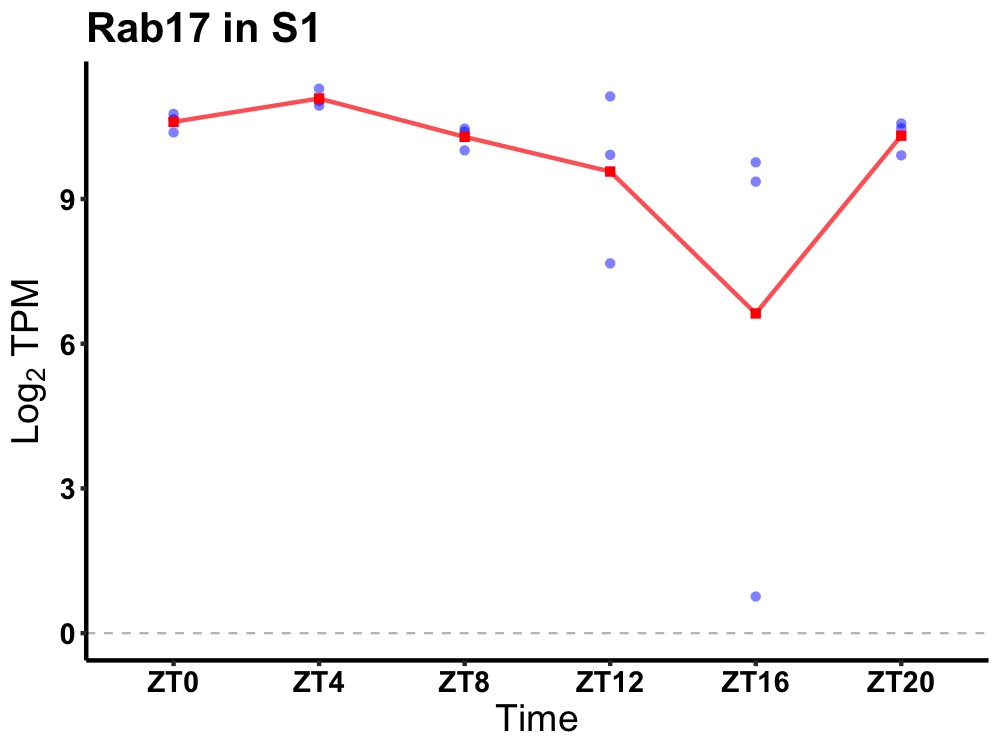 |
RAB17, member RAS oncogene family | 0.010 | 24 | 4 | 0.54 |
| ENSMUSG00000118345 | AC110913.1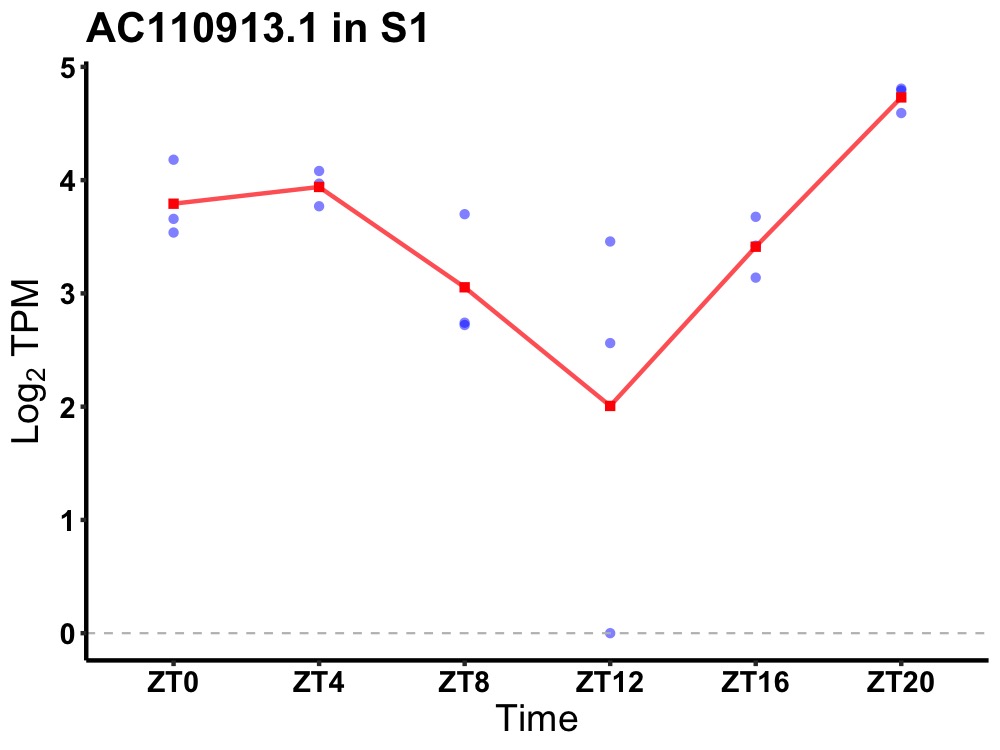 |
none | 0.001 | 20 | 2 | 0.54 |
| ENSMUSG00000083326 | Scnm1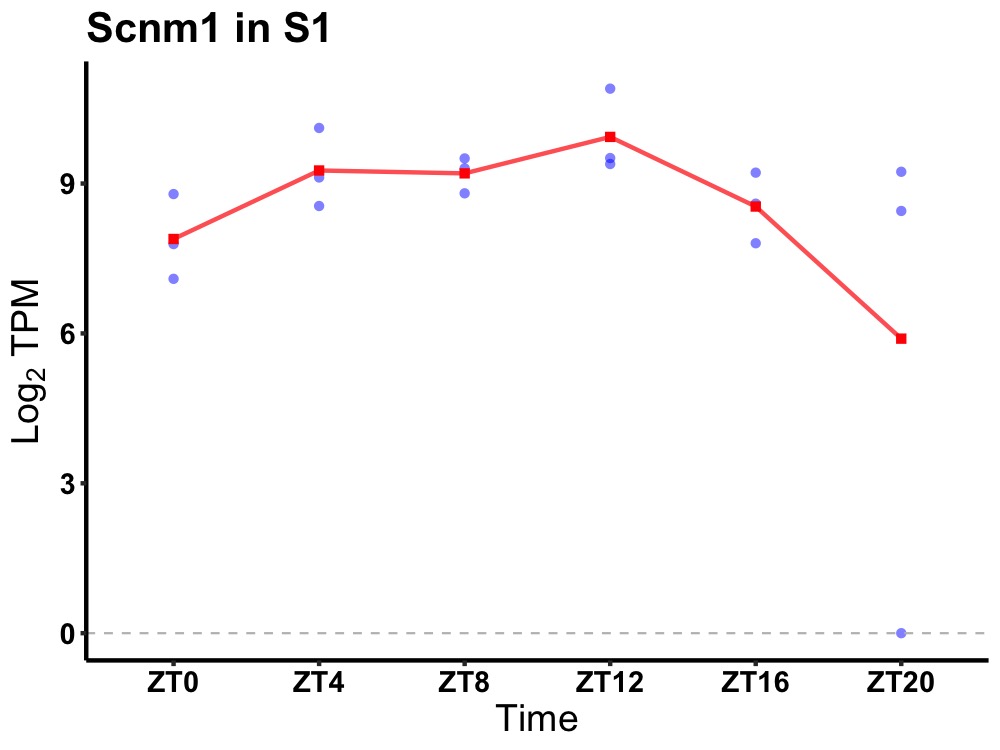 |
sodium channel modifier 1 | 0.036 | 24 | 12 | 0.53 |
| ENSMUSG00000068823 | Gclm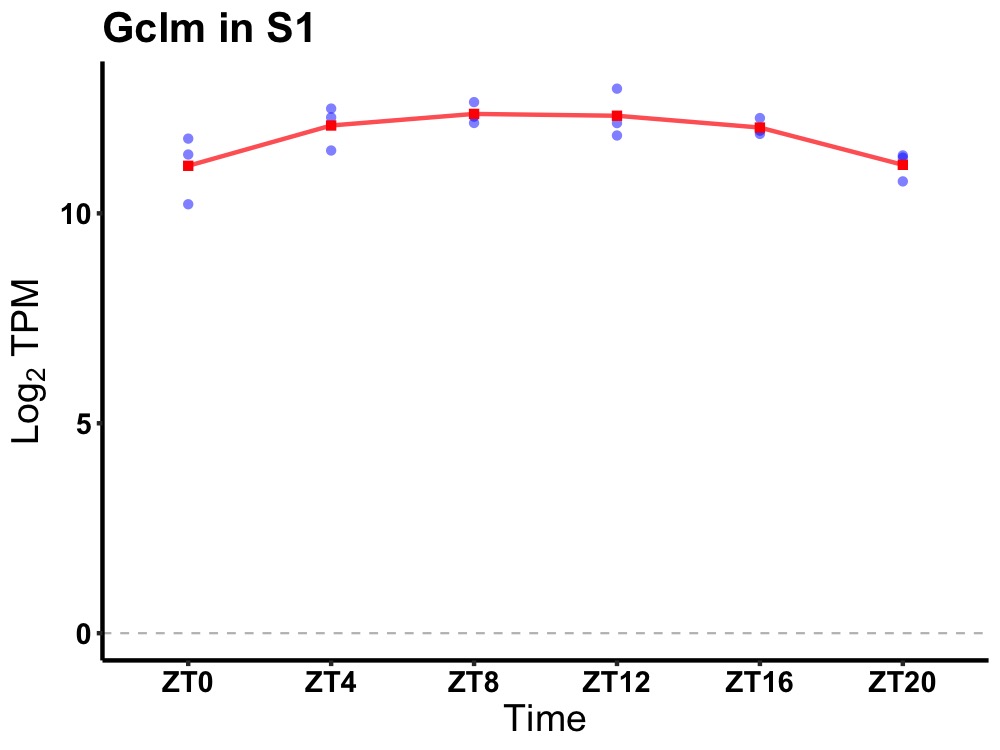 |
glutamate-cysteine ligase, modifier subunit | 0.005 | 24 | 10 | 0.53 |
| ENSMUSG00000036707 | Dgat2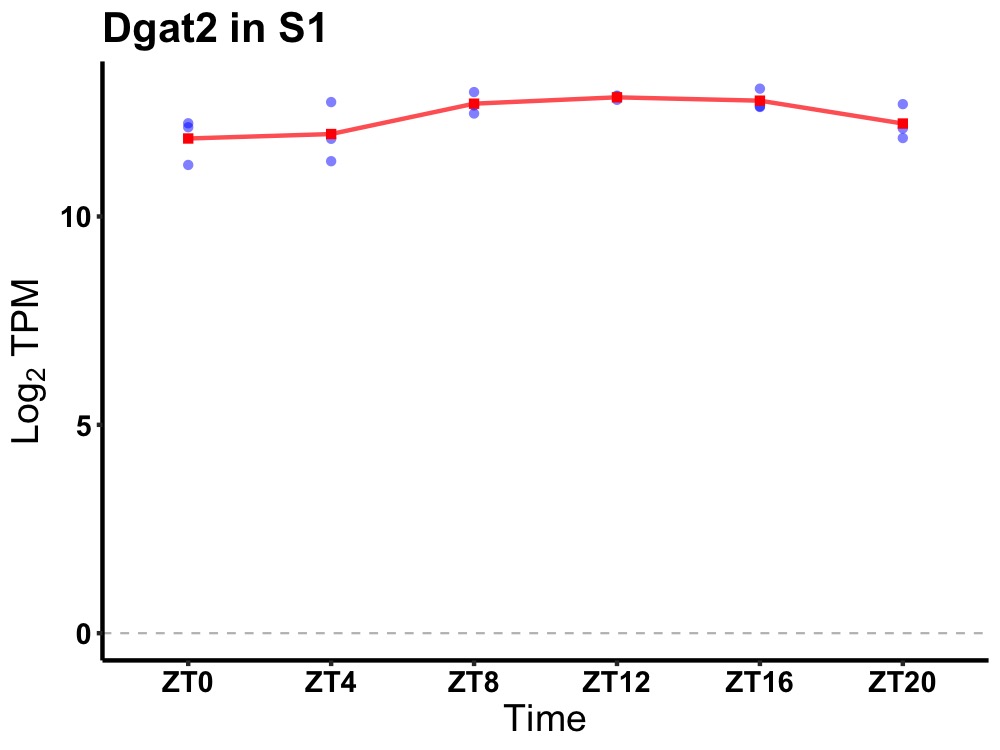 |
diacylglycerol O-acyltransferase 2 | 0.049 | 24 | 14 | 0.53 |
| ENSMUSG00000021814 | Nat8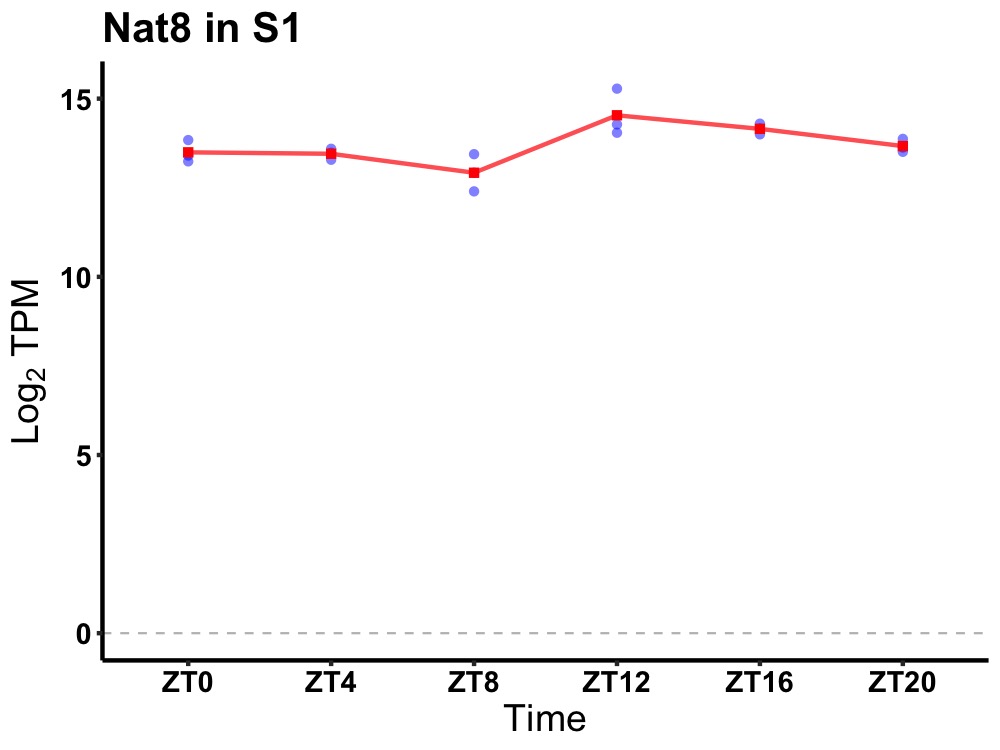 |
N-acetyltransferase 8 (GCN5-related) | 0.049 | 20 | 16 | 0.52 |
| ENSMUSG00000030498 | Gna11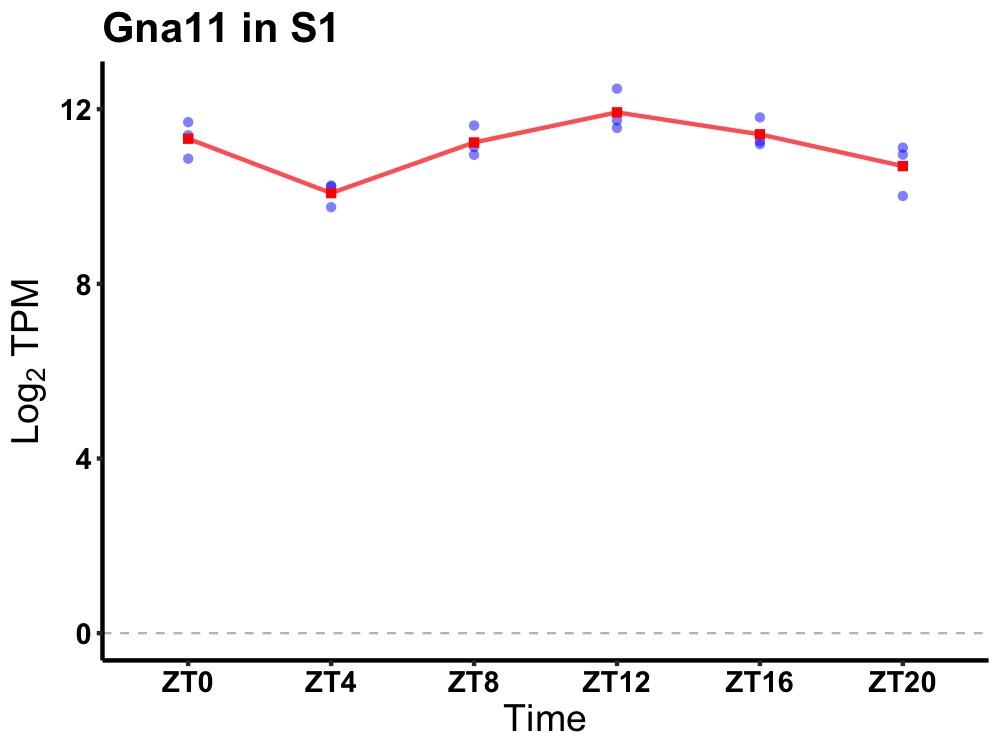 |
guanine nucleotide binding protein, alpha 11 | 0.003 | 20 | 14 | 0.52 |
| ENSMUSG00000027366 | Echdc1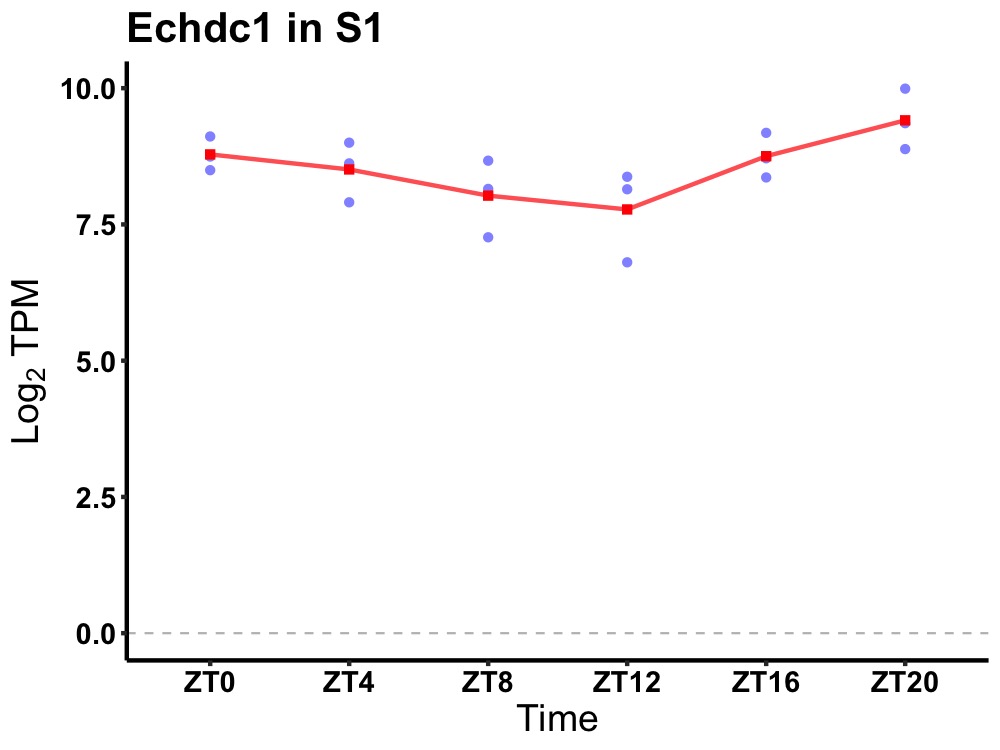 |
enoyl Coenzyme A hydratase domain containing 1 | 0.027 | 24 | 22 | 0.52 |
| ENSMUSG00000103137 | Nae1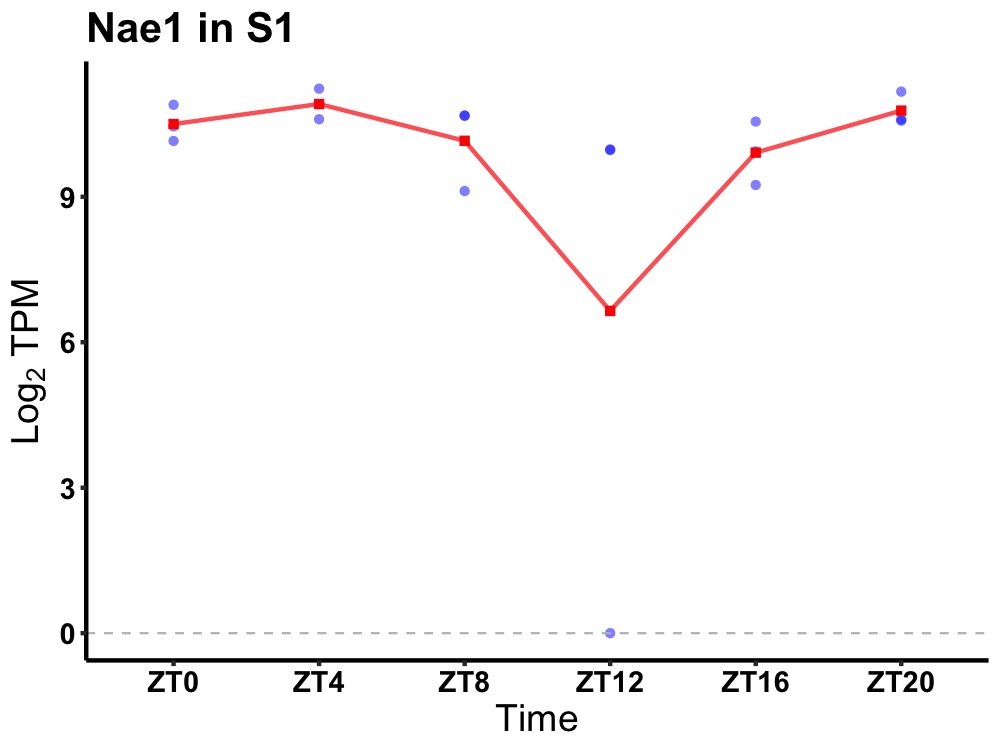 |
NEDD8 activating enzyme E1 subunit 1 | 0.019 | 20 | 4 | 0.52 |
| ENSMUSG00000083261 | Macrod1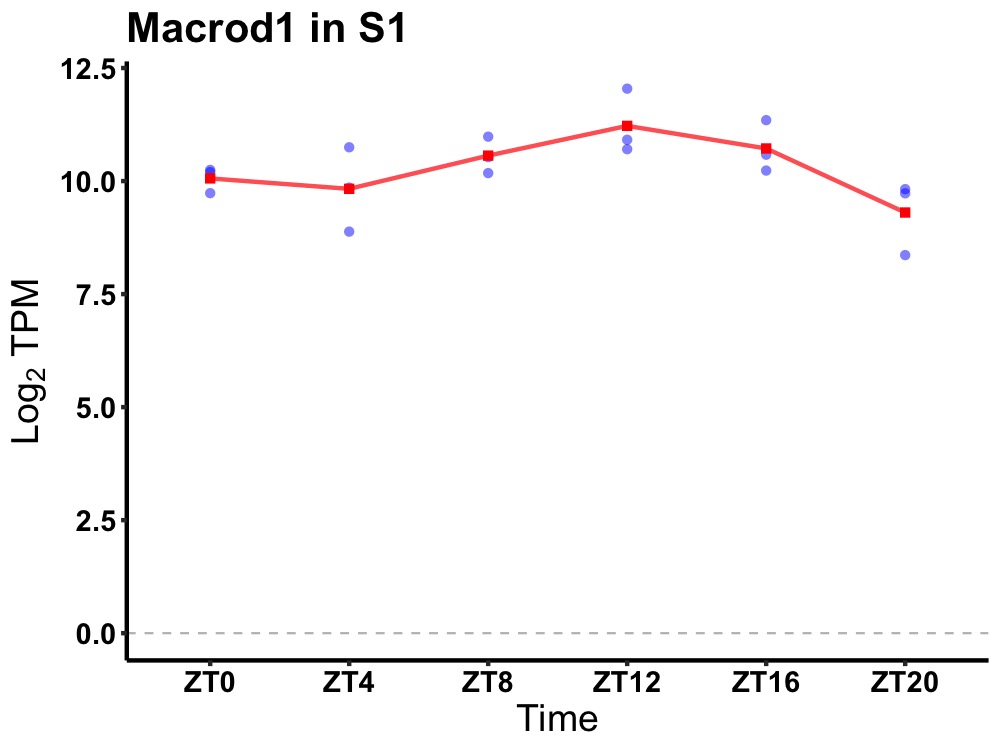 |
MACRO domain containing 1 | 0.019 | 20 | 12 | 0.52 |
| ENSMUSG00000080775 | Gm6368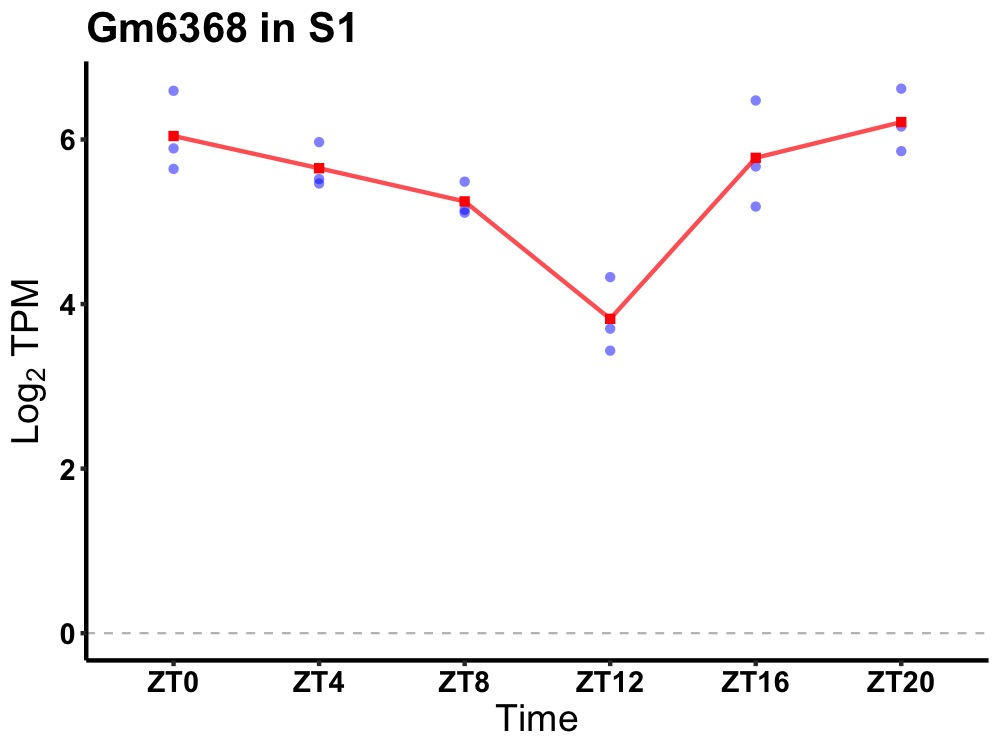 |
predicted gene 6368 | 0.001 | 20 | 2 | 0.52 |
| ENSMUSG00000021302 | Nsdhl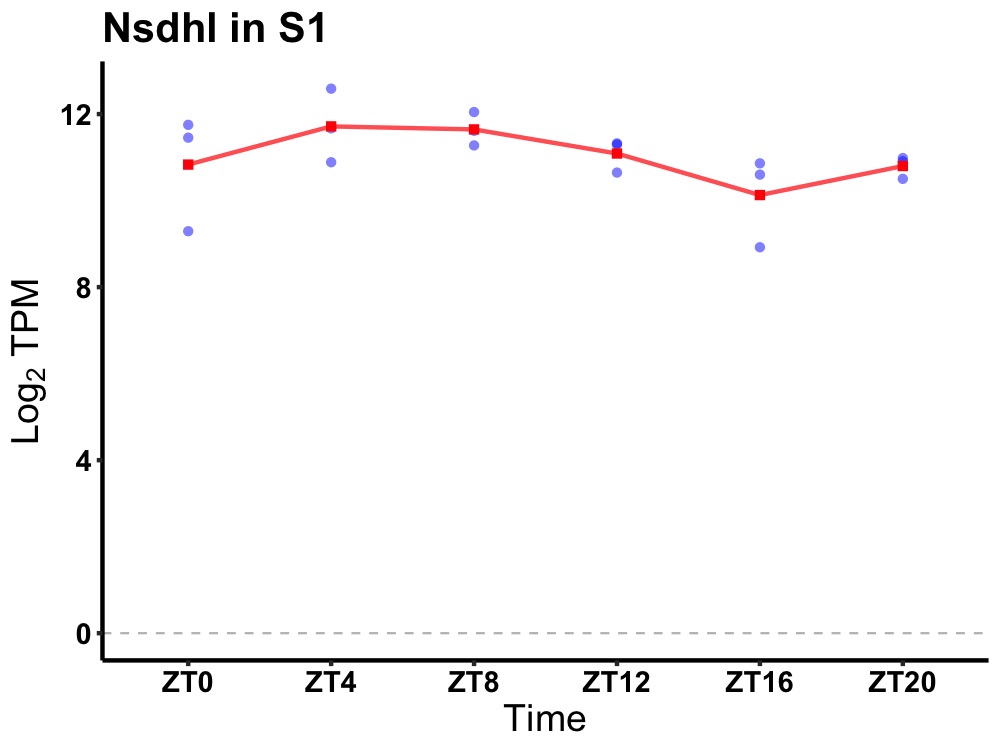 |
NAD(P) dependent steroid dehydrogenase-like | 0.049 | 24 | 6 | 0.52 |
| ENSMUSG00000097245 | Gm5421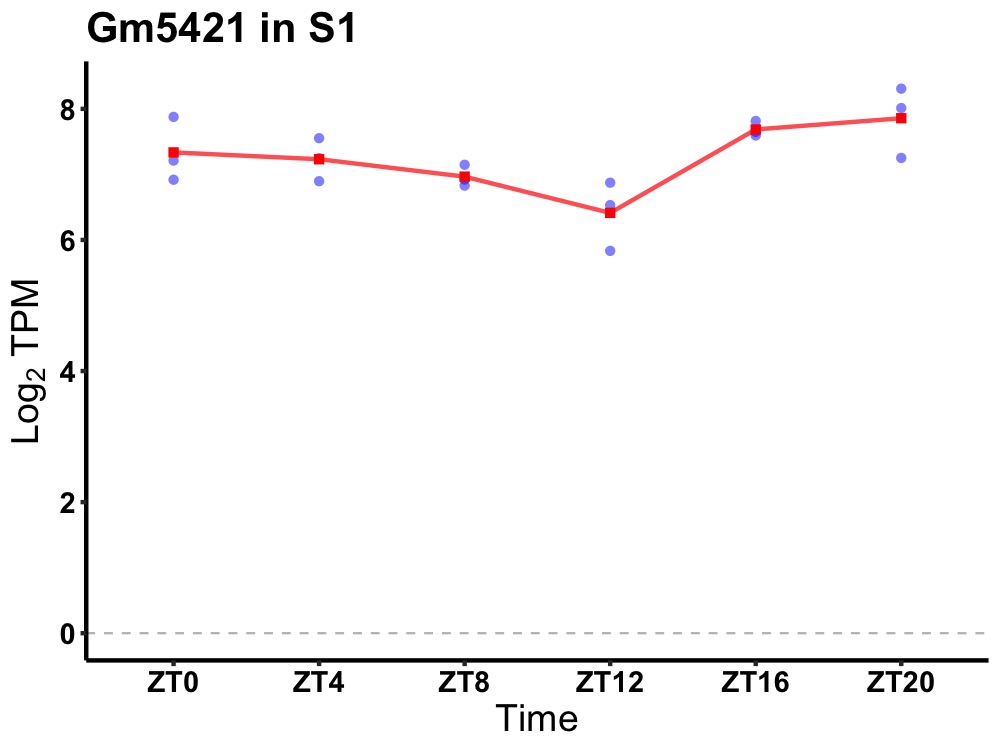 |
predicted gene 5421 | 0.027 | 24 | 22 | 0.51 |
| ENSMUSG00000078193 | Bud23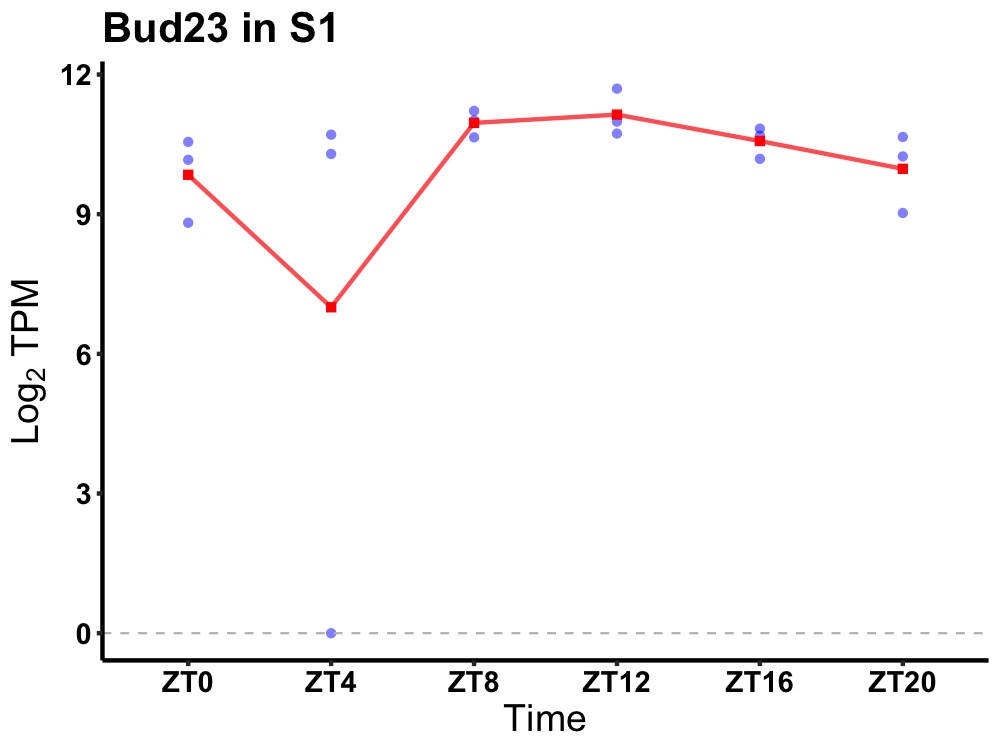 |
BUD23, rRNA methyltransferase and ribosome maturation factor | 0.019 | 24 | 12 | 0.51 |
| ENSMUSG00000039616 | Ptms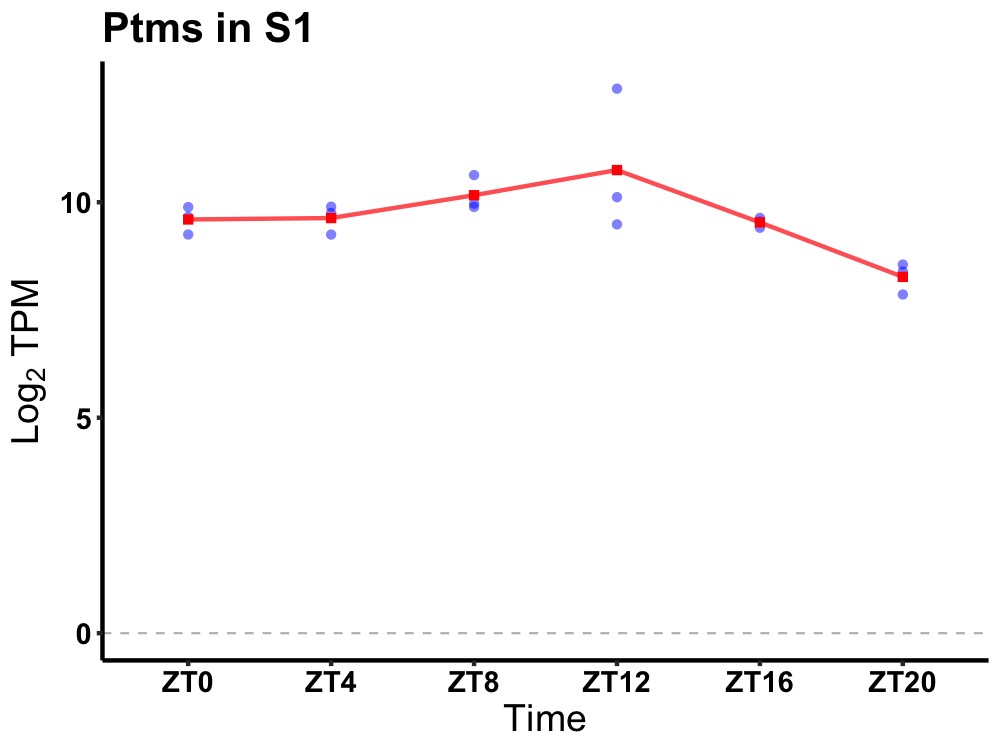 |
parathymosin | 0.014 | 24 | 10 | 0.50 |
| ENSMUSG00000032097 | Slc9a8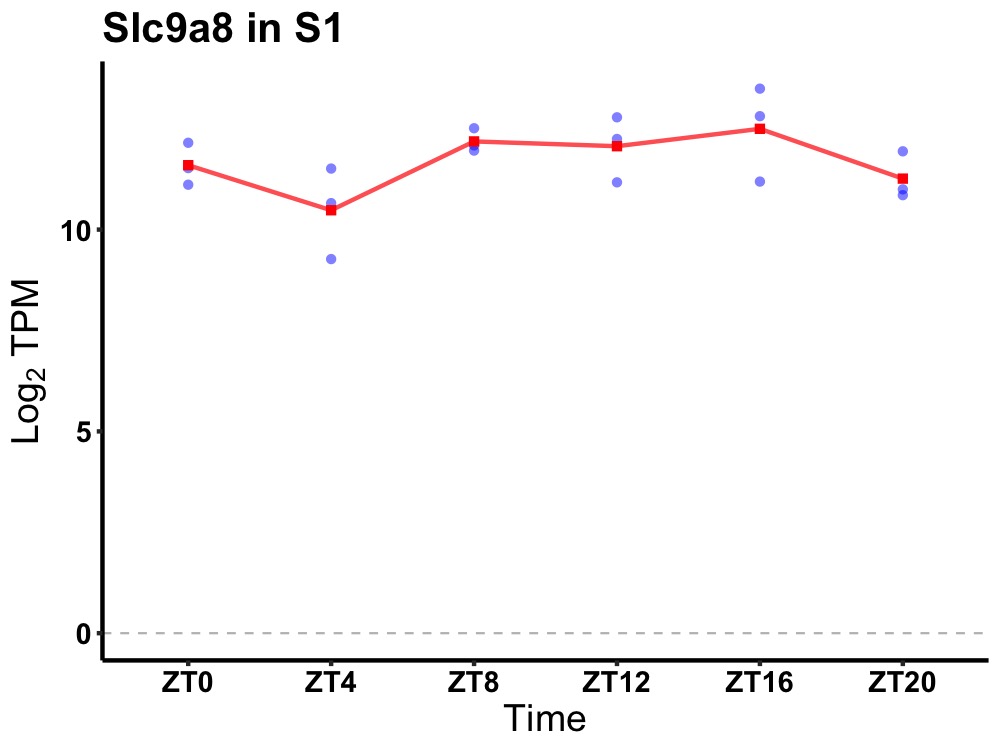 |
solute carrier family 9 (sodium/hydrogen exchanger), member 8 | 0.049 | 20 | 14 | 0.50 |
| ENSMUSG00000017686 | Frg1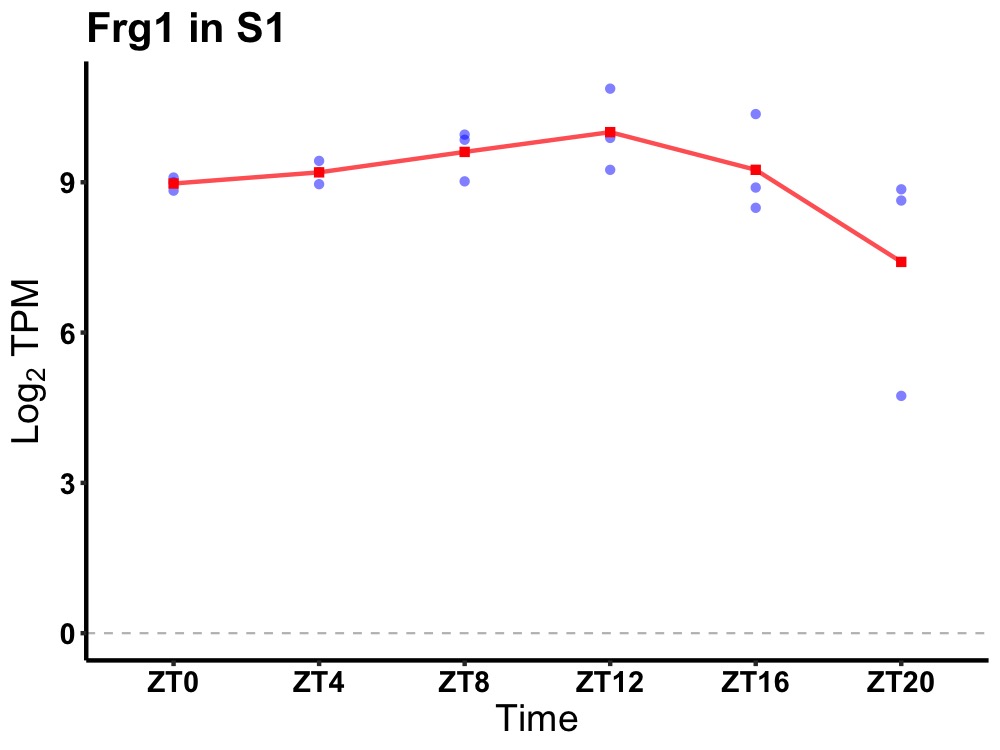 |
FSHD region gene 1 | 0.036 | 24 | 10 | 0.50 |
| ENSMUSG00000033096 | Hsp90aa1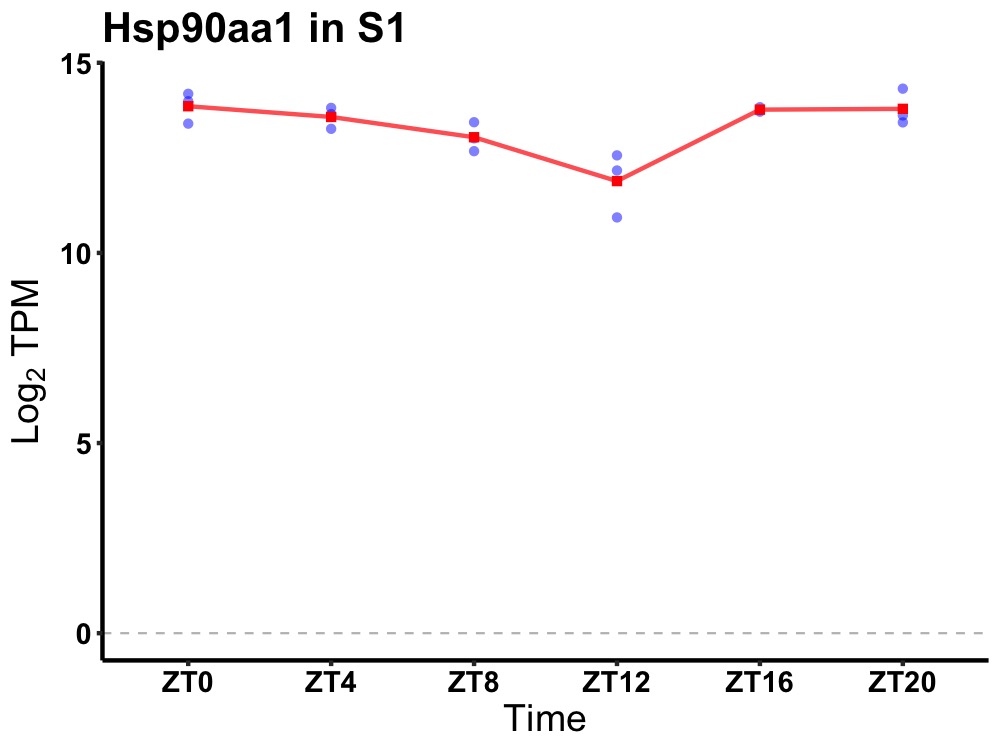 |
heat shock protein 90, alpha (cytosolic), class A member 1 | 0.049 | 20 | 2 | 0.50 |
| ENSMUSG00000036461 | Uba1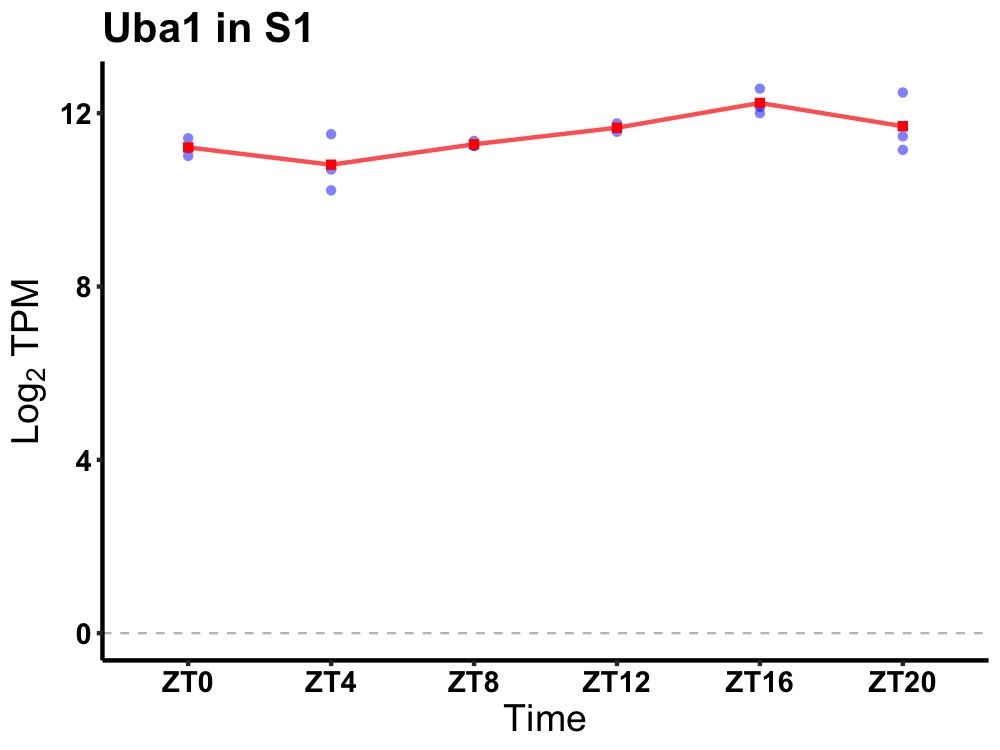 |
ubiquitin-like modifier activating enzyme 1 | 0.003 | 24 | 16 | 0.50 |
| ENSMUSG00000029836 | Cbx3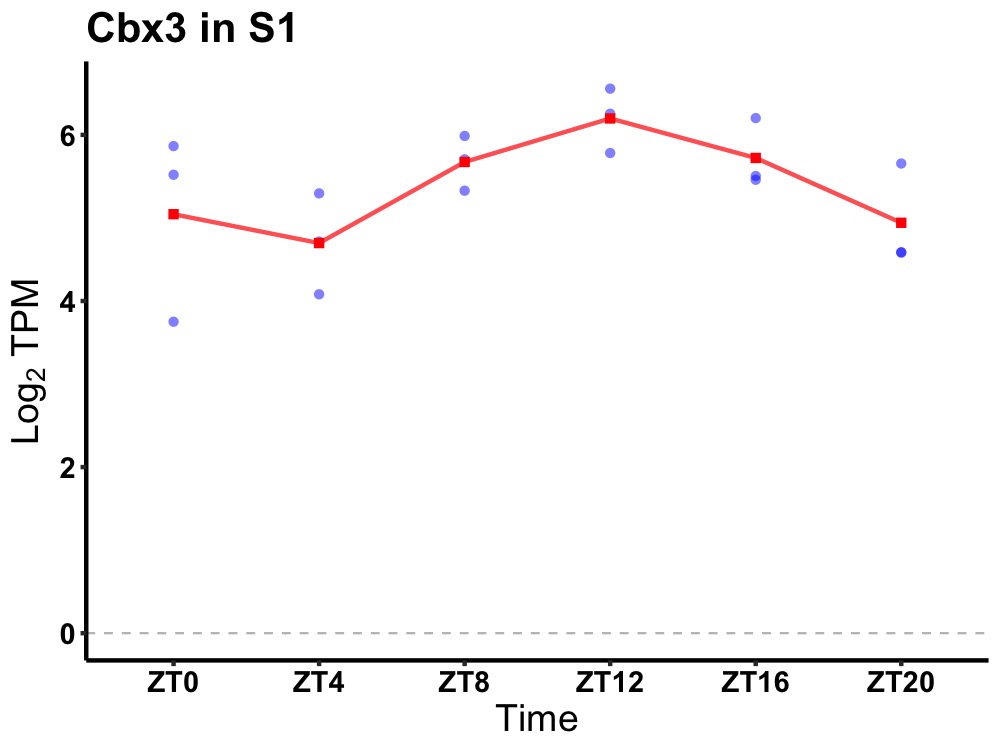 |
chromobox 3 | 0.036 | 20 | 14 | 0.50 |
| ENSMUSG00000093402 | Gm18588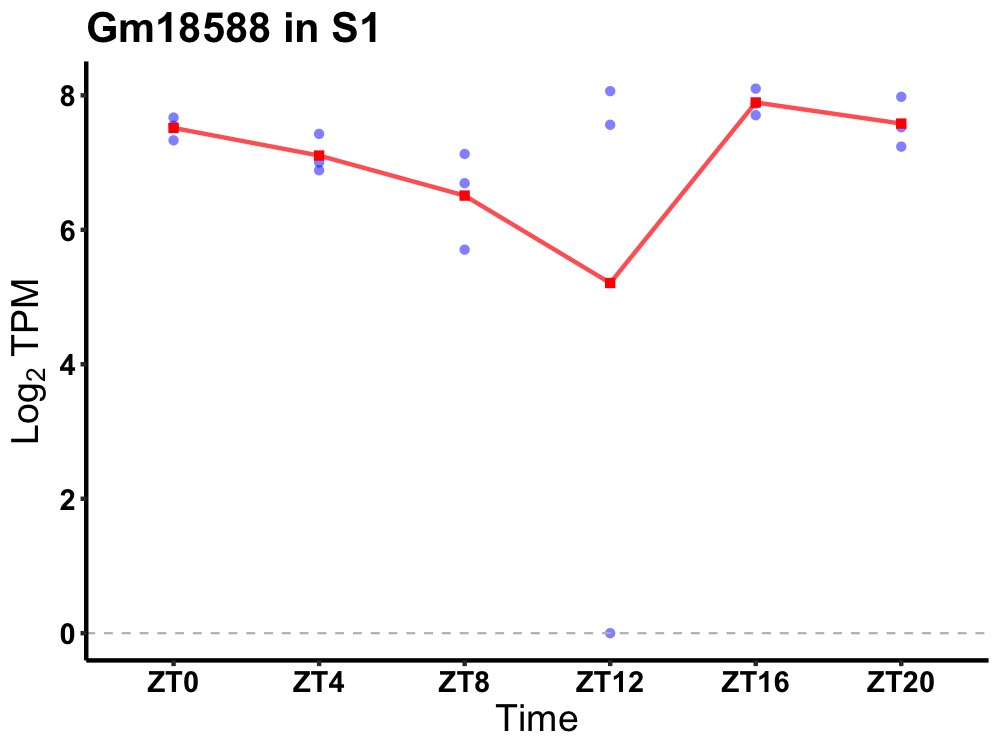 |
predicted gene, 18588 | 0.036 | 20 | 18 | 0.50 |
| ENSMUSG00000083820 | Apopt1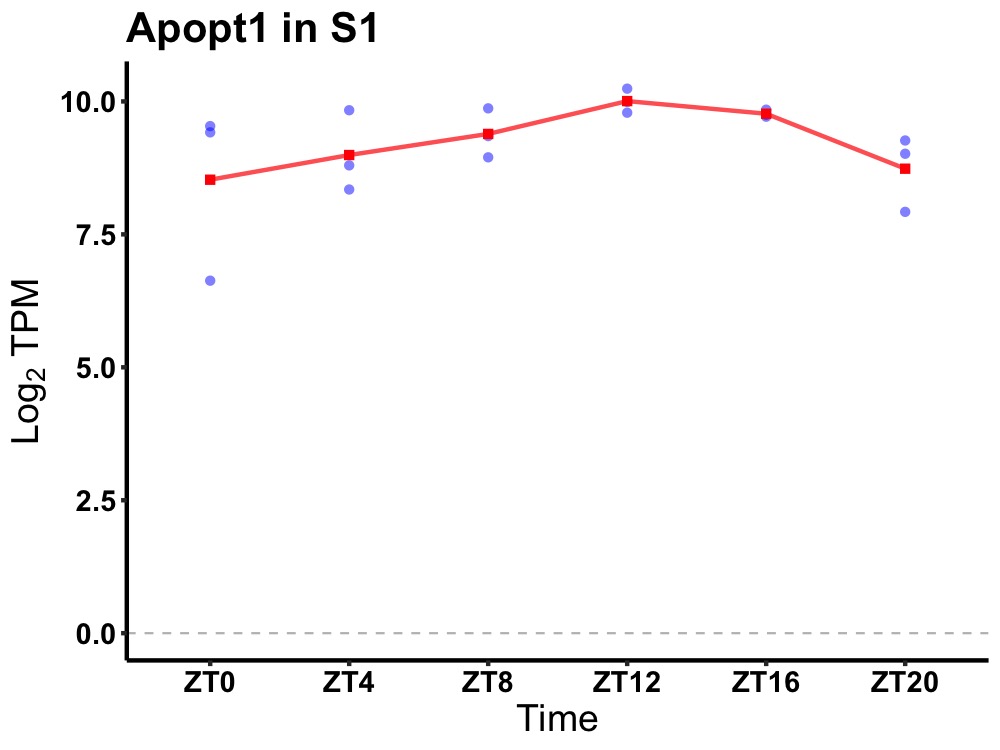 |
apoptogenic, mitochondrial 1 | 0.049 | 20 | 14 | 0.49 |
| ENSMUSG00000024661 | Trak2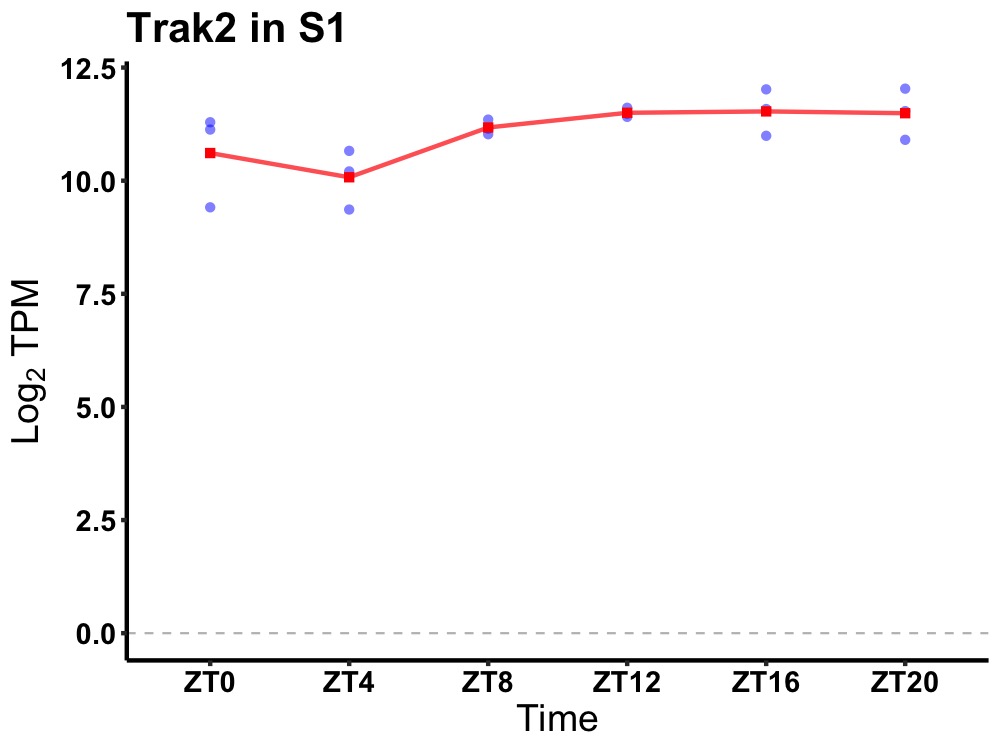 |
trafficking protein, kinesin binding 2 | 0.036 | 24 | 16 | 0.49 |
| ENSMUSG00000107950 | Dmac2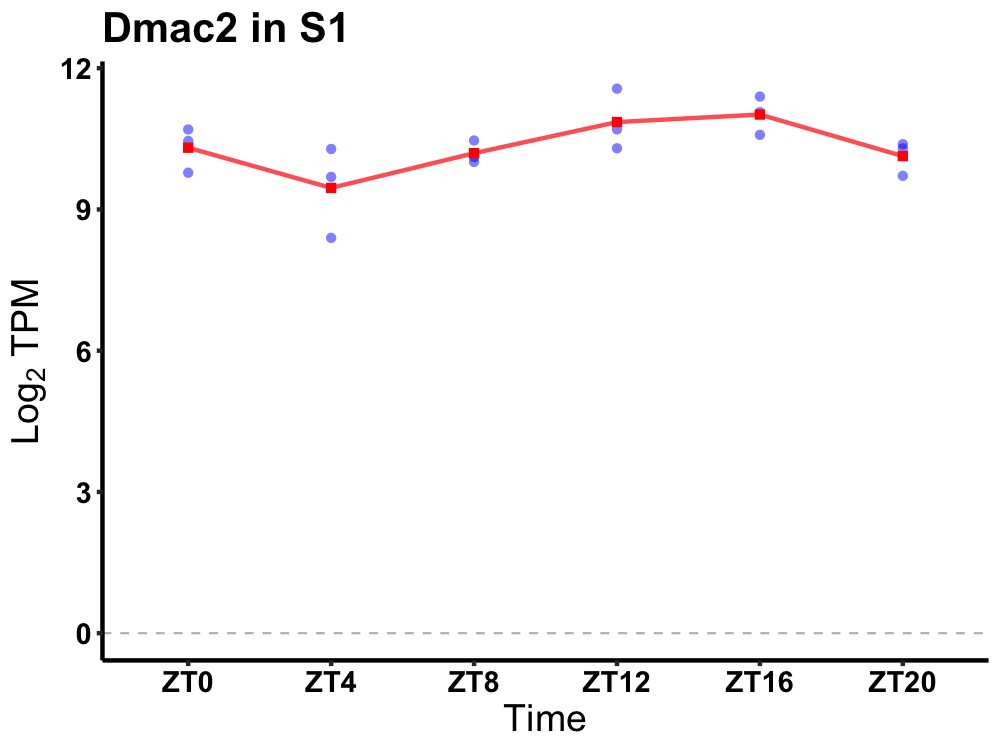 |
distal membrane arm assembly complex 2 | 0.010 | 20 | 16 | 0.49 |
| ENSMUSG00000059565 | Lyplal1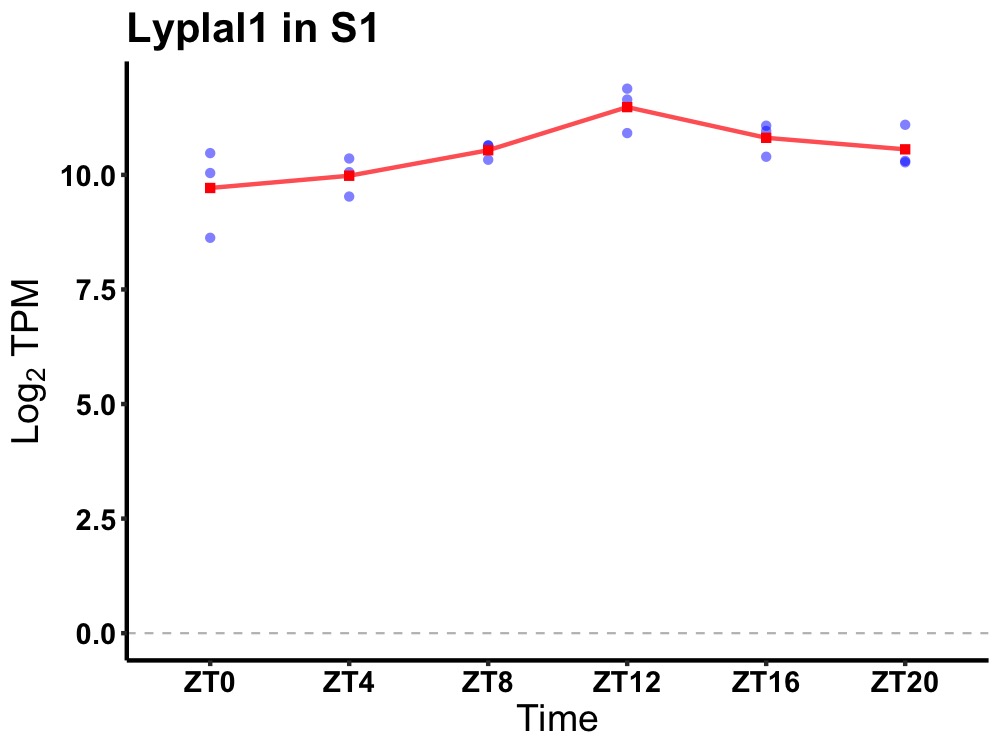 |
lysophospholipase-like 1 | 0.005 | 24 | 14 | 0.48 |
| ENSMUSG00000046934 | Csl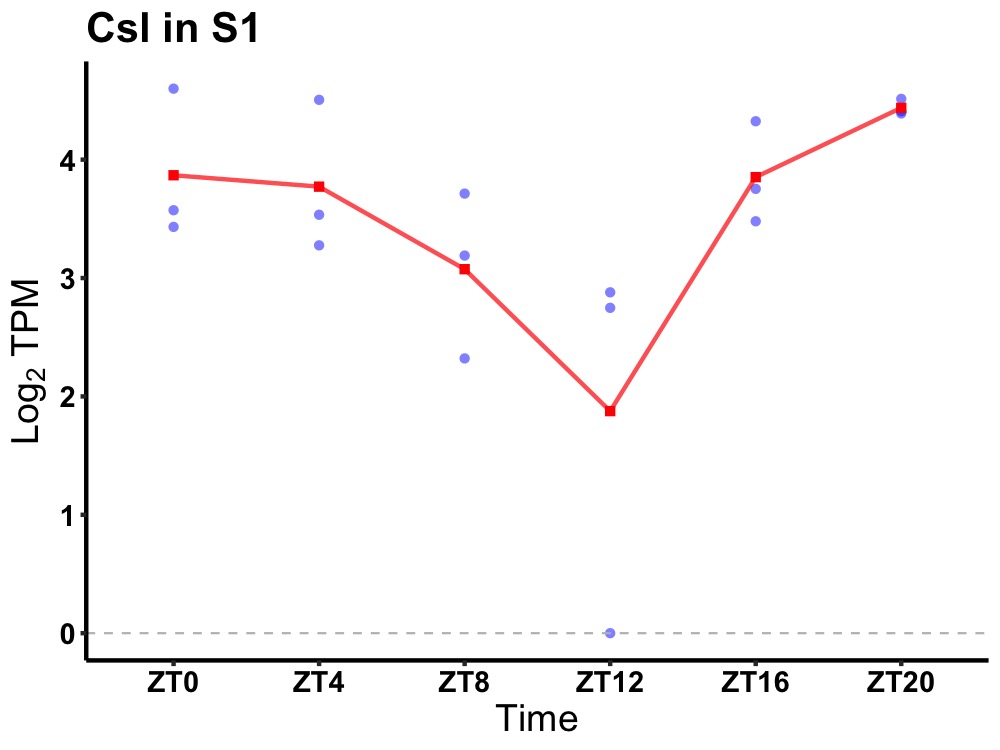 |
citrate synthase like | 0.014 | 20 | 2 | 0.48 |
| ENSMUSG00000067194 | Def8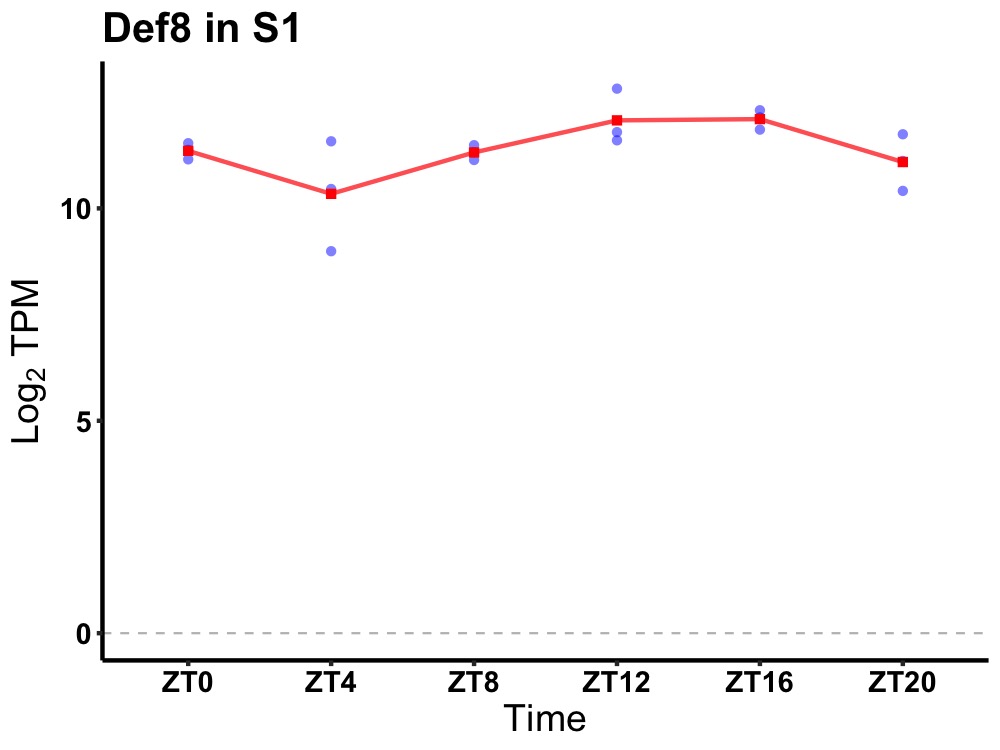 |
differentially expressed in FDCP 8 | 0.014 | 20 | 16 | 0.48 |
| ENSMUSG00000096255 | Dynlt1b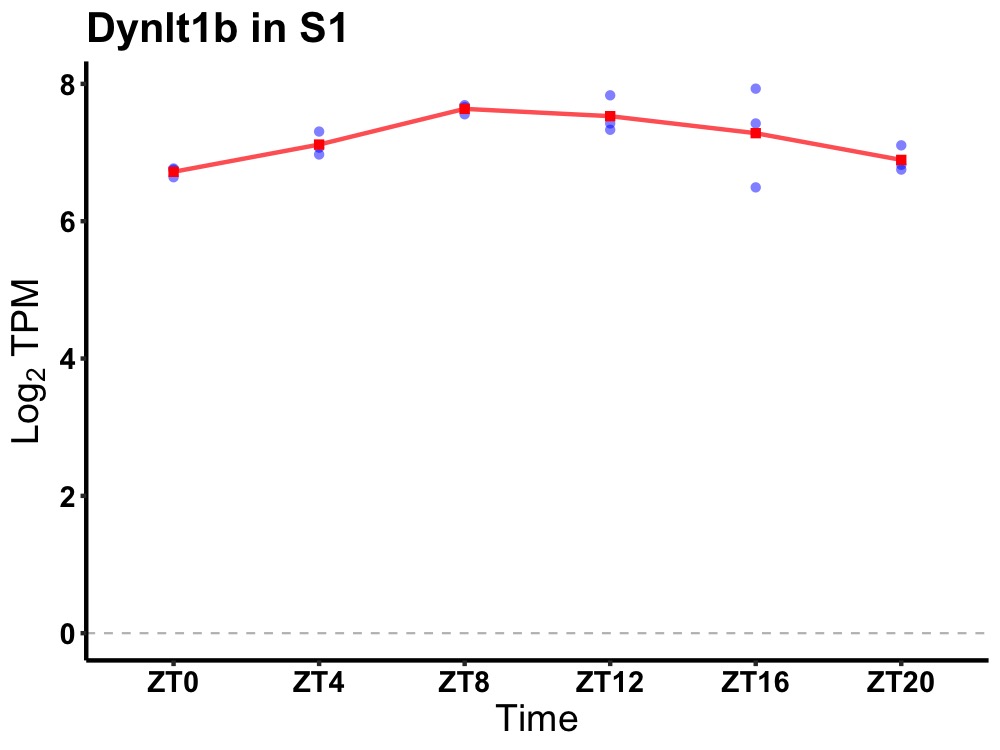 |
dynein light chain Tctex-type 1B | 0.014 | 24 | 12 | 0.47 |
| ENSMUSG00000066592 | Gm7589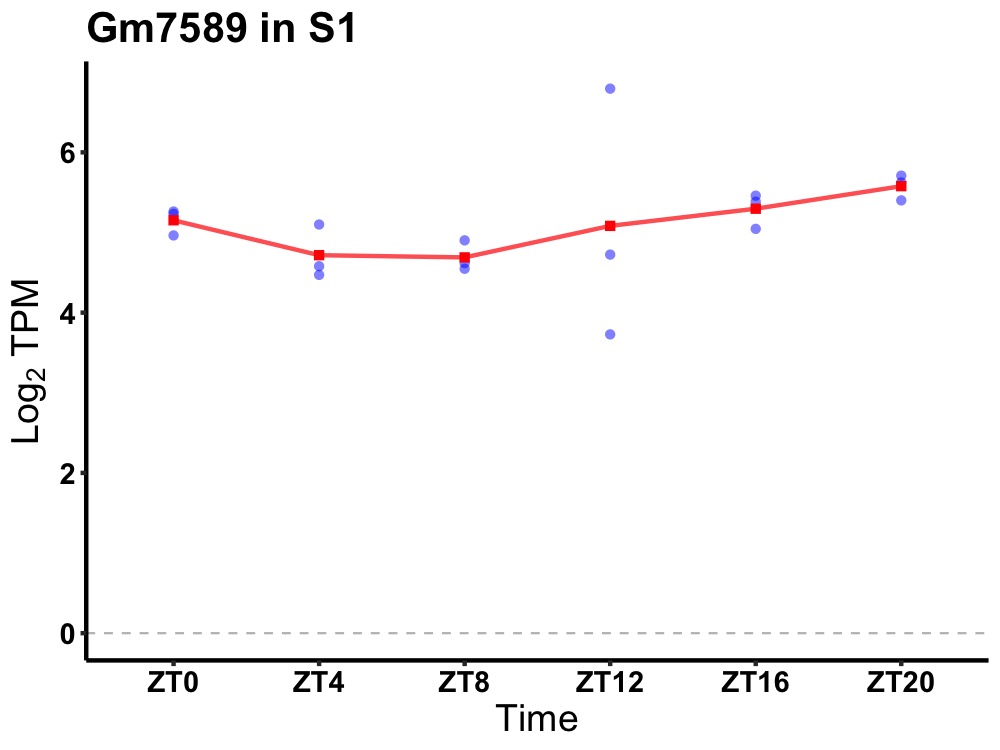 |
predicted gene 7589 | 0.027 | 24 | 20 | 0.46 |
| ENSMUSG00000027199 | Ccar1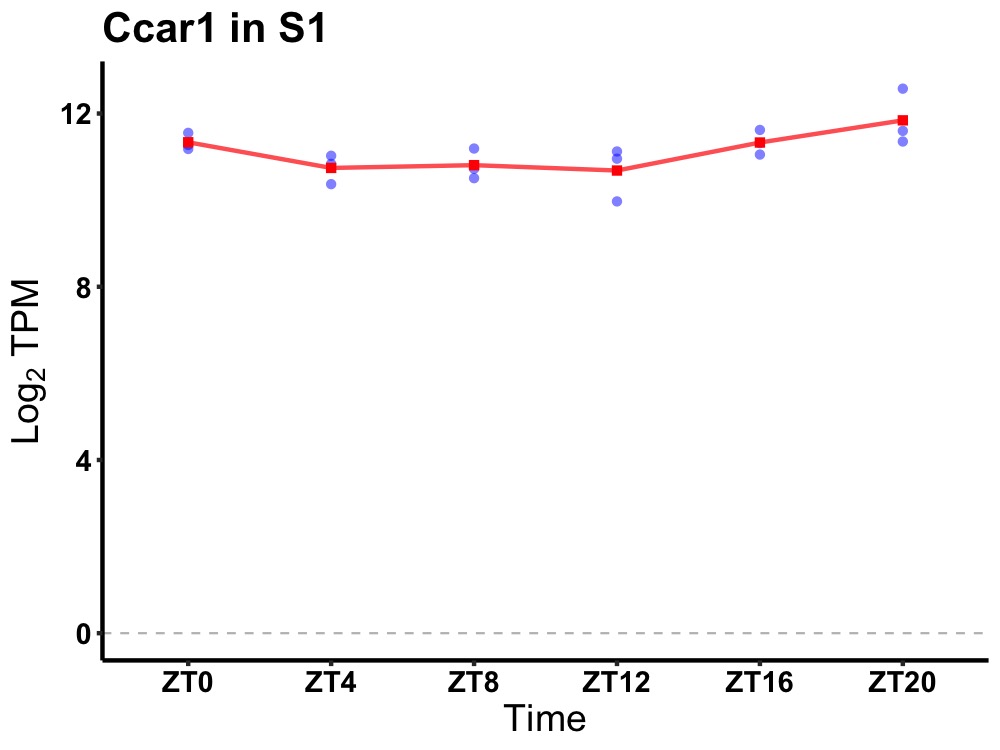 |
cell division cycle and apoptosis regulator 1 | 0.010 | 24 | 20 | 0.45 |
| ENSMUSG00000021951 | Alad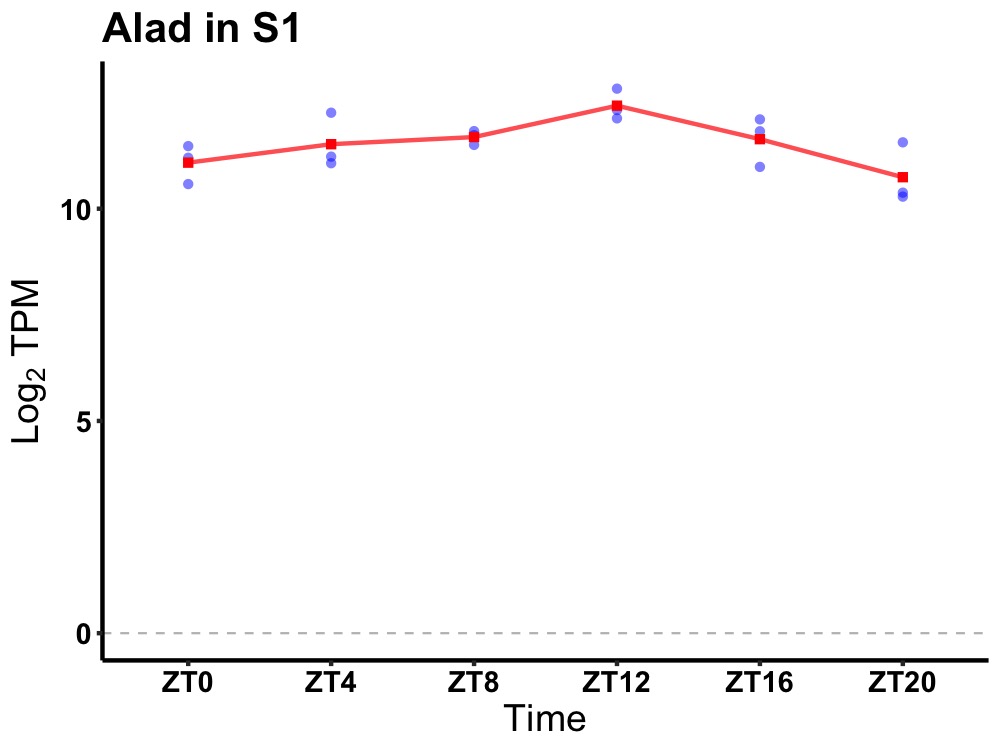 |
aminolevulinate, delta-, dehydratase | 0.019 | 20 | 12 | 0.44 |
| ENSMUSG00000048709 | Gm8666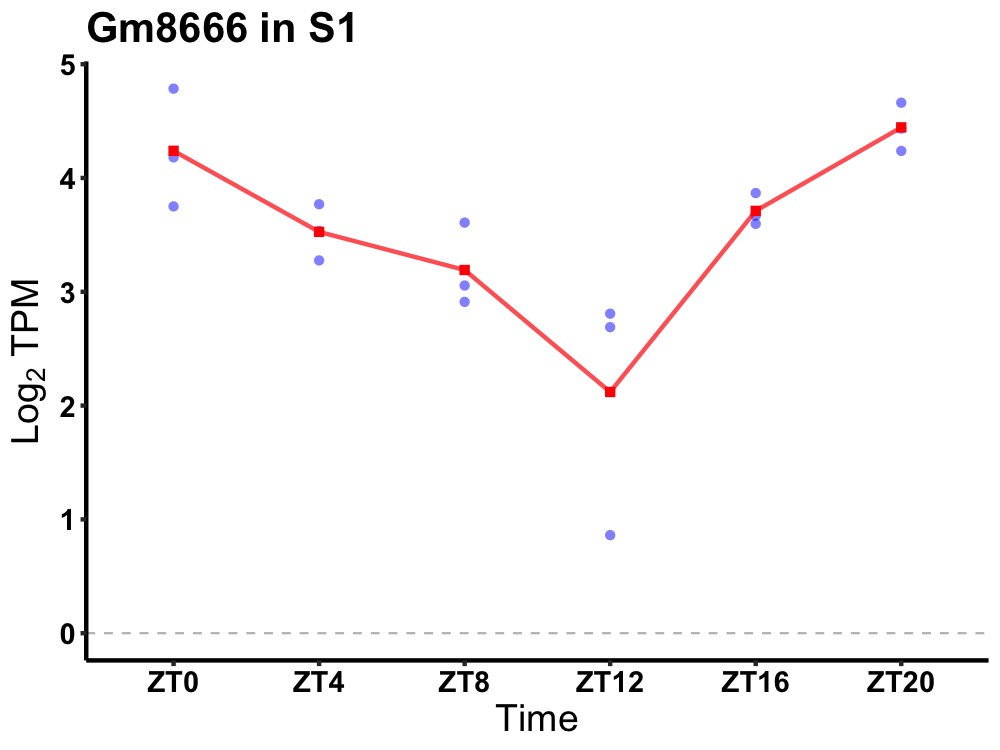 |
predicted gene 8666 | 0.000 | 20 | 2 | 0.44 |
| ENSMUSG00000115448 | Gm21178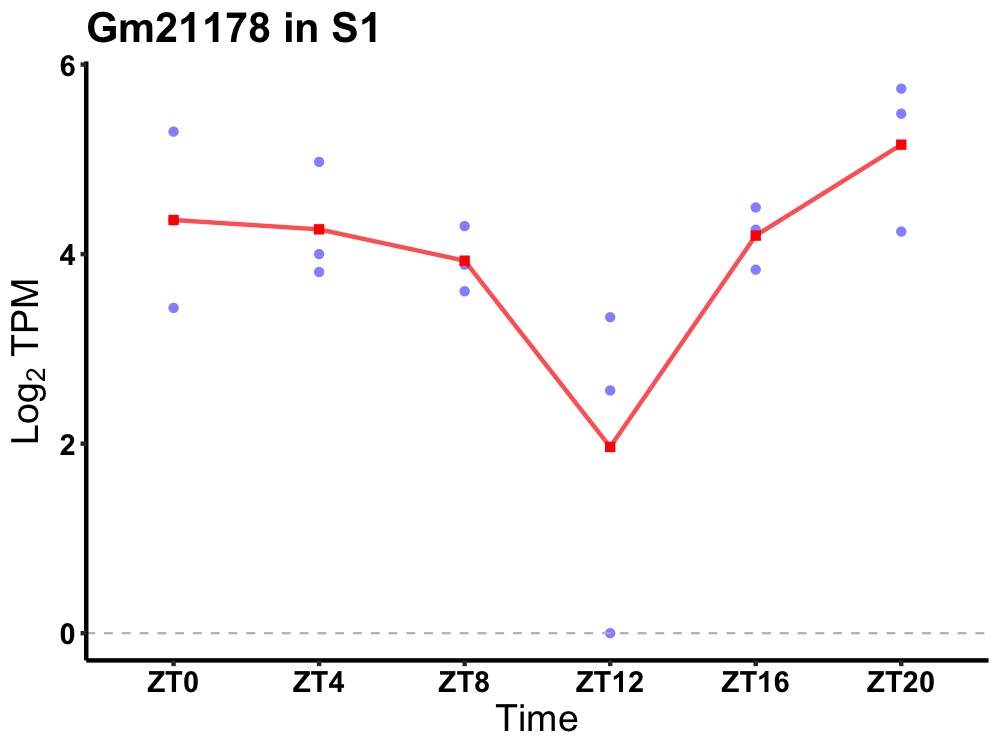 |
predicted gene, 21178 | 0.036 | 20 | 2 | 0.43 |
| ENSMUSG00000000938 | Oxa1l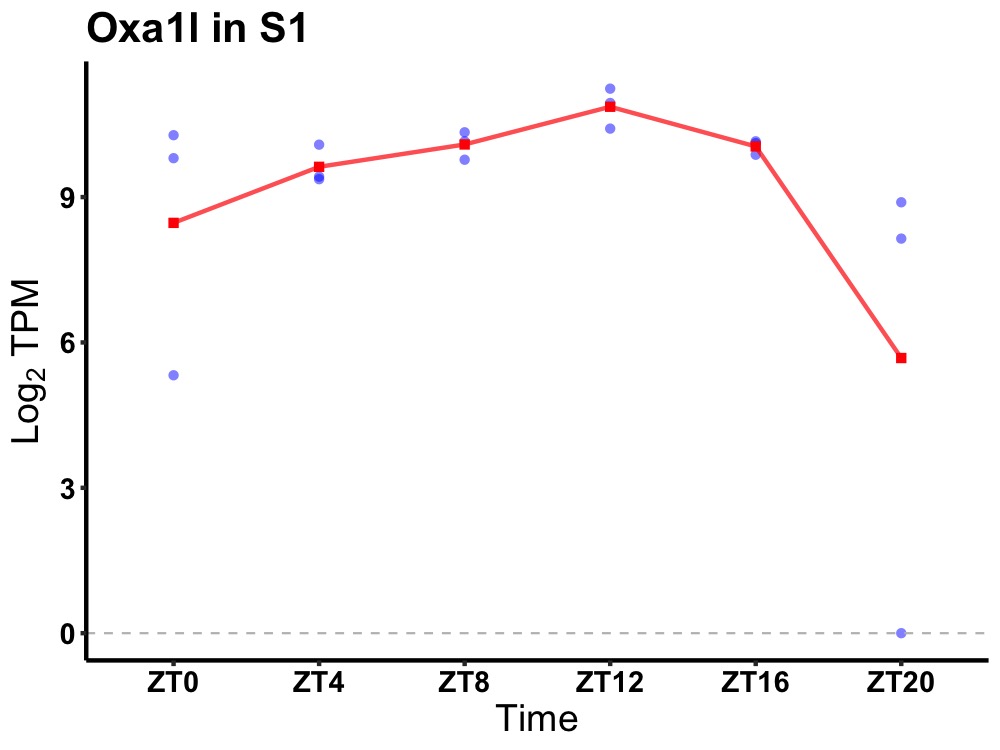 |
oxidase assembly 1-like | 0.001 | 20 | 12 | 0.43 |
| ENSMUSG00000106106 | Slc25a42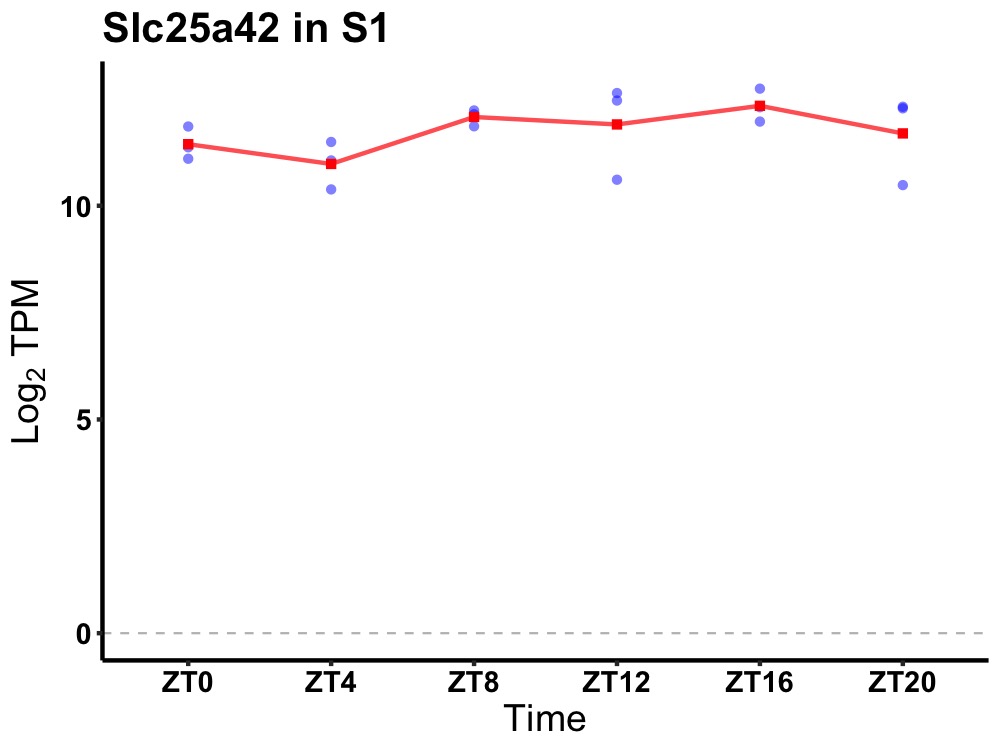 |
solute carrier family 25, member 42 | 0.049 | 24 | 16 | 0.43 |
| ENSMUSG00000019857 | Ubc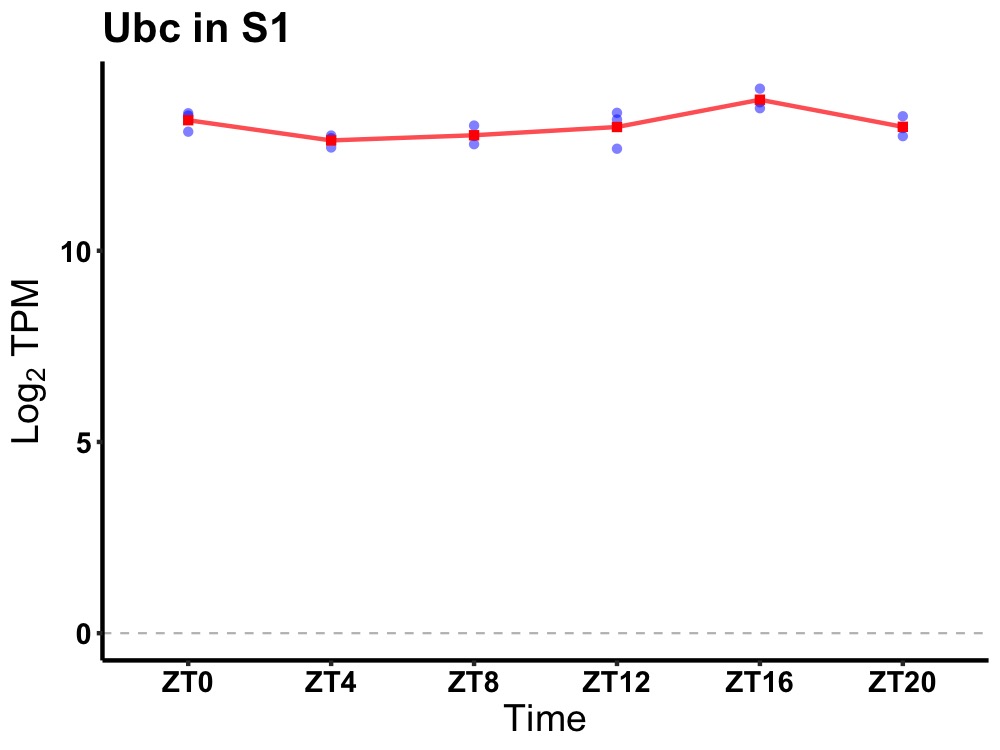 |
ubiquitin C | 0.014 | 20 | 16 | 0.43 |
| ENSMUSG00000021417 | Psmd14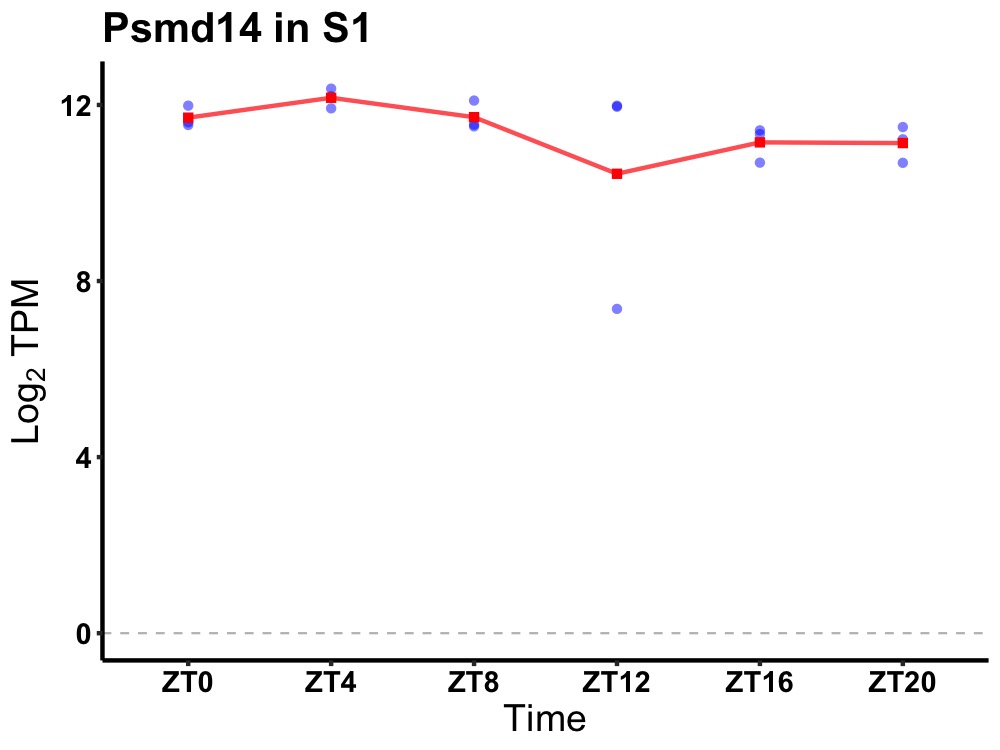 |
proteasome (prosome, macropain) 26S subunit, non-ATPase, 14 | 0.036 | 24 | 6 | 0.42 |
| ENSMUSG00000026490 | Tmem19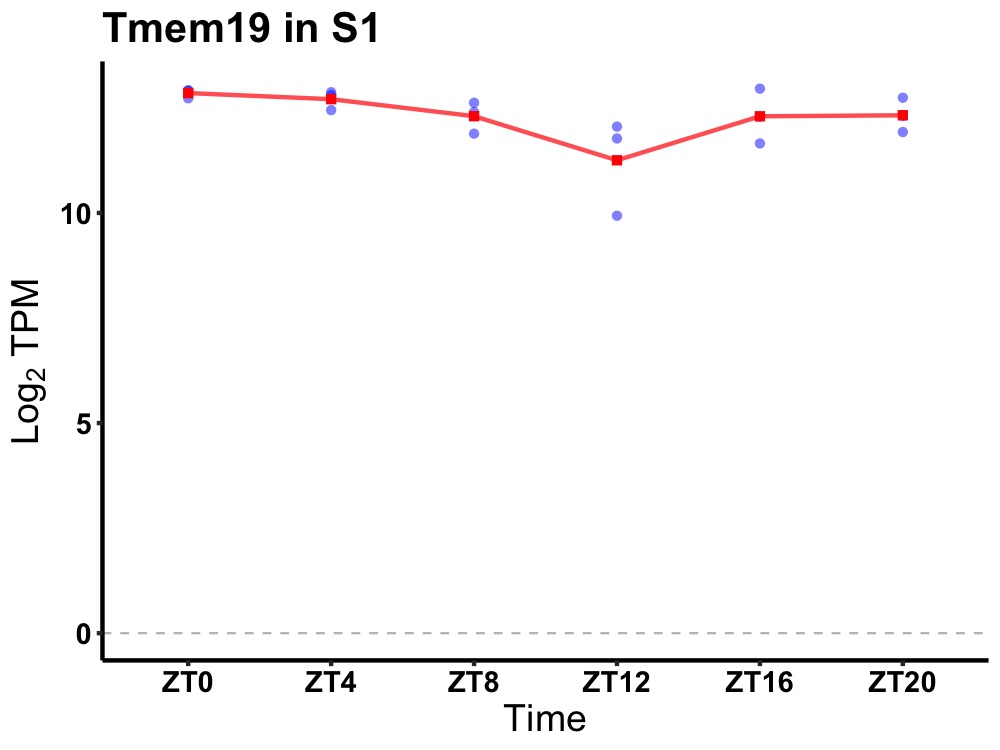 |
transmembrane protein 19 | 0.027 | 24 | 2 | 0.41 |
| ENSMUSG00000081607 | Gm15294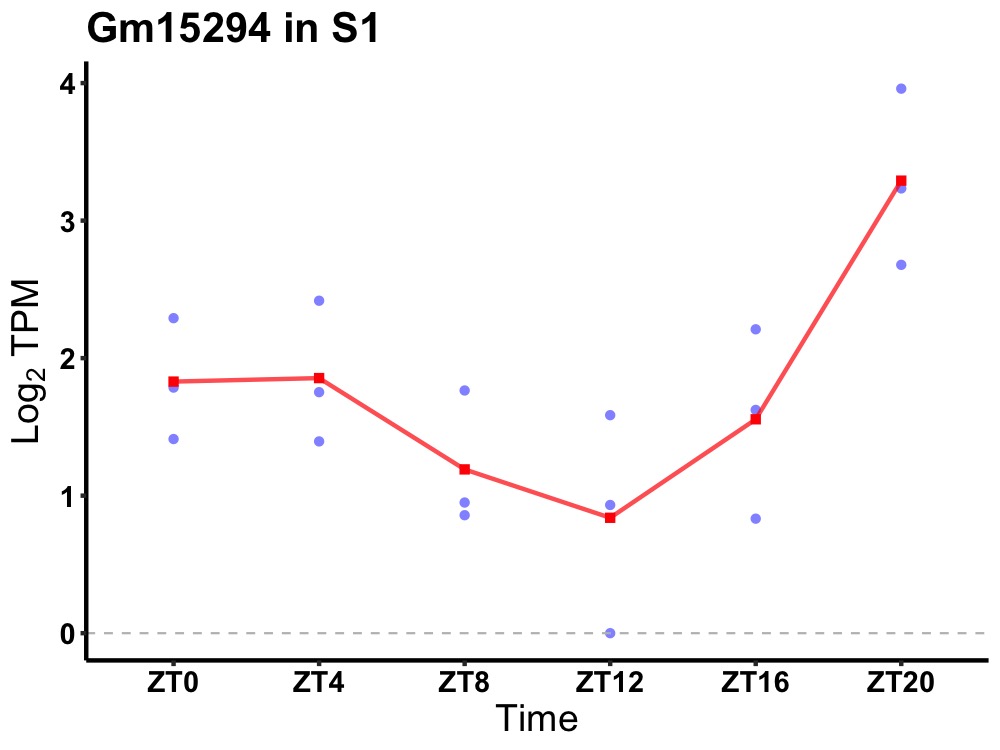 |
none | 0.010 | 20 | 2 | 0.41 |
| ENSMUSG00000029610 | Gstt2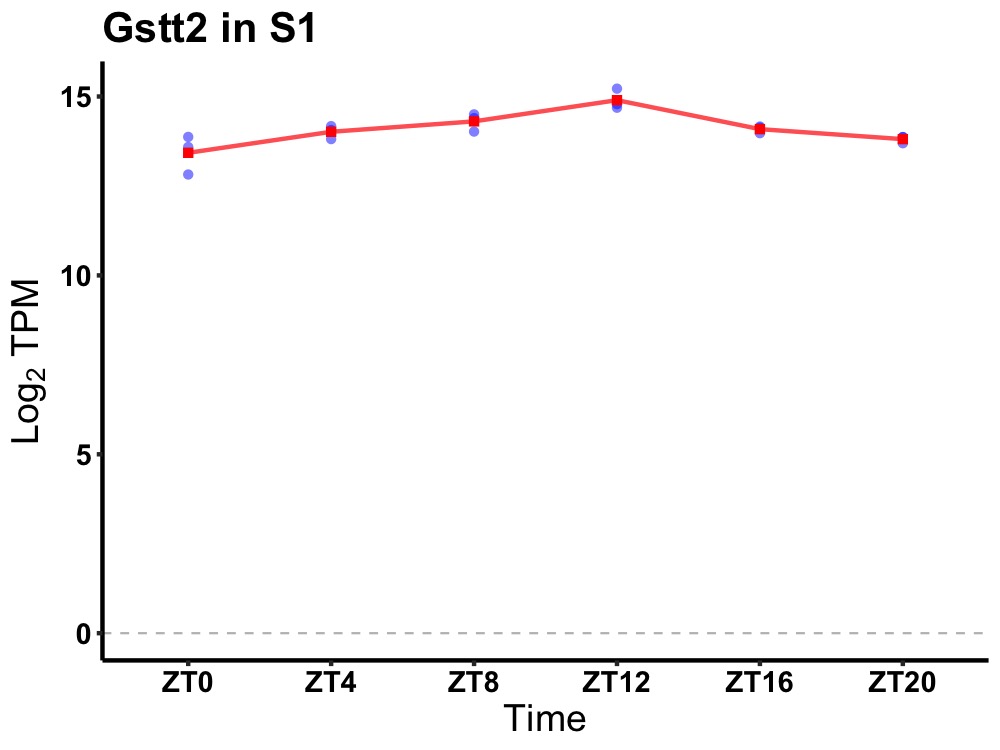 |
glutathione S-transferase, theta 2 | 0.000 | 24 | 12 | 0.40 |
| ENSMUSG00000083431 | Mlx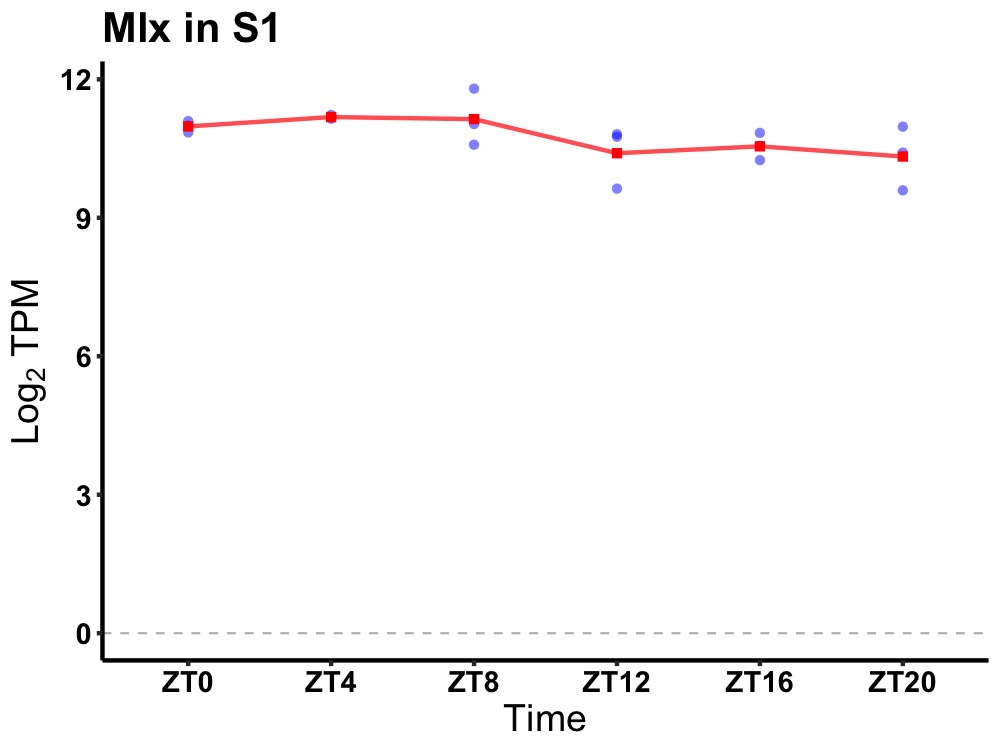 |
MAX-like protein X | 0.019 | 24 | 6 | 0.40 |
| ENSMUSG00000100615 | Gm5511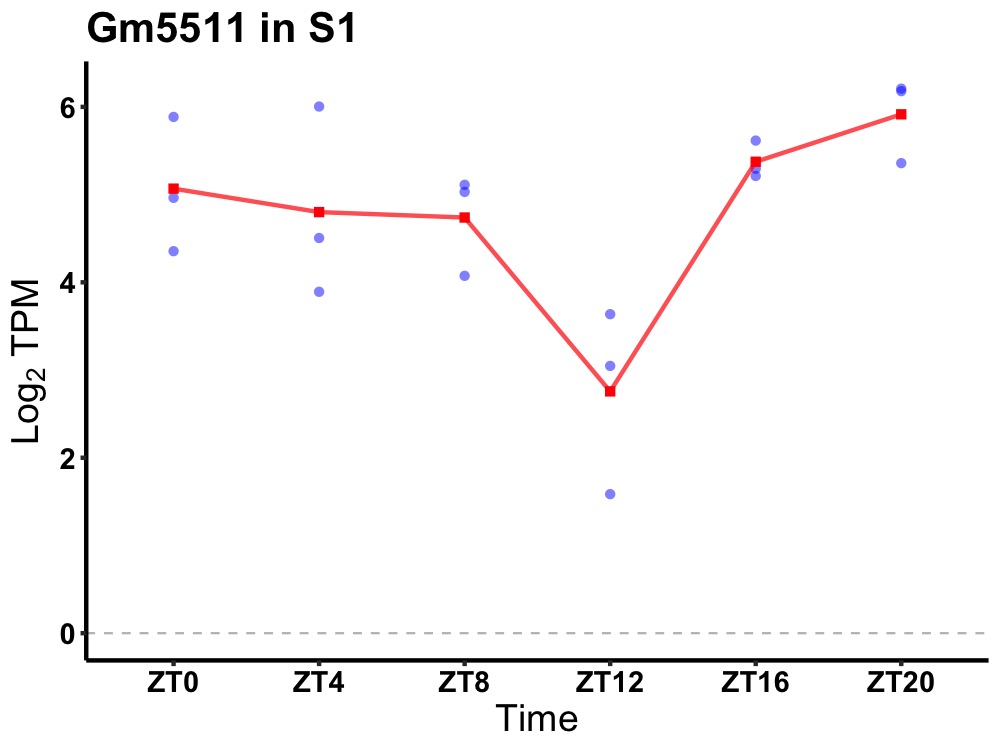 |
predicted gene 5511 | 0.036 | 20 | 2 | 0.40 |
| ENSMUSG00000029580 | Chpt1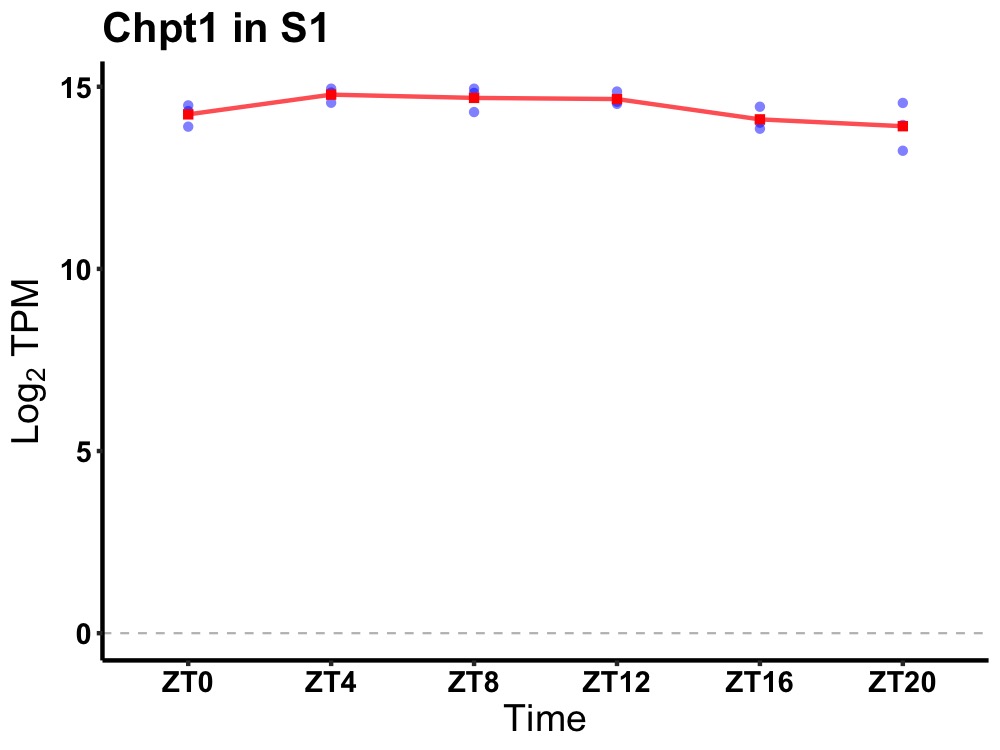 |
choline phosphotransferase 1 | 0.049 | 24 | 8 | 0.40 |
| ENSMUSG00000106558 | Gm5552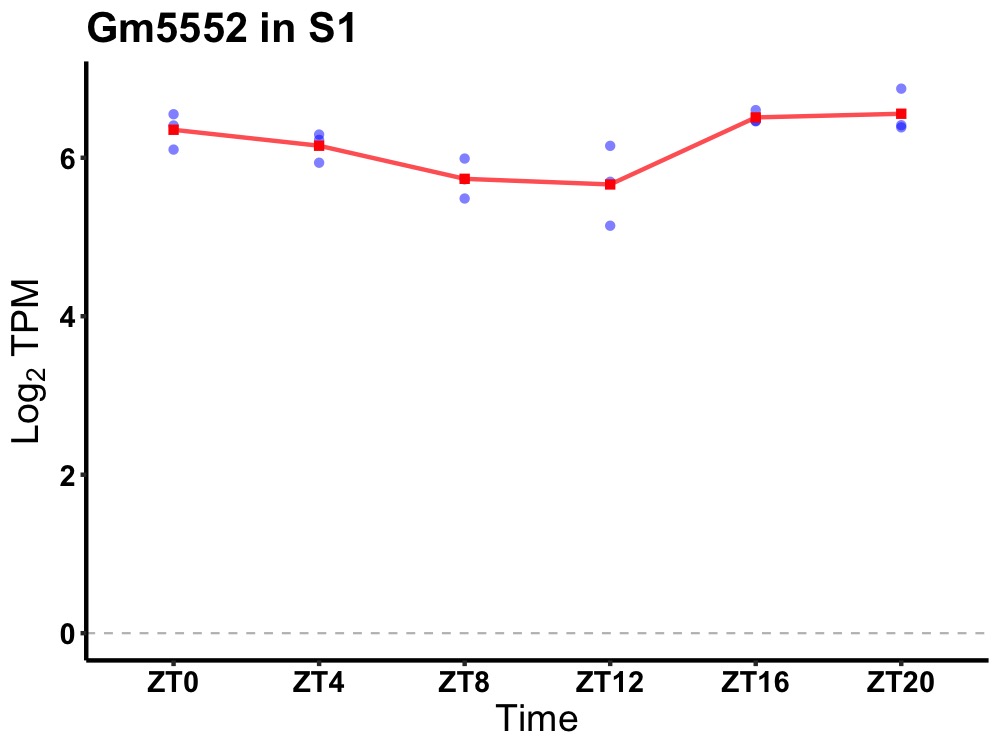 |
predicted gene 5552 | 0.019 | 24 | 22 | 0.40 |
| ENSMUSG00000026278 | Acy1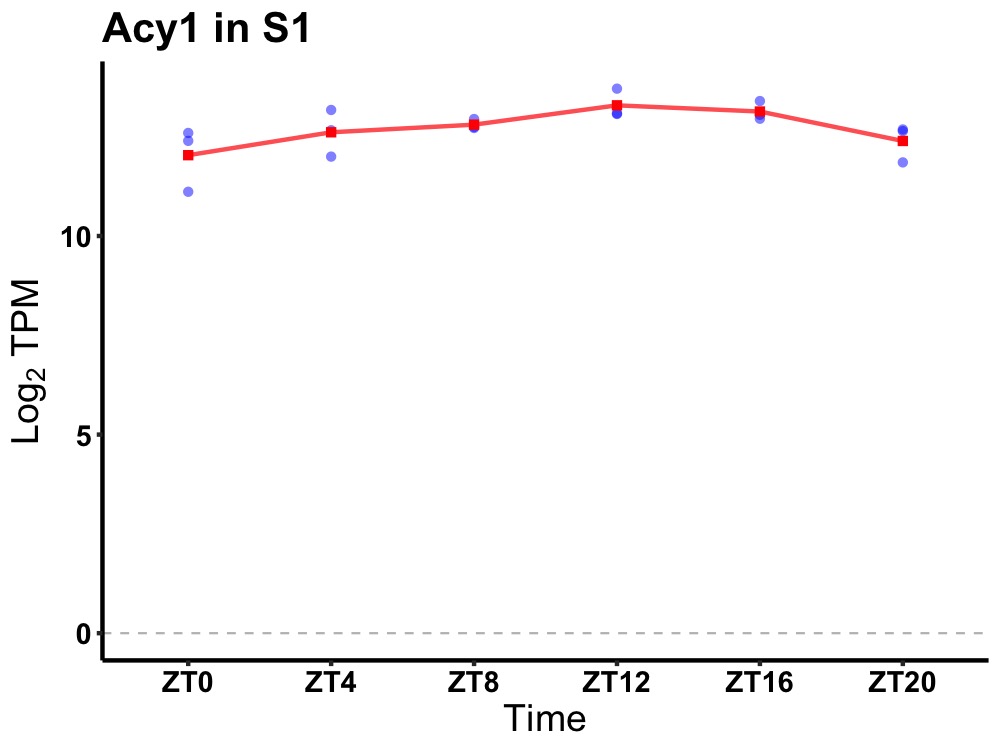 |
aminoacylase 1 | 0.001 | 24 | 14 | 0.39 |
| ENSMUSG00000106190 | Isca1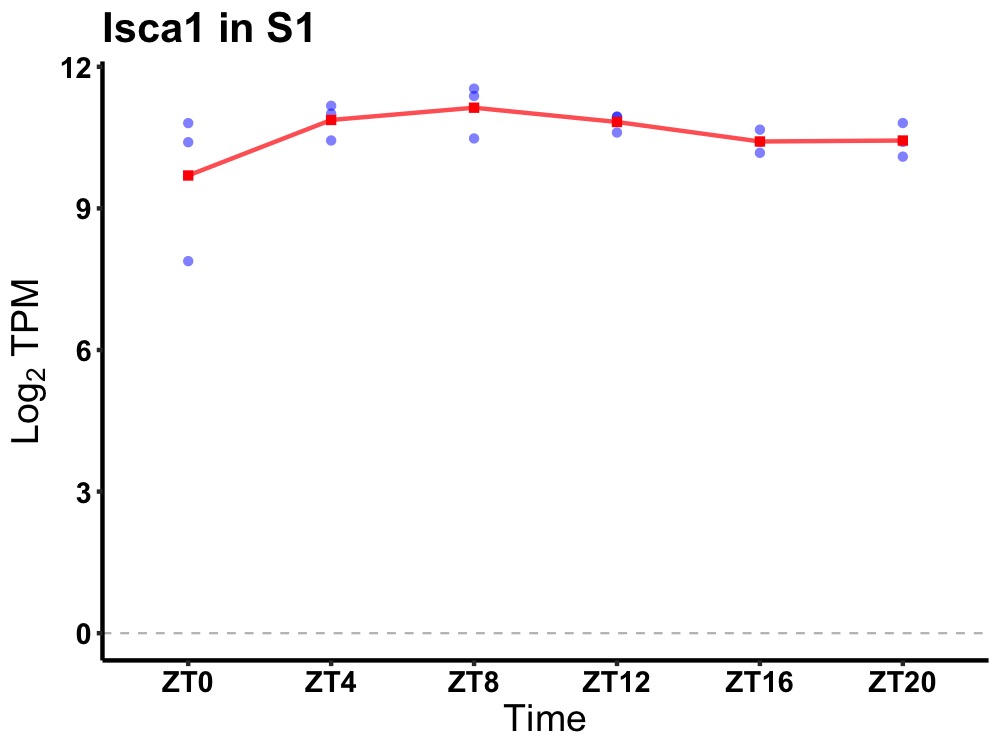 |
iron-sulfur cluster assembly 1 | 0.049 | 20 | 8 | 0.39 |
| ENSMUSG00000026095 | Dnaja1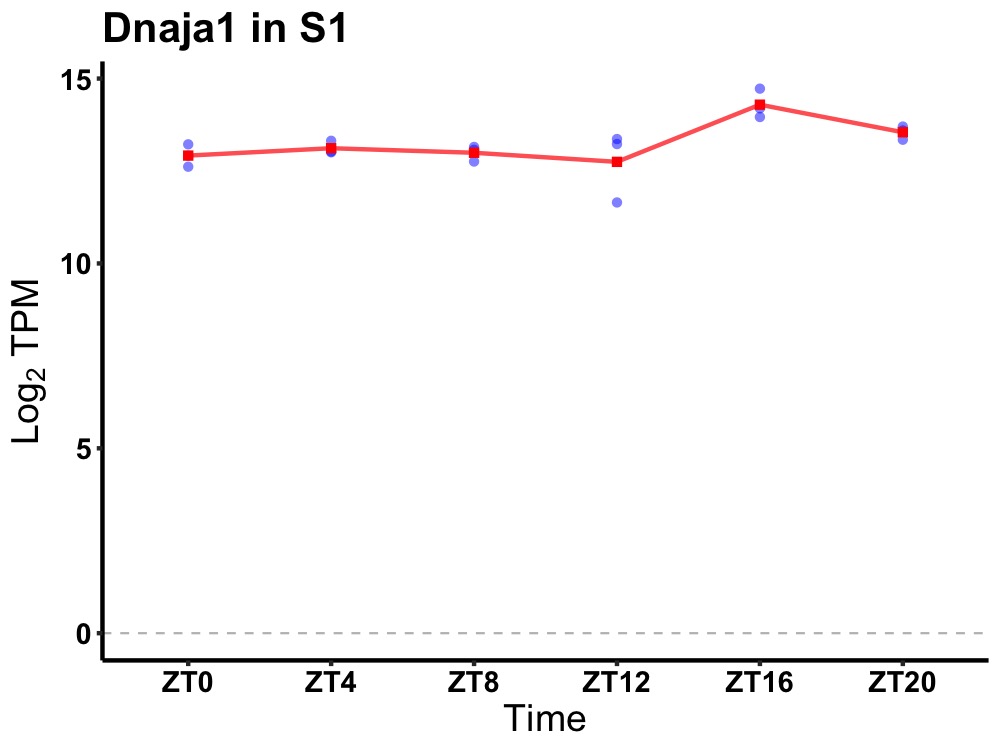 |
DnaJ heat shock protein family (Hsp40) member A1 | 0.036 | 24 | 18 | 0.39 |
| ENSMUSG00000022124 | Slc25a30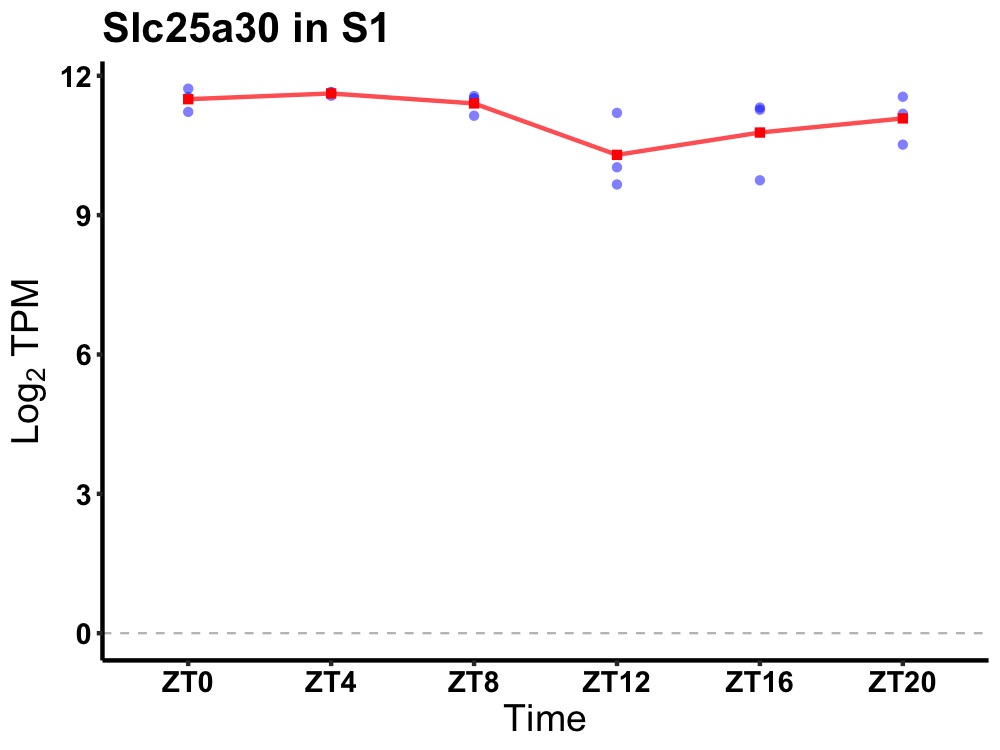 |
solute carrier family 25, member 30 | 0.049 | 24 | 4 | 0.38 |
| ENSMUSG00000091803 | Cox16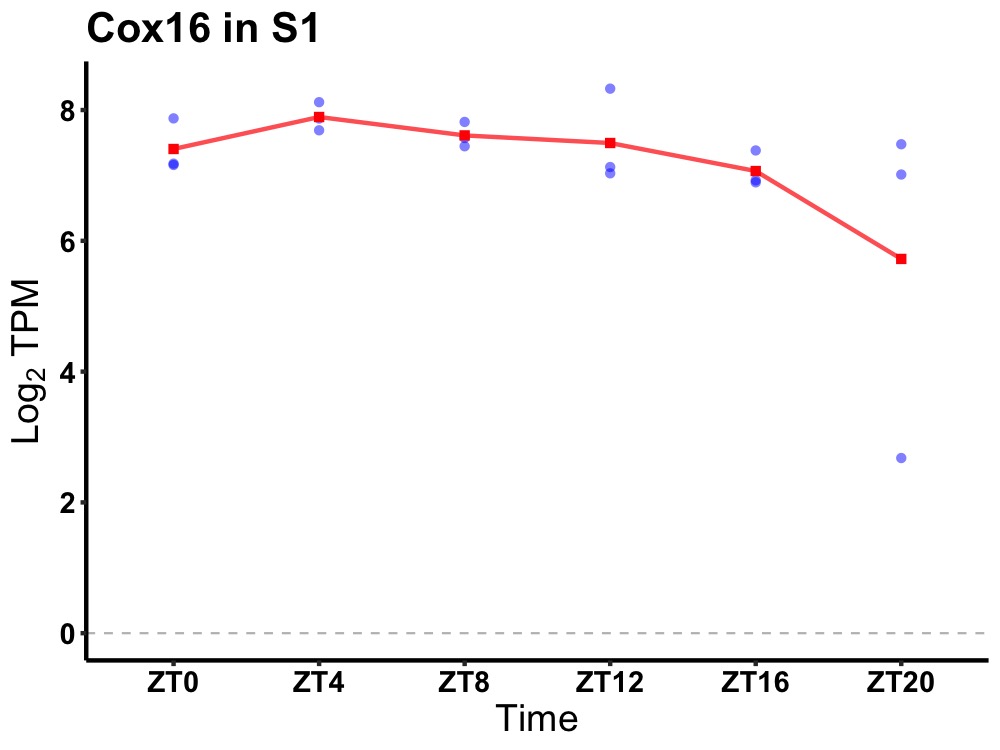 |
cytochrome c oxidase assembly protein 16 | 0.036 | 24 | 6 | 0.38 |
| ENSMUSG00000117359 | Gm7466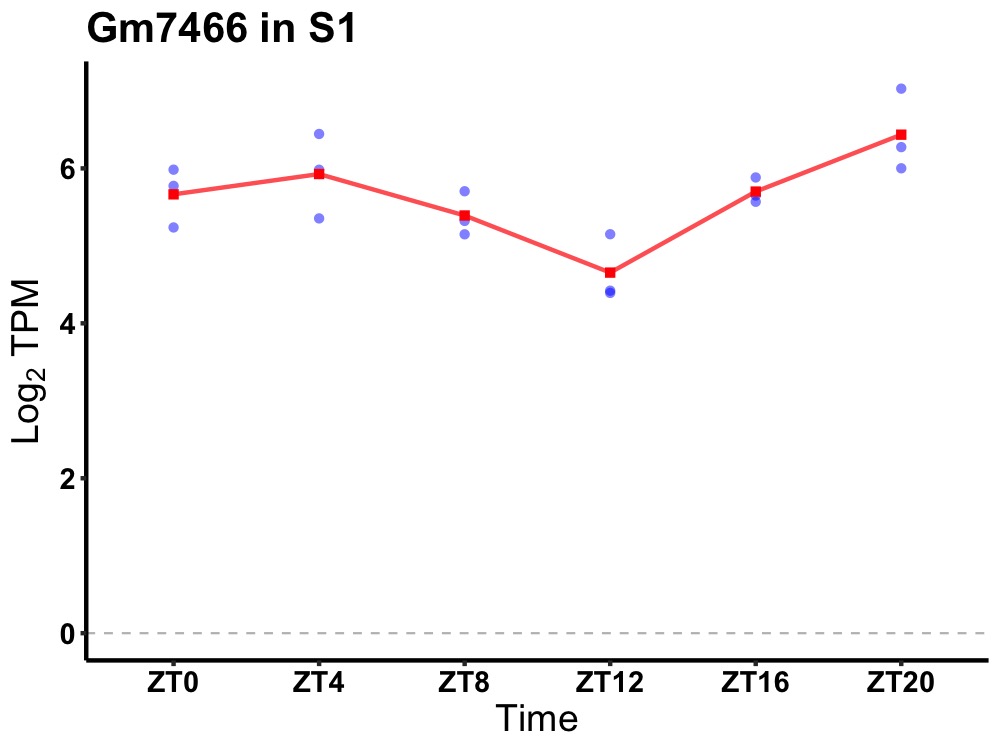 |
none | 0.003 | 20 | 2 | 0.38 |
| ENSMUSG00000035530 | Unc119b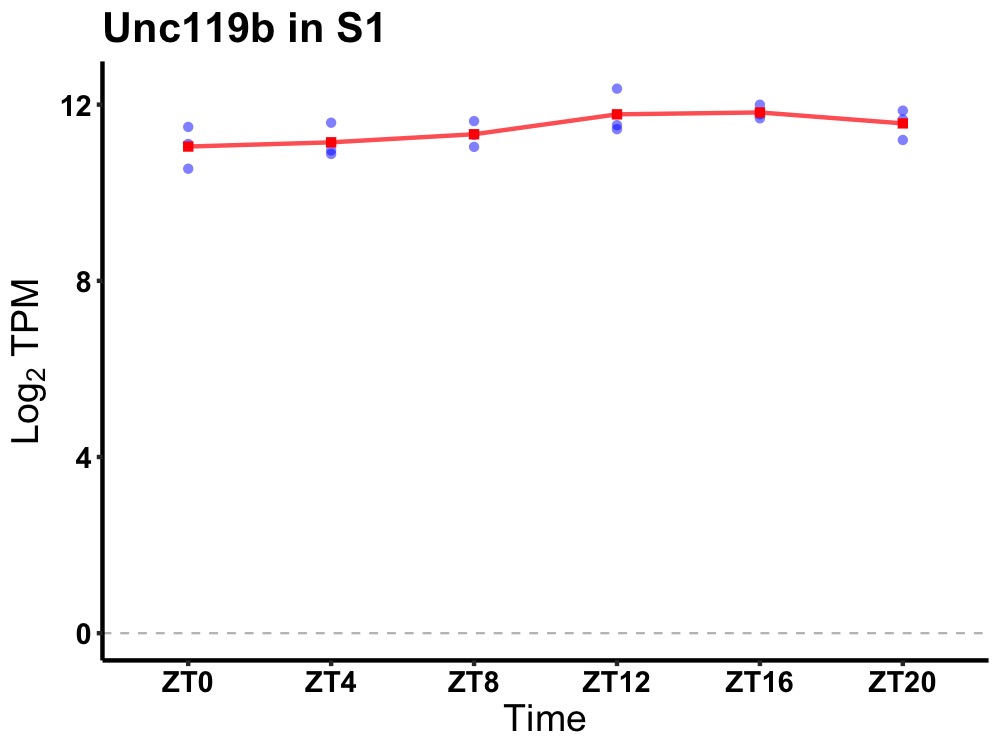 |
unc-119 lipid binding chaperone B | 0.036 | 24 | 16 | 0.38 |
| ENSMUSG00000022210 | Adrm1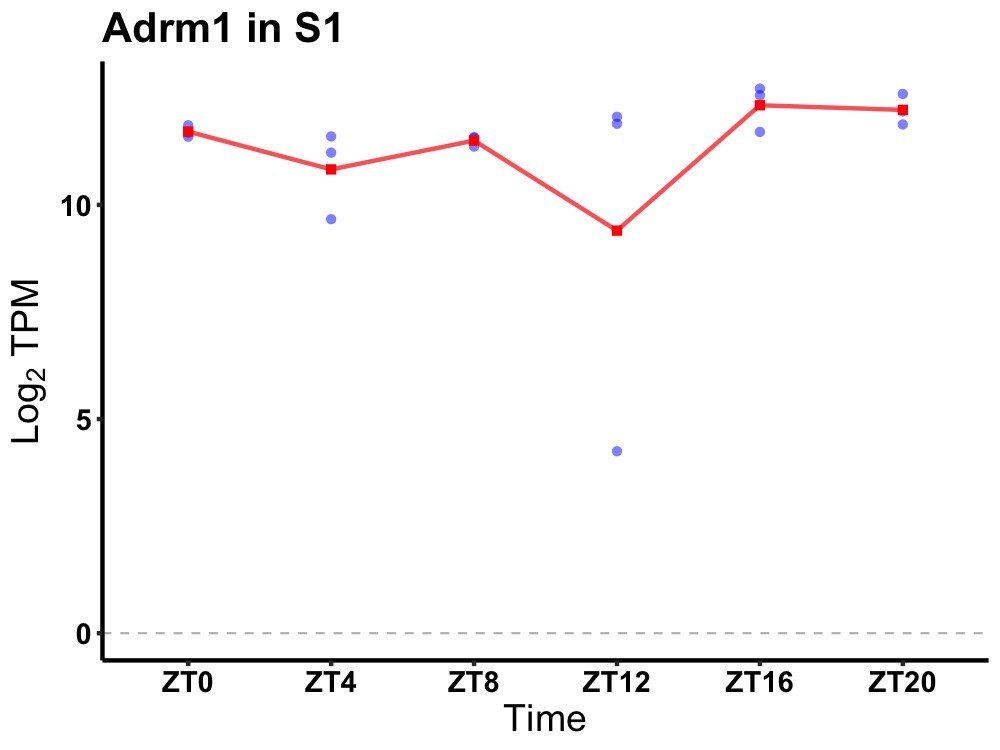 |
adhesion regulating molecule 1 | 0.005 | 24 | 18 | 0.38 |
| ENSMUSG00000091549 | Gm6548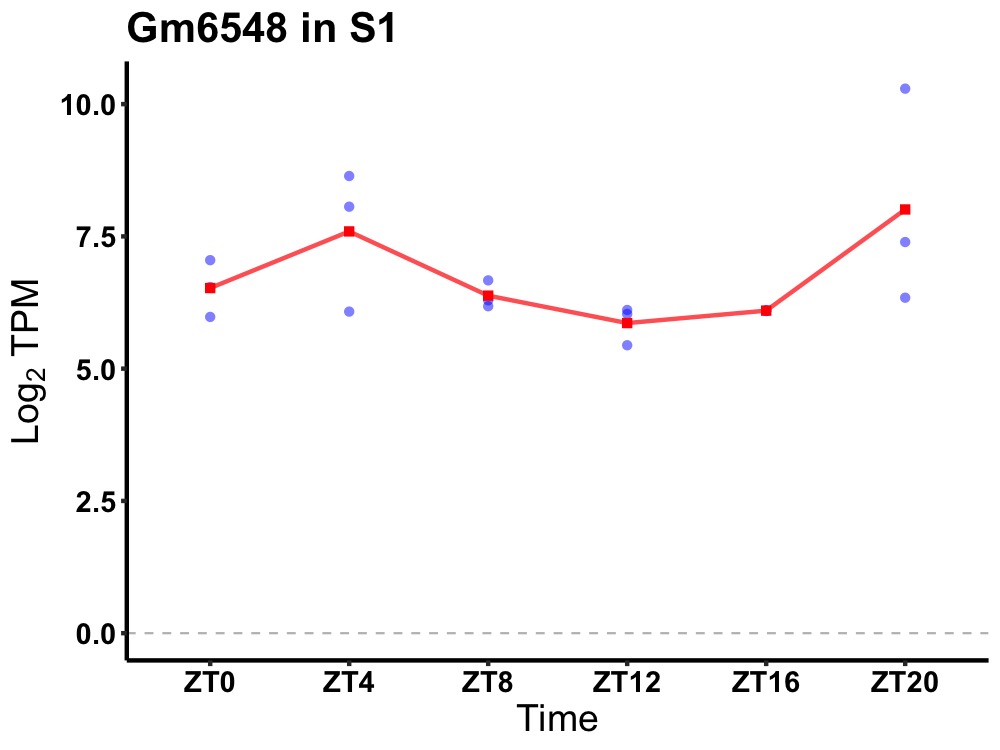 |
predicted gene 6548 | 0.049 | 20 | 4 | 0.38 |
| ENSMUSG00000027810 | Sirt3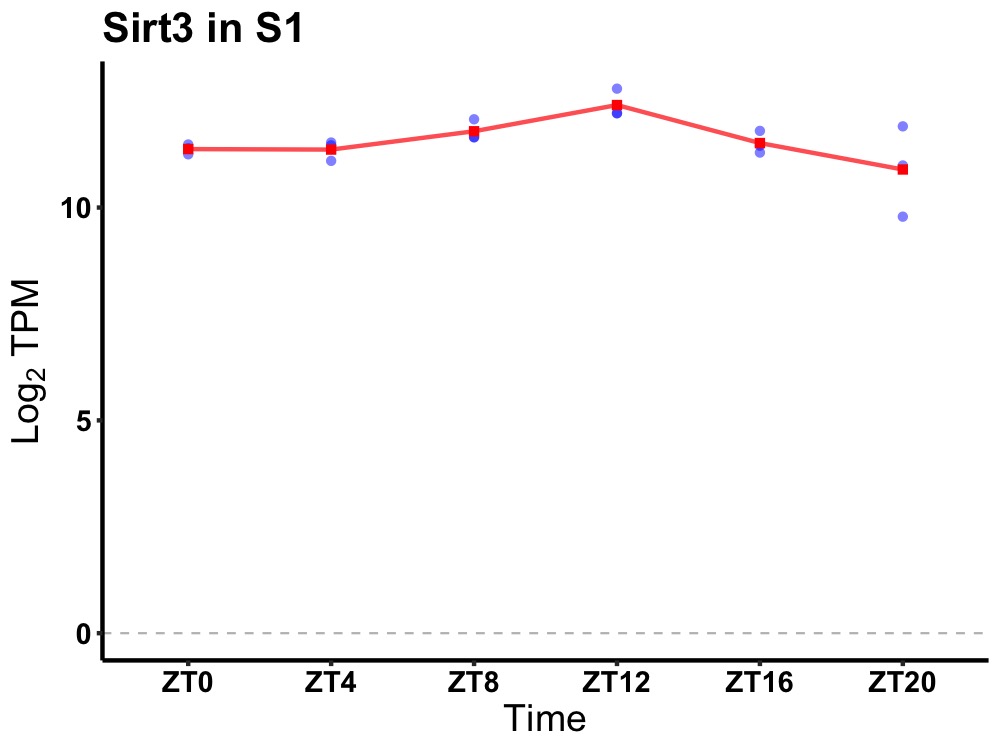 |
sirtuin 3 | 0.010 | 20 | 12 | 0.37 |
| ENSMUSG00000117504 | Sarnp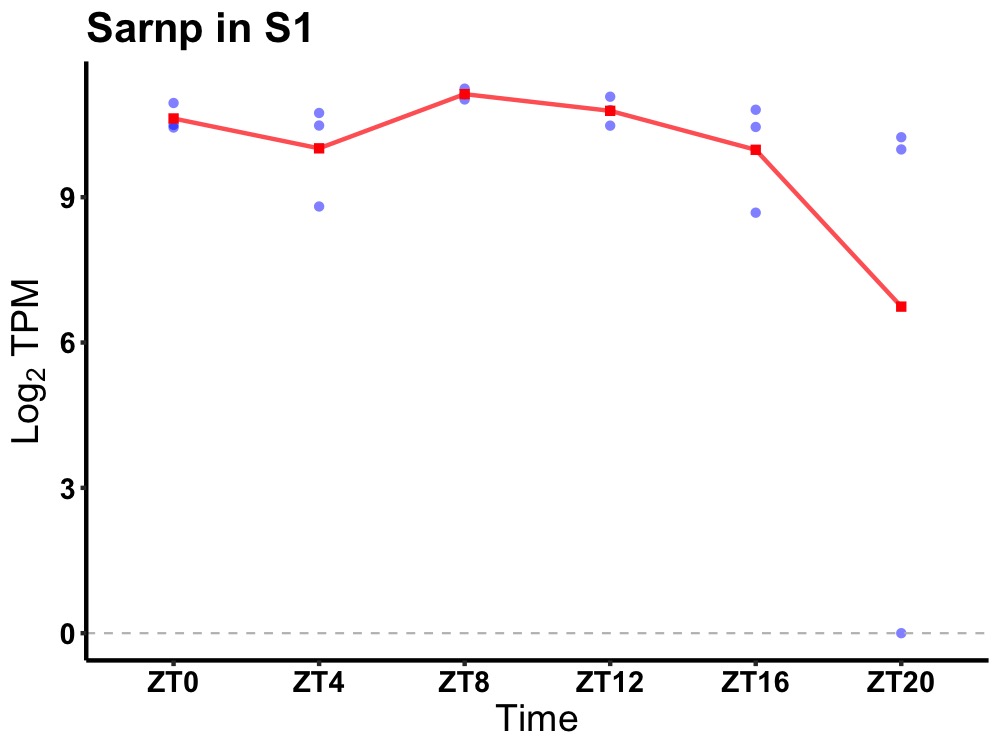 |
SAP domain containing ribonucleoprotein | 0.036 | 24 | 10 | 0.37 |
| ENSMUSG00000082896 | Aph1c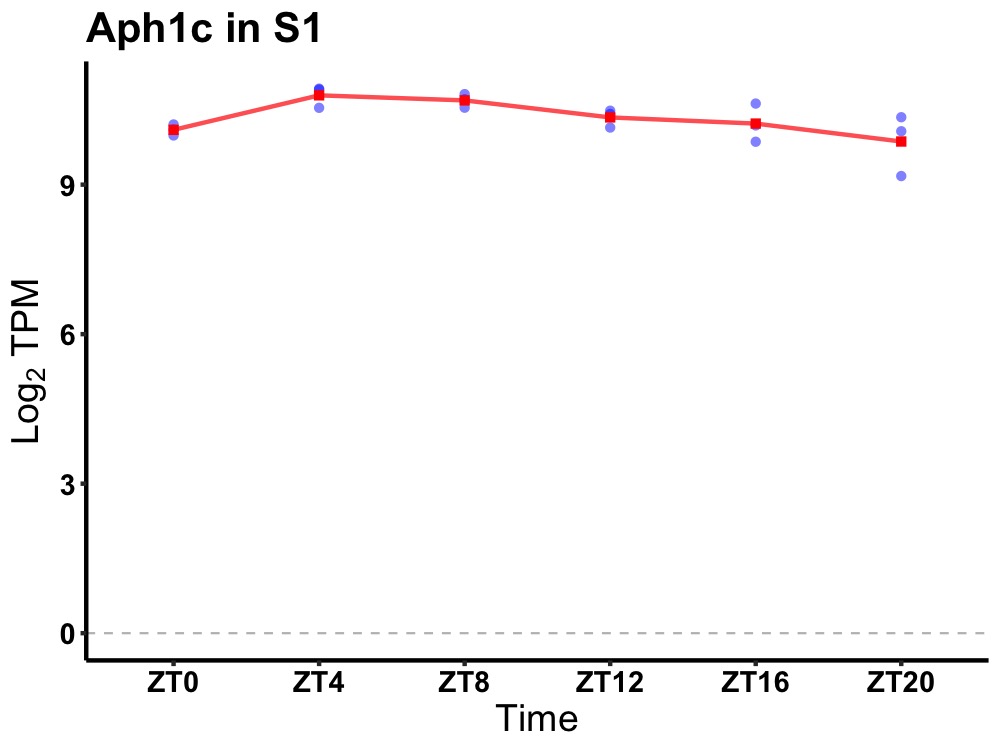 |
aph1 homolog C, gamma secretase subunit | 0.019 | 24 | 8 | 0.37 |
| ENSMUSG00000028248 | Dmac1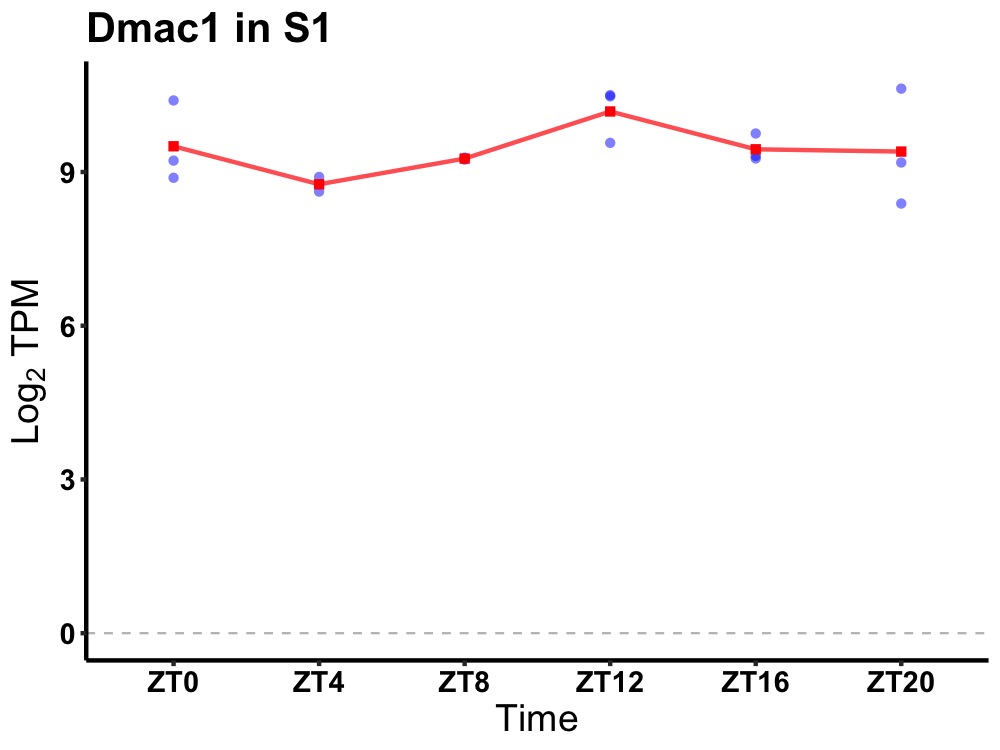 |
distal membrane arm assembly complex 1 | 0.019 | 20 | 14 | 0.37 |
| ENSMUSG00000067148 | Rbks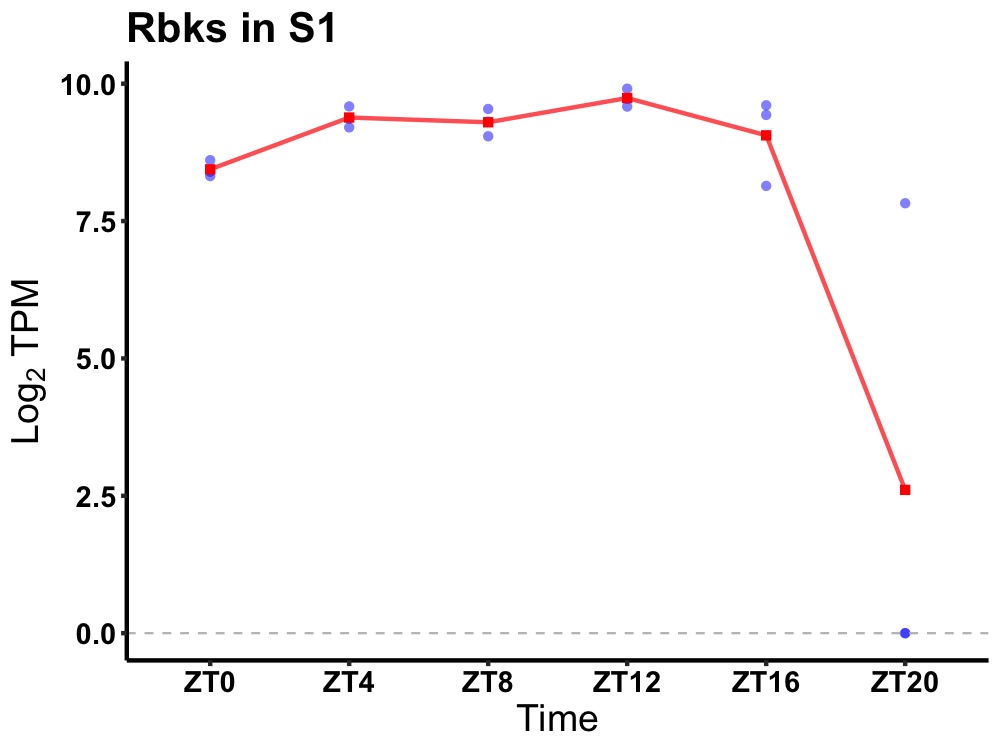 |
ribokinase | 0.002 | 20 | 12 | 0.36 |
| ENSMUSG00000083004 | Rpsa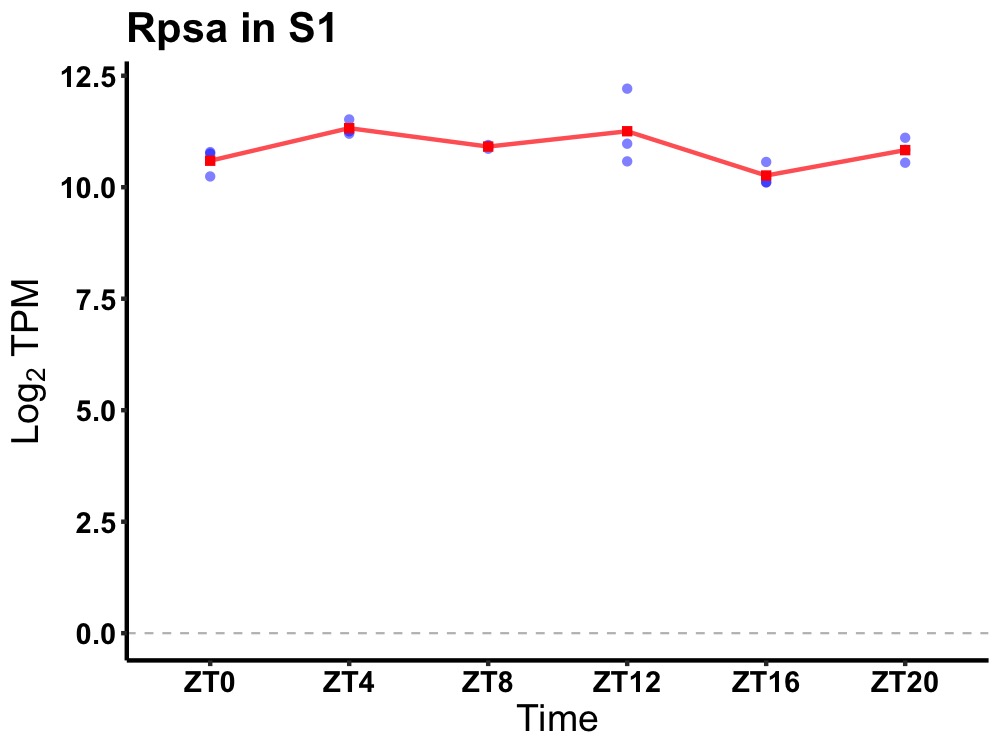 |
ribosomal protein SA | 0.049 | 20 | 6 | 0.35 |
| ENSMUSG00000096979 | Plscr1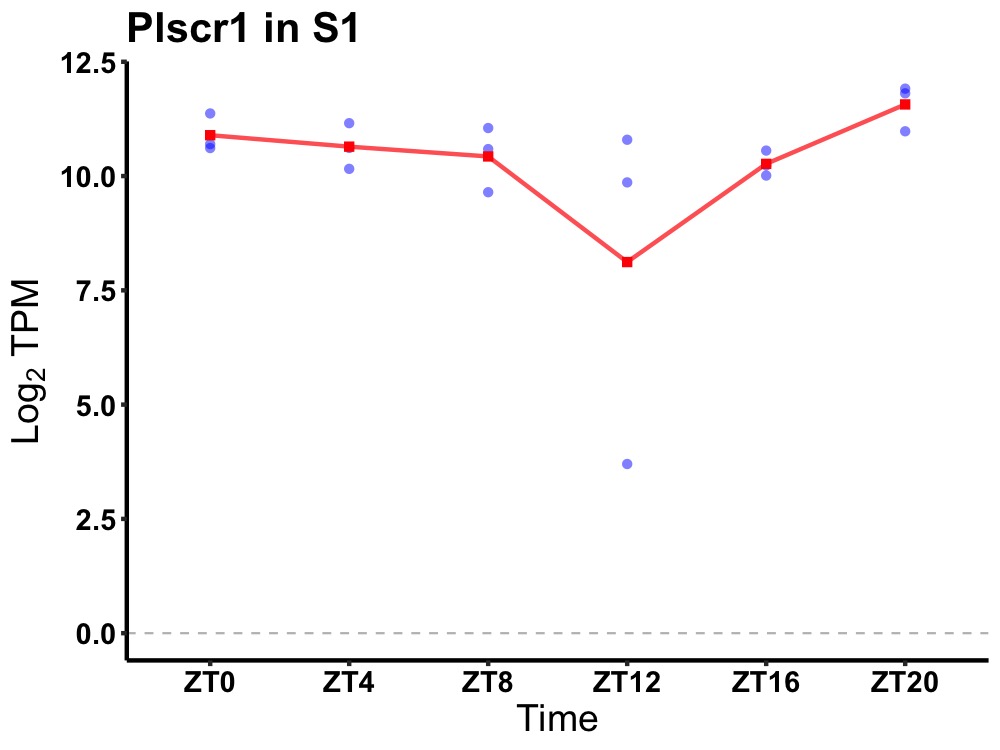 |
phospholipid scramblase 1 | 0.049 | 20 | 2 | 0.35 |
| ENSMUSG00000048118 | H3f3b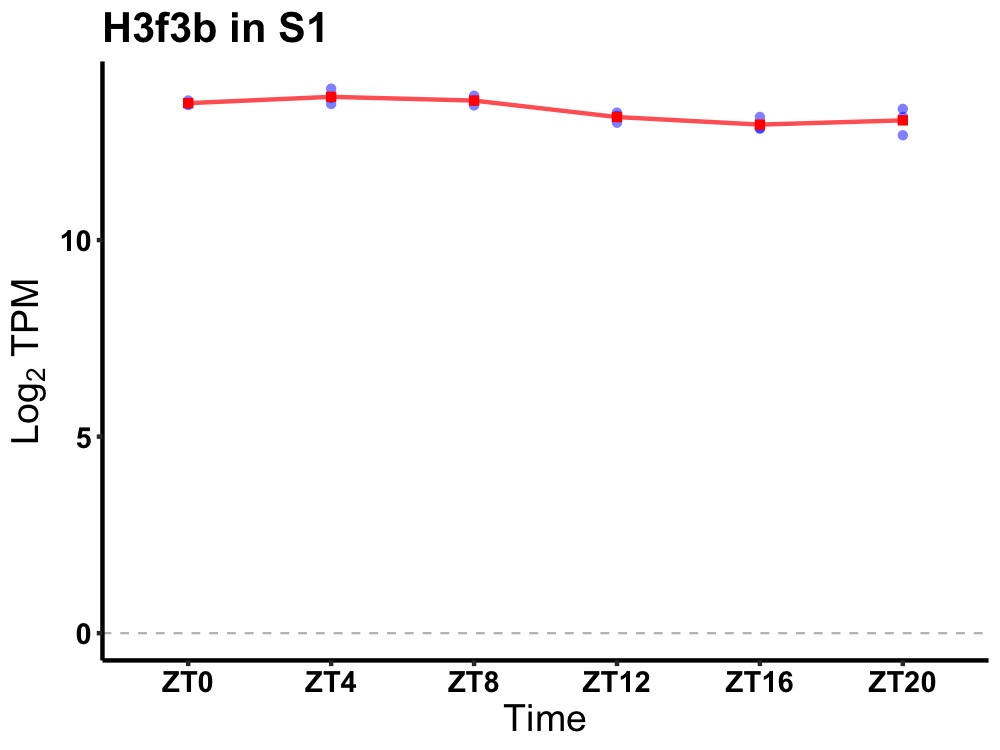 |
H3 histone, family 3B | 0.001 | 24 | 6 | 0.35 |
| ENSMUSG00000058355 | mt-Nd5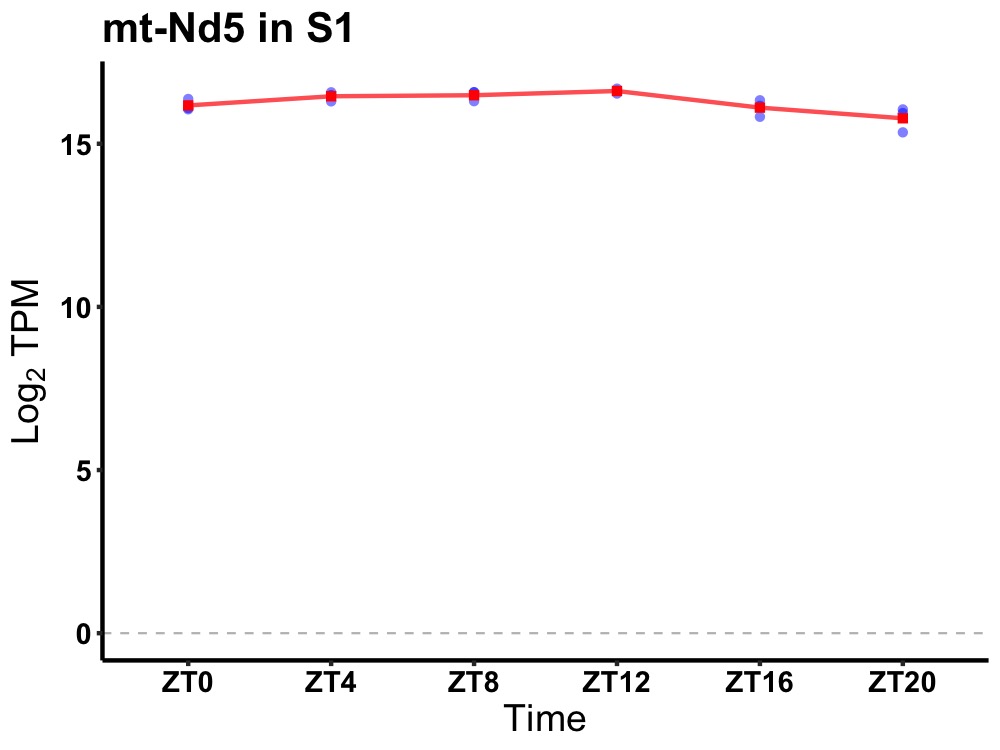 |
mitochondrially encoded NADH dehydrogenase 5 | 0.003 | 24 | 10 | 0.34 |
| ENSMUSG00000084259 | Rps8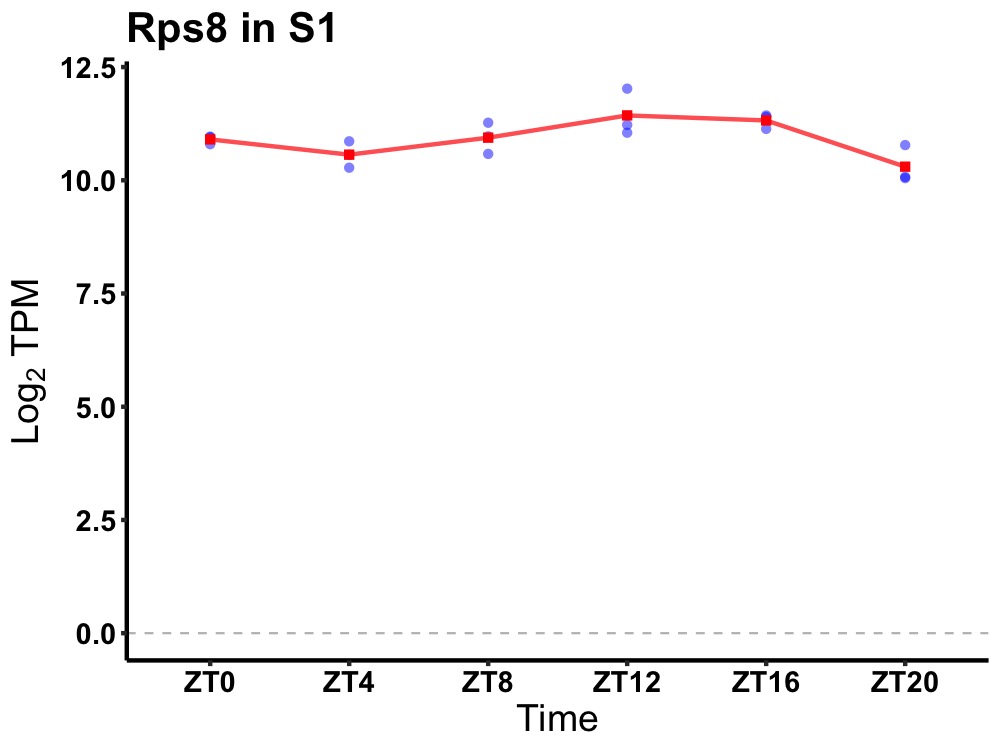 |
ribosomal protein S8 | 0.003 | 20 | 14 | 0.34 |
| ENSMUSG00000084844 | Trmt112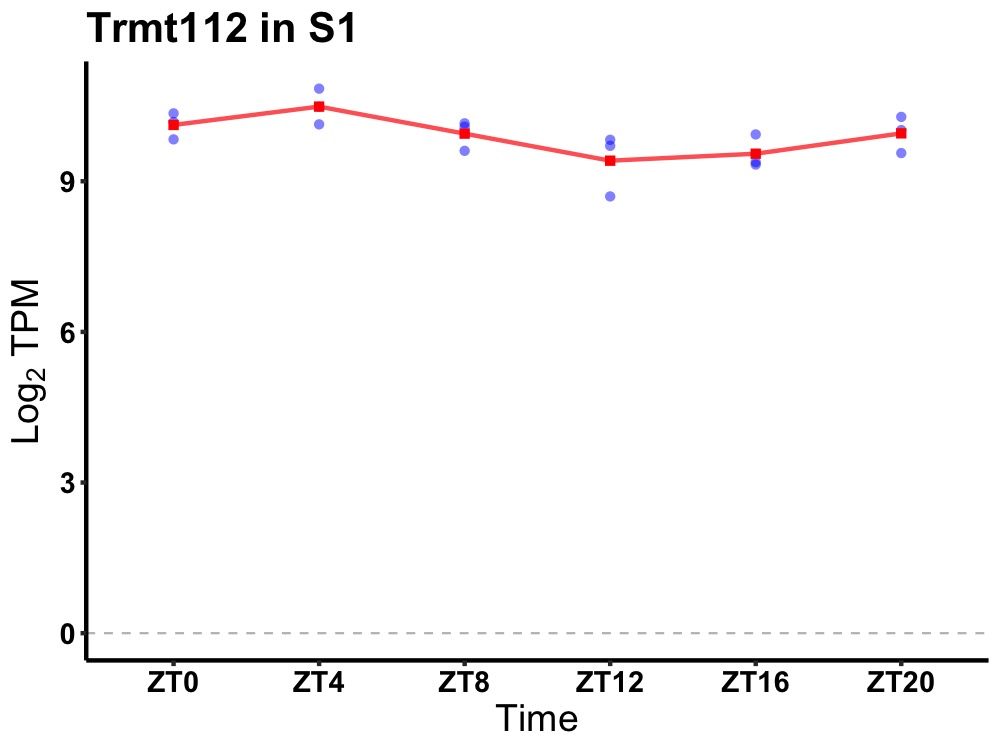 |
tRNA methyltransferase 11-2 | 0.014 | 24 | 4 | 0.34 |
| ENSMUSG00000019947 | Calr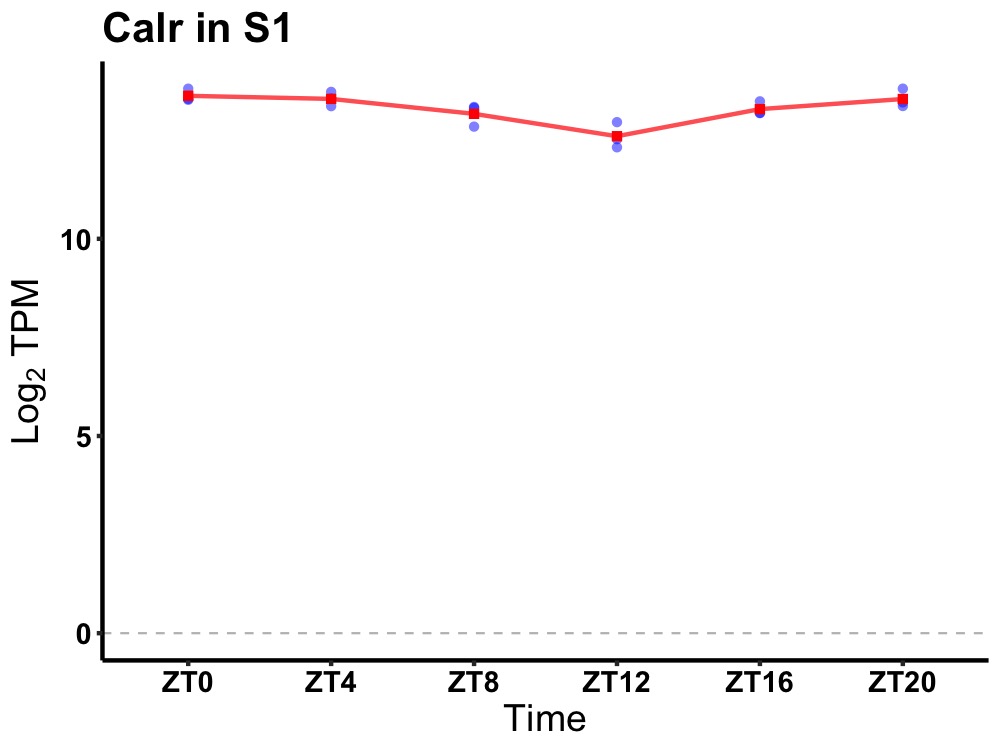 |
calreticulin | 0.001 | 24 | 0 | 0.34 |
| ENSMUSG00000053329 | Stard3nl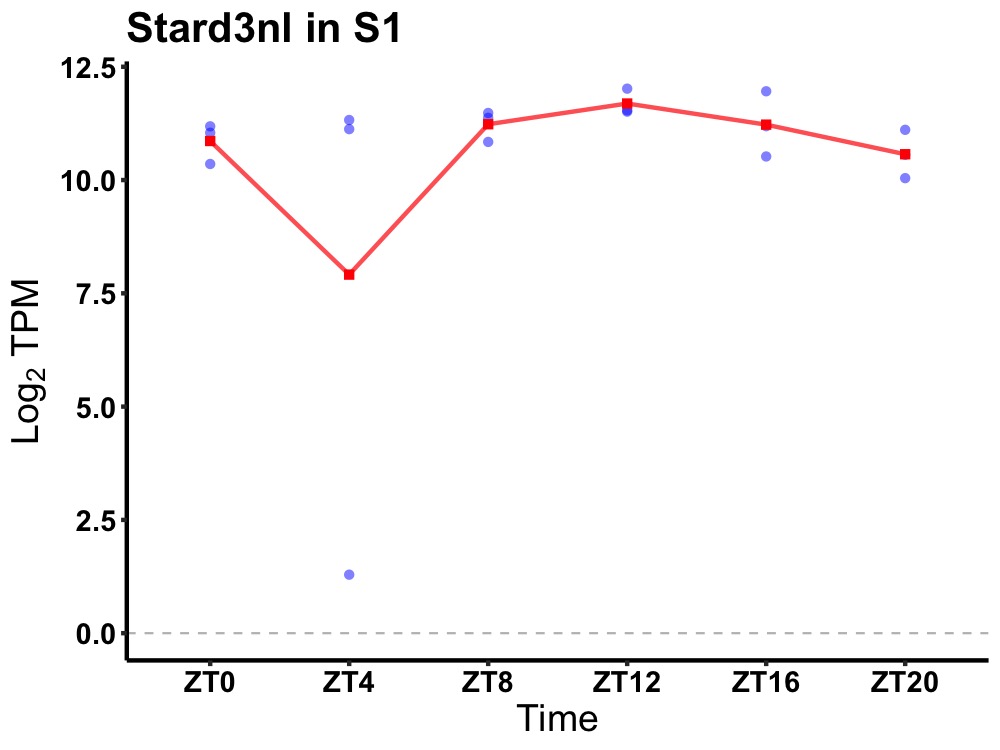 |
STARD3 N-terminal like | 0.036 | 20 | 12 | 0.33 |
| ENSMUSG00000112605 | Fads3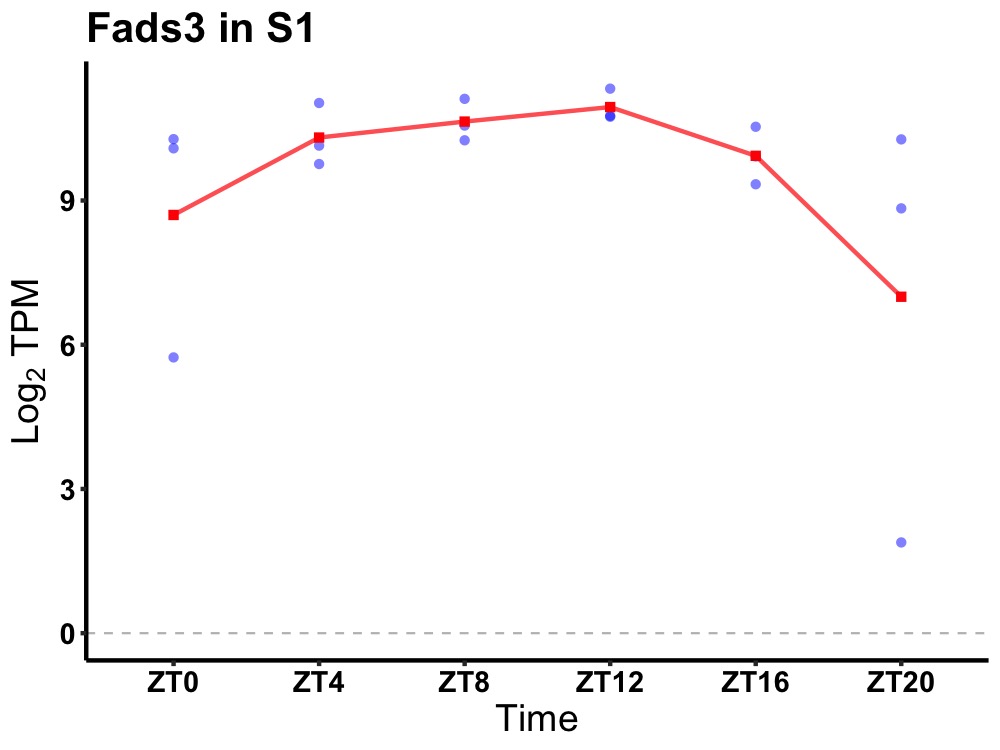 |
fatty acid desaturase 3 | 0.049 | 20 | 12 | 0.33 |
| ENSMUSG00000004364 | Elovl1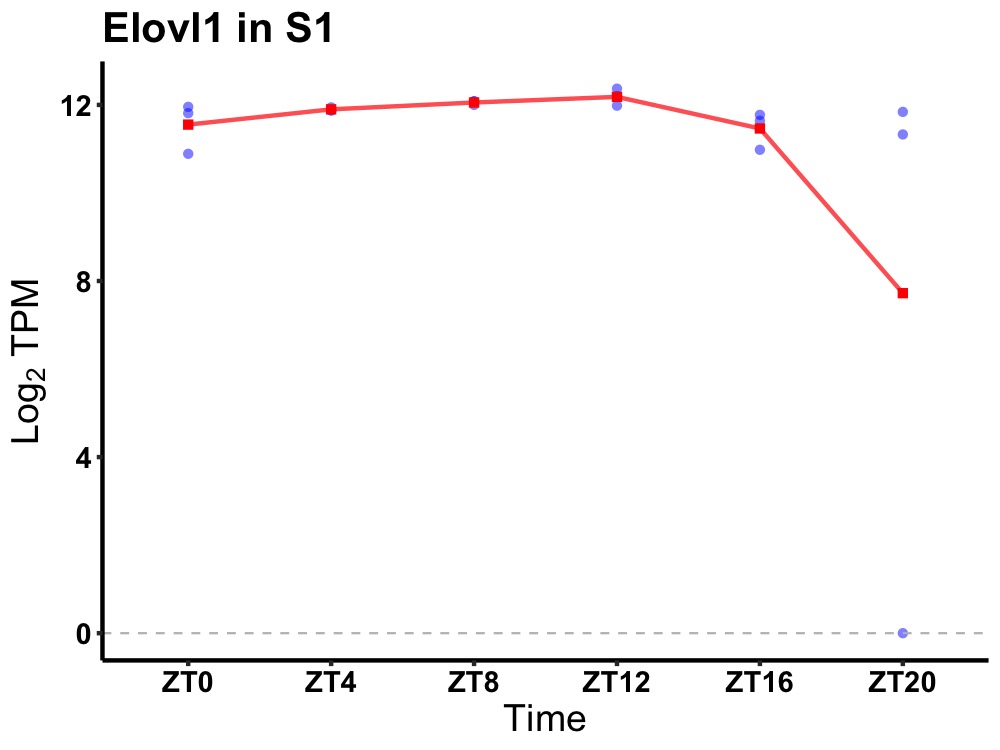 |
elongation of very long chain fatty acids (FEN1/Elo2, SUR4/Elo3, yeast)-like 1 | 0.005 | 24 | 10 | 0.33 |
| ENSMUSG00000068290 | Ywhaq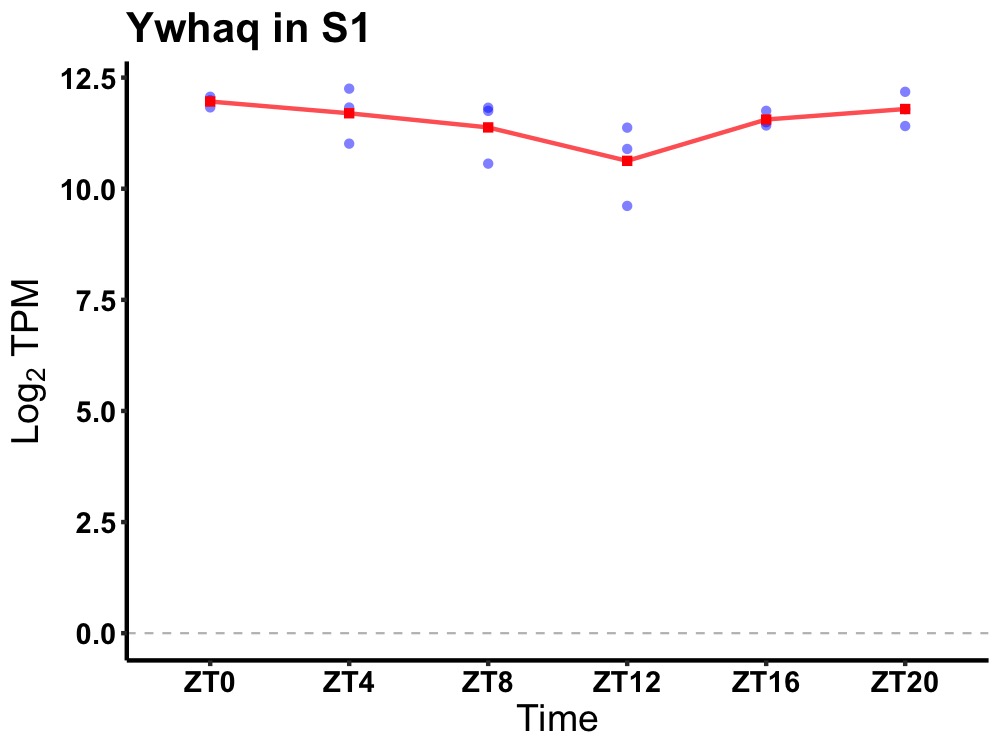 |
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein theta | 0.036 | 24 | 2 | 0.32 |
| ENSMUSG00000037772 | Gatc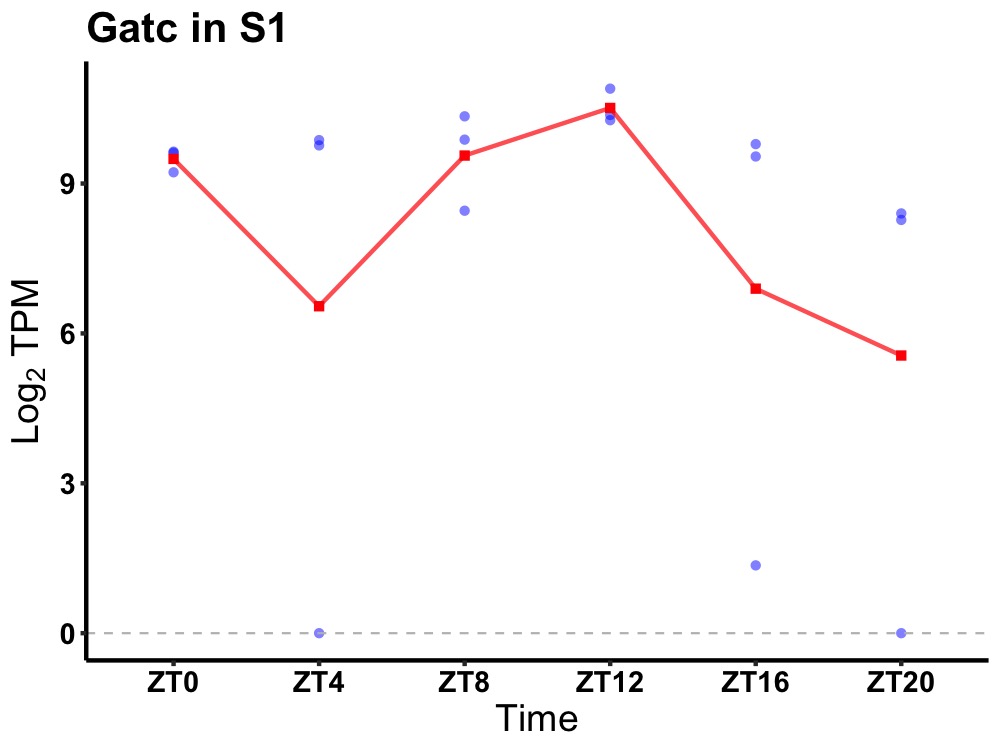 |
glutamyl-tRNA(Gln) amidotransferase subunit C, mitochondrial | 0.016 | 20 | 12 | 0.32 |
| ENSMUSG00000037686 | Eif5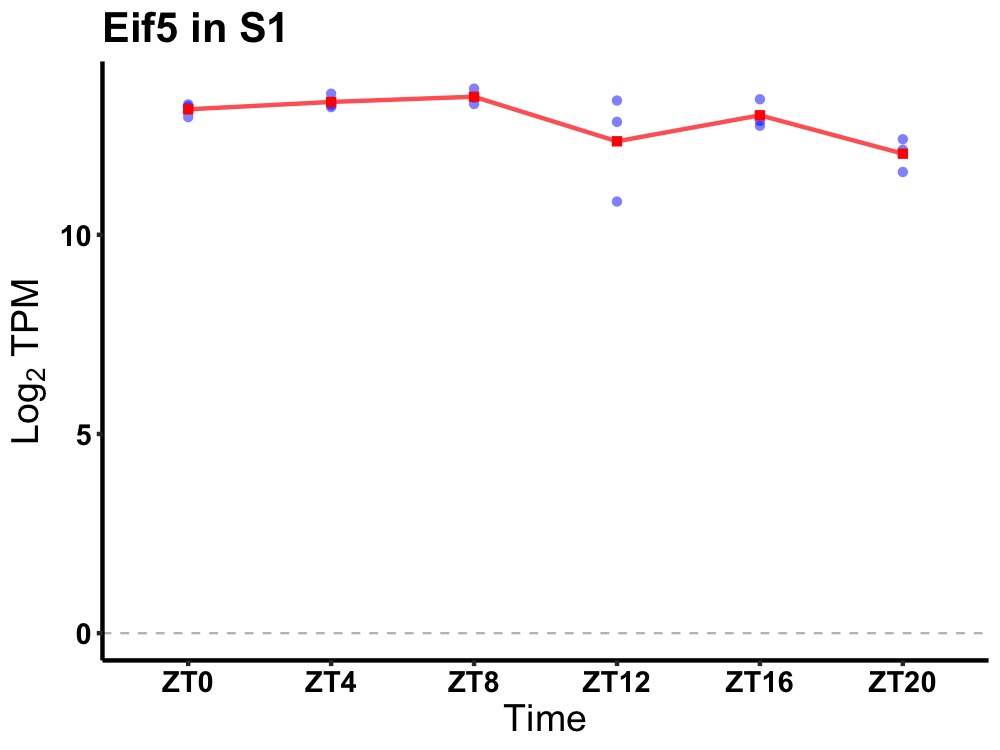 |
eukaryotic translation initiation factor 5 | 0.049 | 24 | 8 | 0.31 |
| ENSMUSG00000015575 | Cers2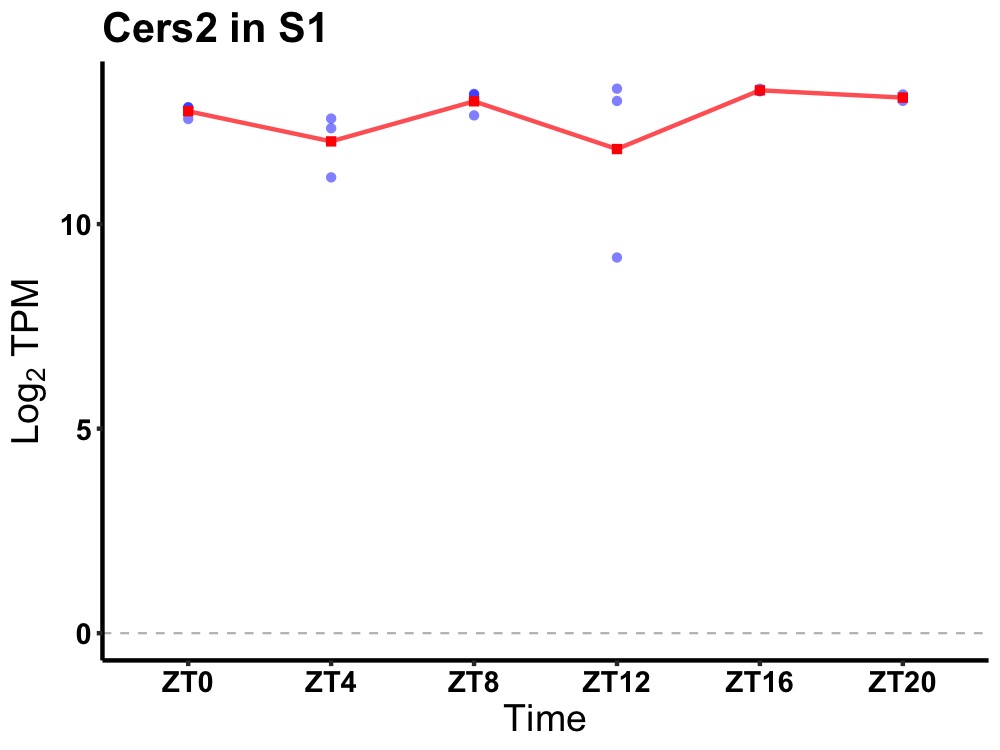 |
ceramide synthase 2 | 0.036 | 24 | 16 | 0.31 |
| ENSMUSG00000095512 | Picalm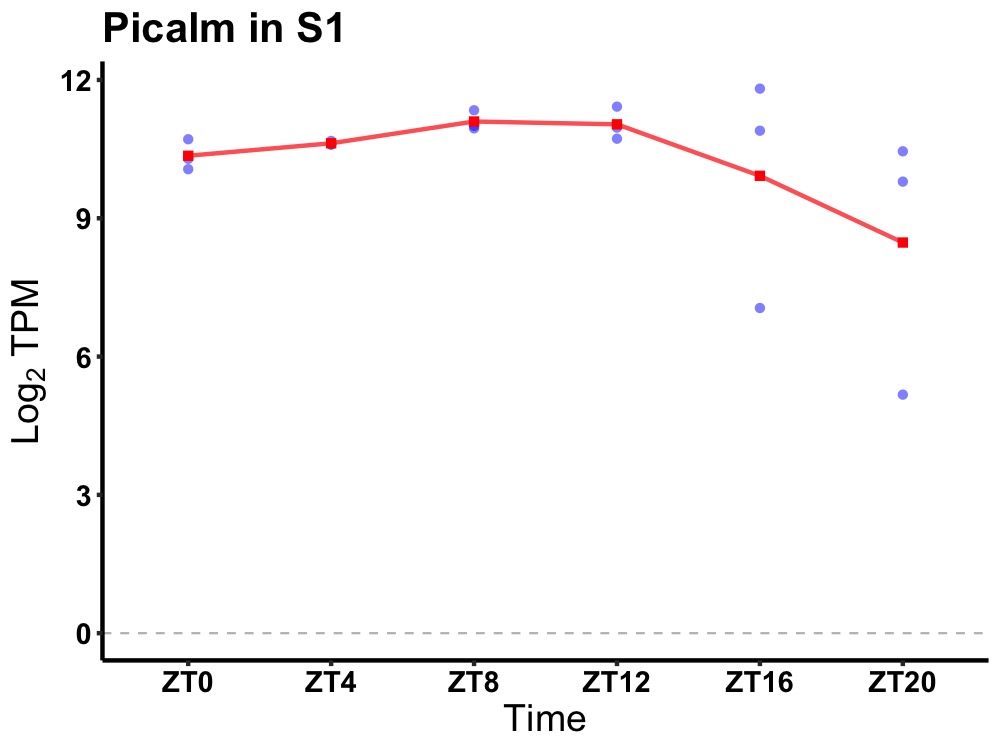 |
phosphatidylinositol binding clathrin assembly protein | 0.014 | 20 | 12 | 0.30 |
| ENSMUSG00000036323 | Snrpg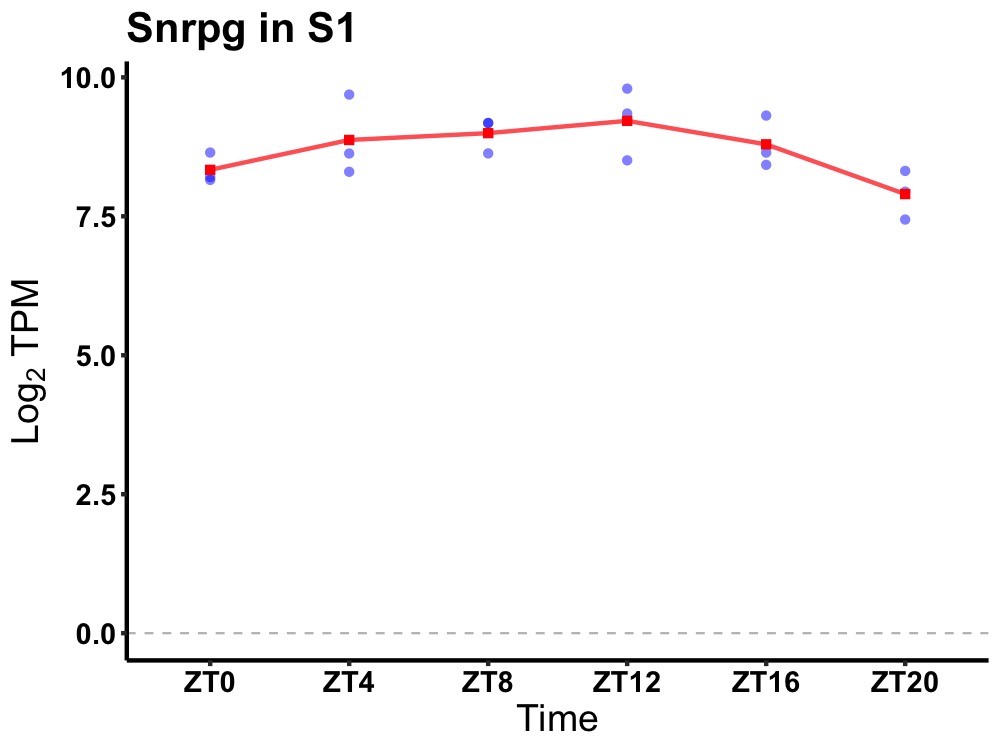 |
small nuclear ribonucleoprotein polypeptide G | 0.036 | 20 | 12 | 0.30 |
| ENSMUSG00000022185 | Hspa5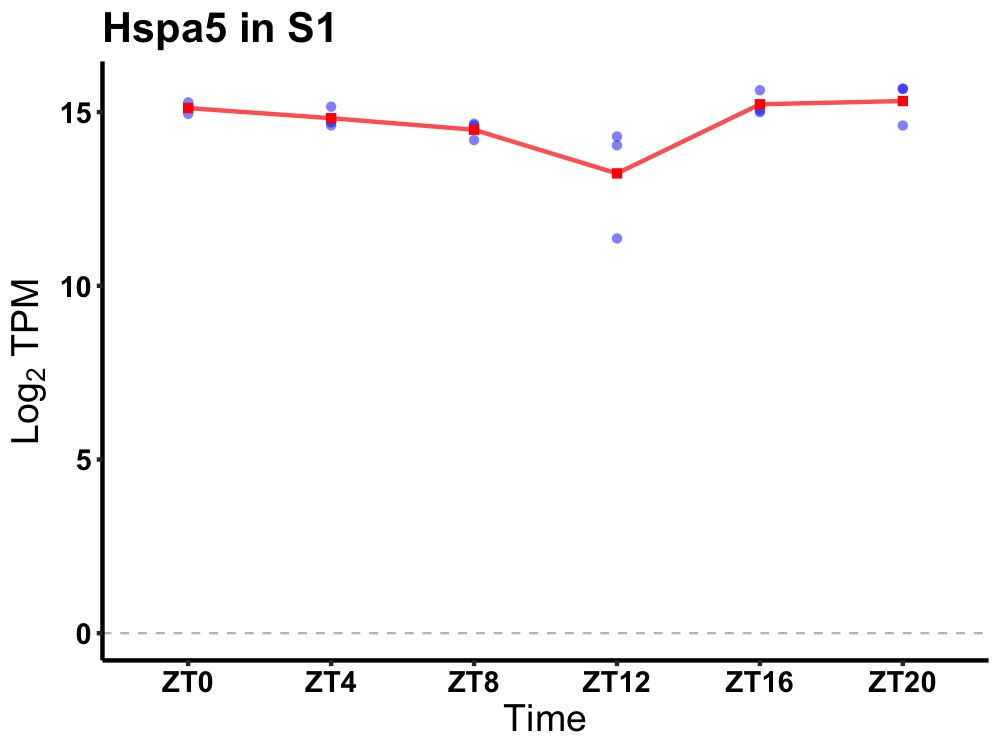 |
heat shock protein 5 | 0.036 | 20 | 2 | 0.30 |
| ENSMUSG00000003809 | Set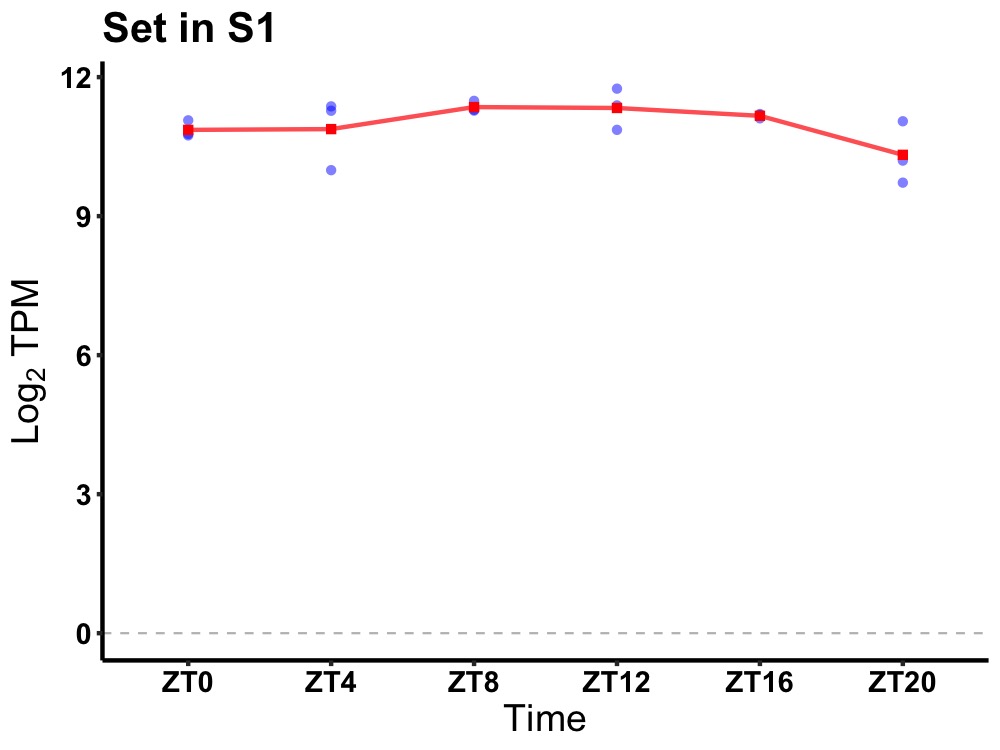 |
SET nuclear oncogene | 0.010 | 24 | 10 | 0.29 |
| ENSMUSG00000030861 | Hsp90ab1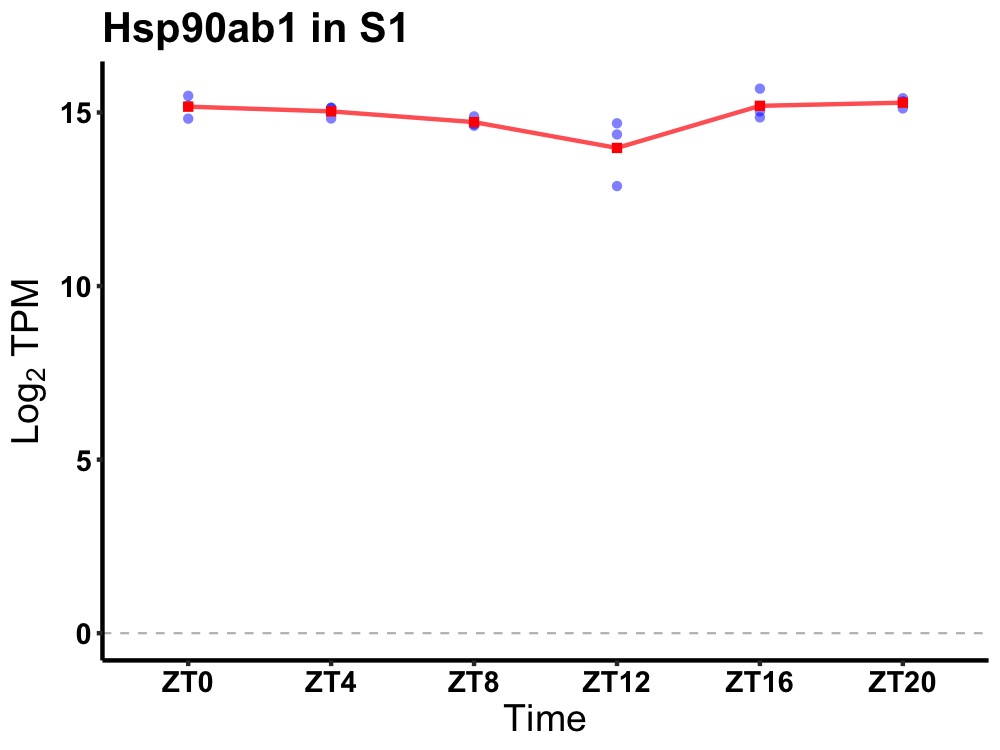 |
heat shock protein 90 alpha (cytosolic), class B member 1 | 0.027 | 20 | 2 | 0.28 |
| ENSMUSG00000042558 | Selenbp1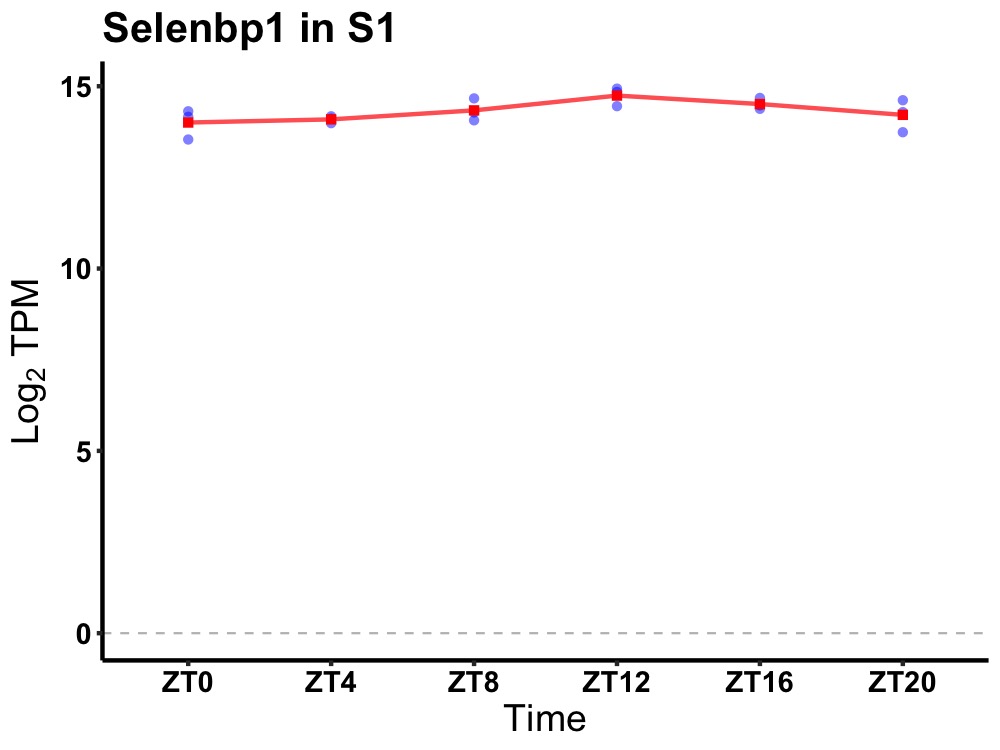 |
selenium binding protein 1 | 0.036 | 24 | 14 | 0.28 |
| ENSMUSG00000084215 | Gm11814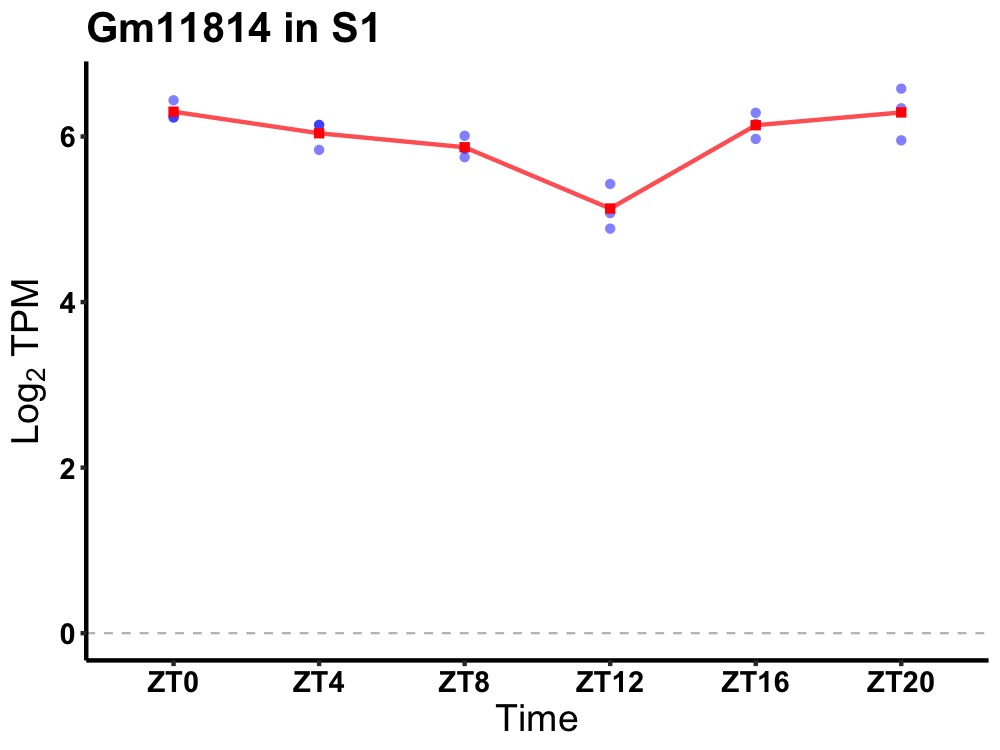 |
predicted gene 11814 | 0.005 | 20 | 2 | 0.28 |
| ENSMUSG00000023075 | Ddx5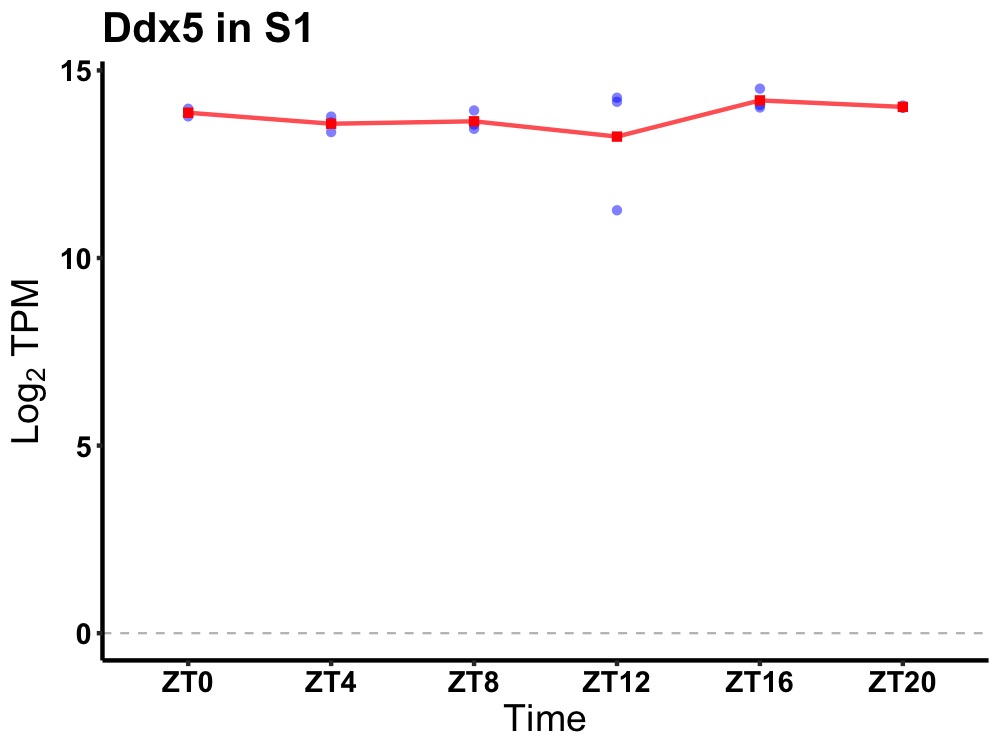 |
DEAD (Asp-Glu-Ala-Asp) box polypeptide 5 | 0.019 | 24 | 18 | 0.27 |
| ENSMUSG00000083813 | Gm15502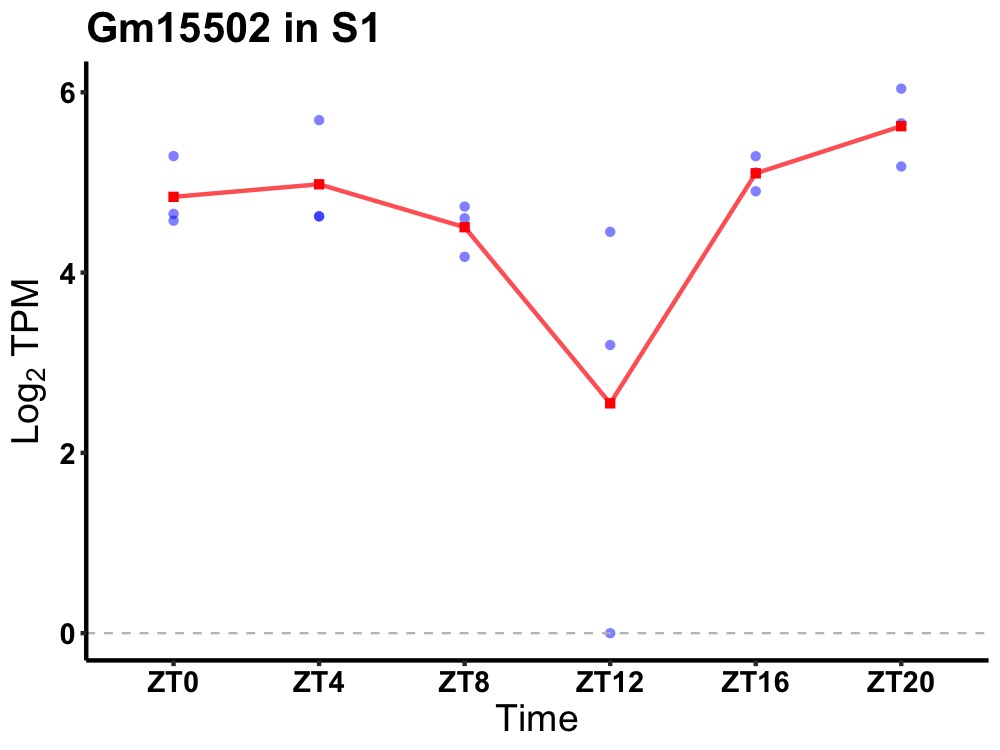 |
predicted gene 15502 | 0.027 | 20 | 2 | 0.27 |
| ENSMUSG00000094683 | Atp5g3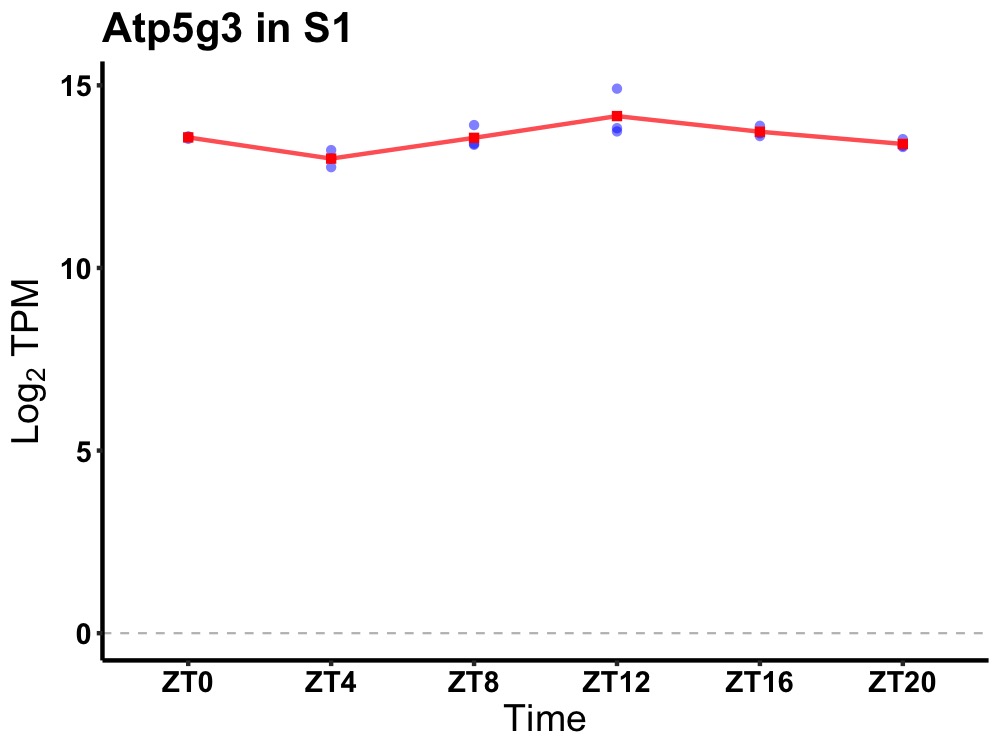 |
ATP synthase, H+ transporting, mitochondrial F0 complex, subunit C3 (subunit 9) | 0.000 | 20 | 14 | 0.27 |
| ENSMUSG00000014313 | Agps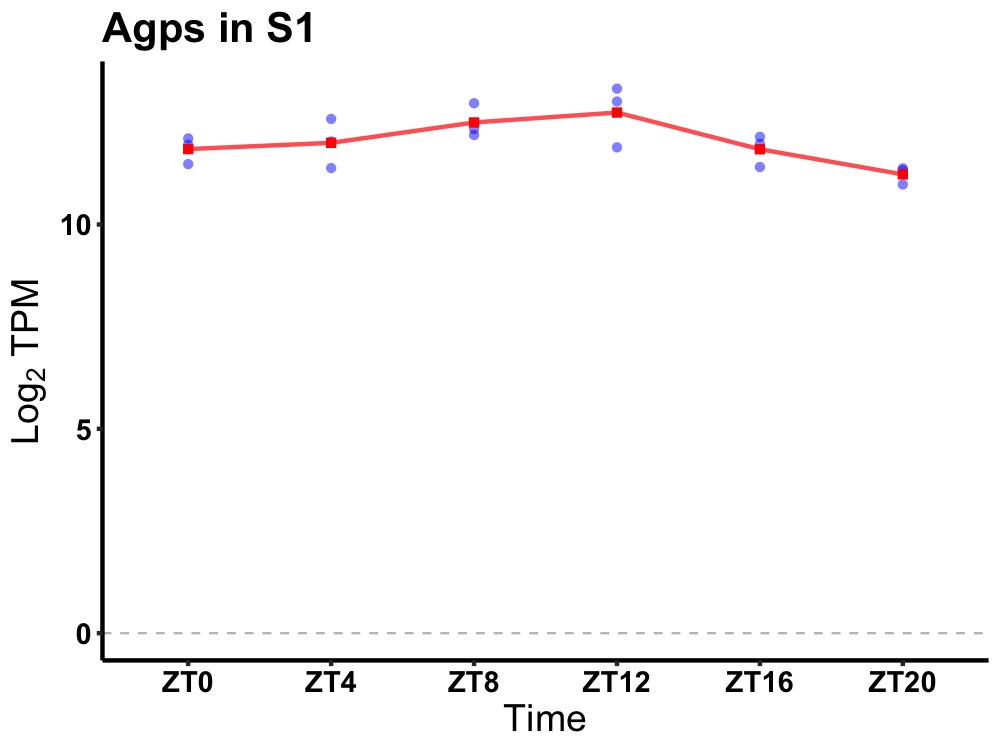 |
alkylglycerone phosphate synthase | 0.010 | 20 | 12 | 0.27 |
| ENSMUSG00000083011 | Decr2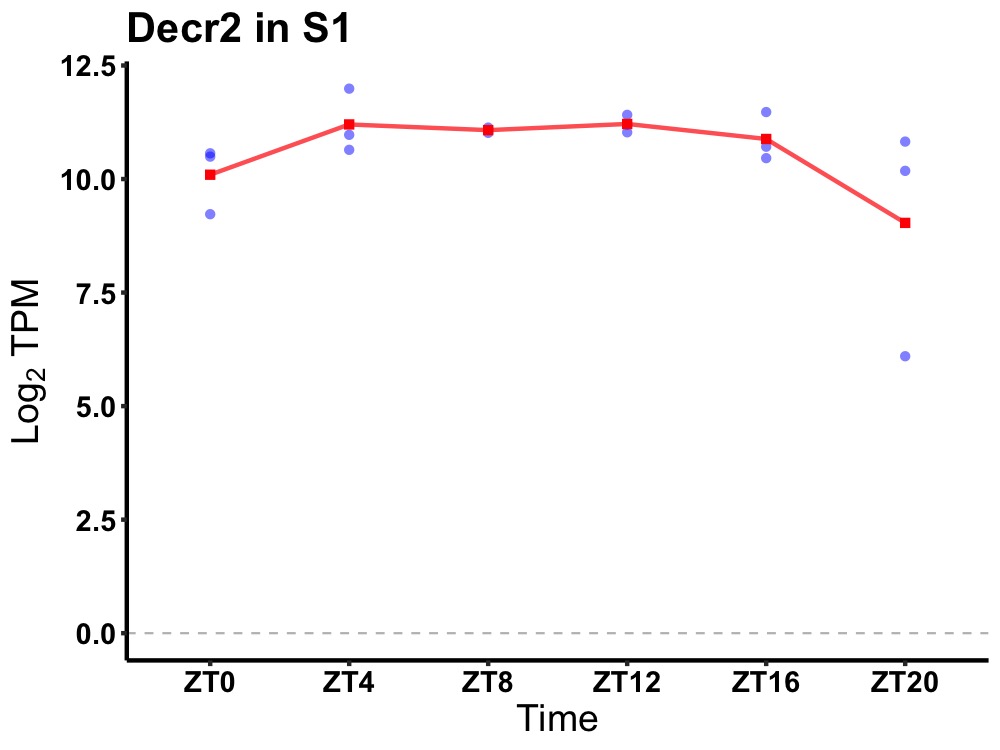 |
2-4-dienoyl-Coenzyme A reductase 2, peroxisomal | 0.049 | 20 | 12 | 0.27 |
| ENSMUSG00000082632 | Gm13365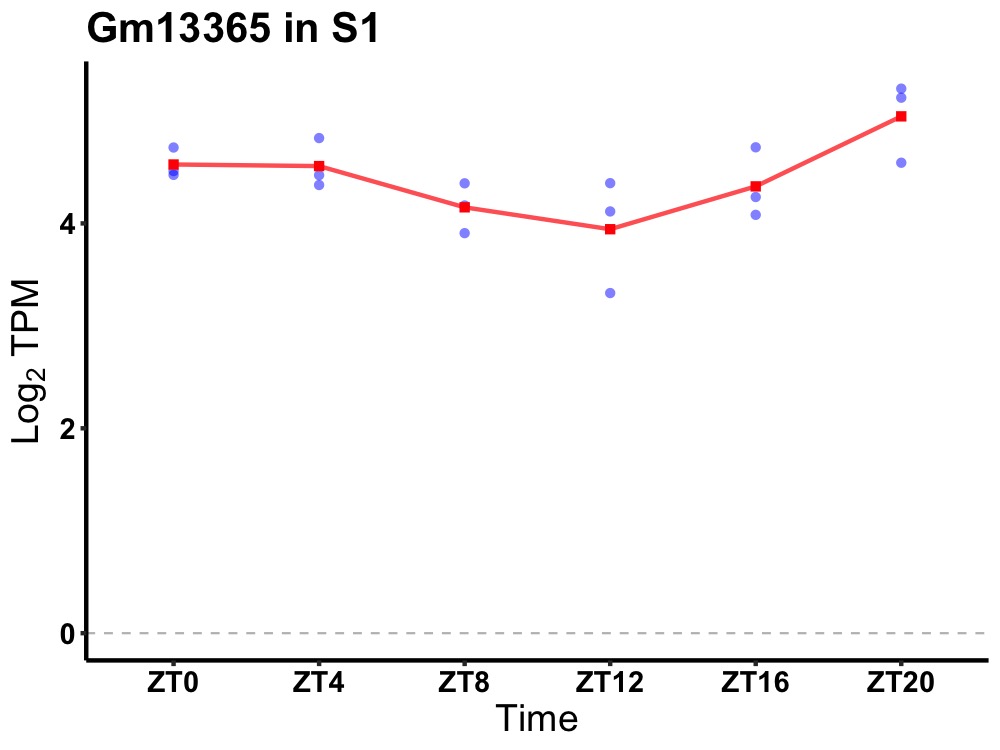 |
predicted gene 13365 | 0.007 | 20 | 2 | 0.26 |
| ENSMUSG00000015357 | Susd3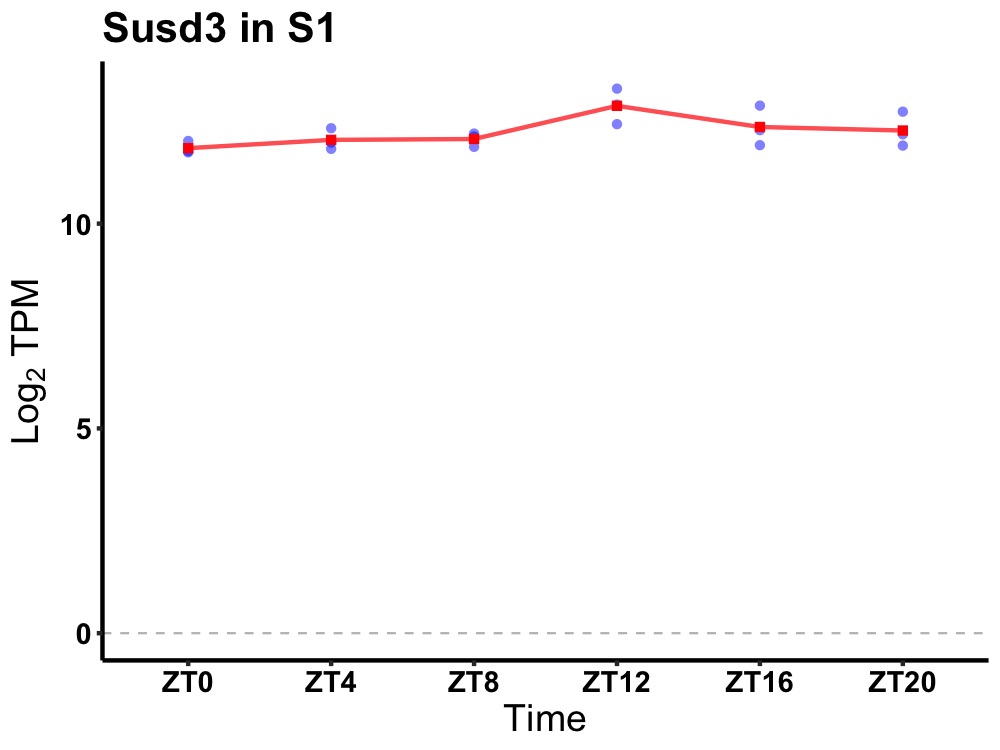 |
sushi domain containing 3 | 0.036 | 24 | 14 | 0.26 |
| ENSMUSG00000083429 | Slc46a1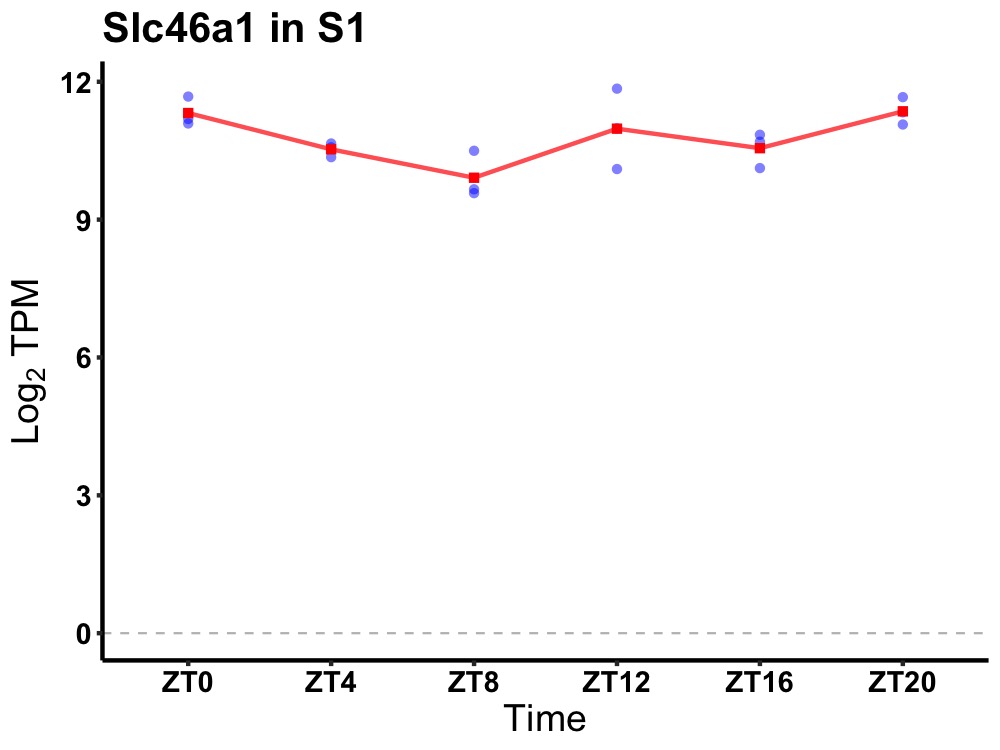 |
solute carrier family 46, member 1 | 0.036 | 20 | 0 | 0.26 |
| ENSMUSG00000082536 | Paqr9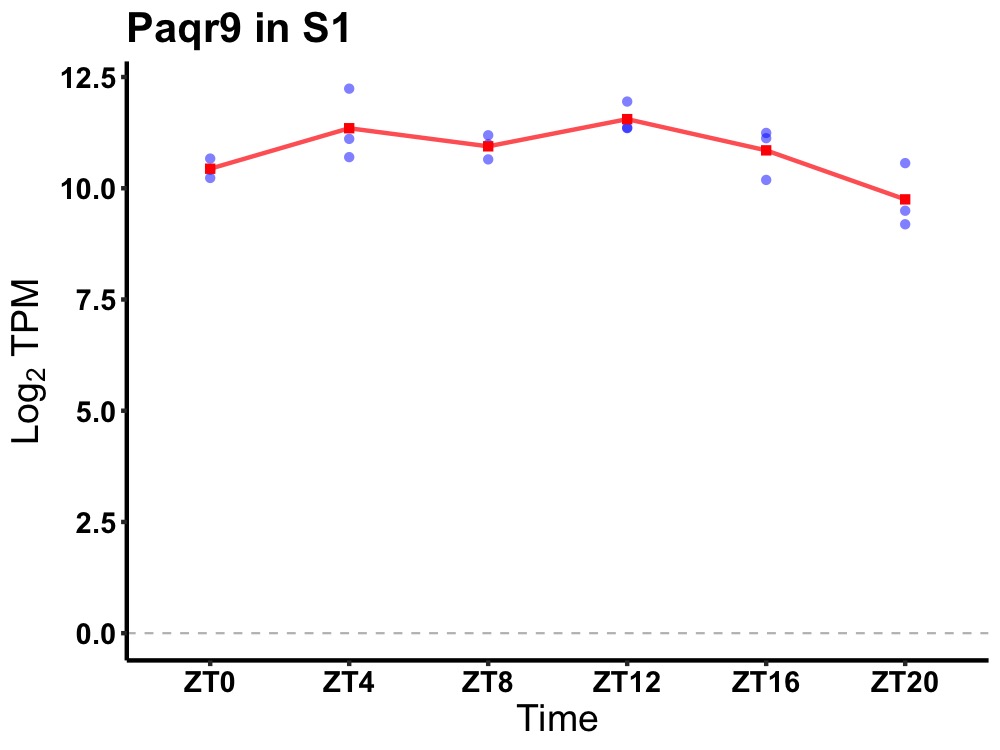 |
progestin and adipoQ receptor family member IX | 0.019 | 20 | 12 | 0.26 |
| ENSMUSG00000081453 | Gm6767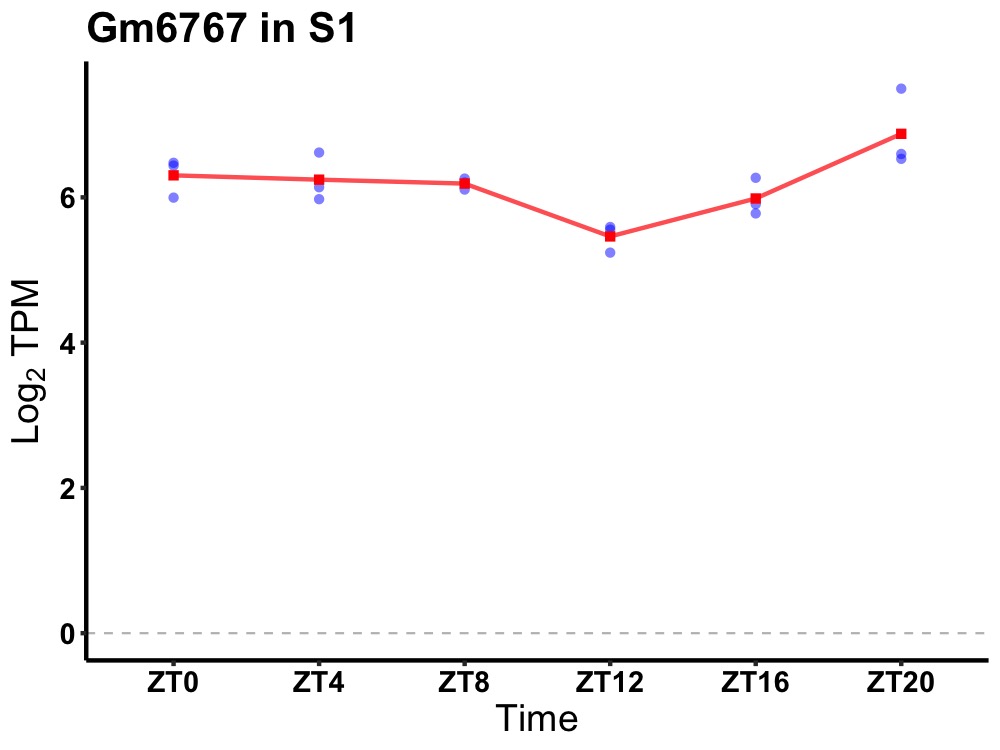 |
predicted gene 6767 | 0.007 | 20 | 2 | 0.25 |
| ENSMUSG00000068328 | Sod1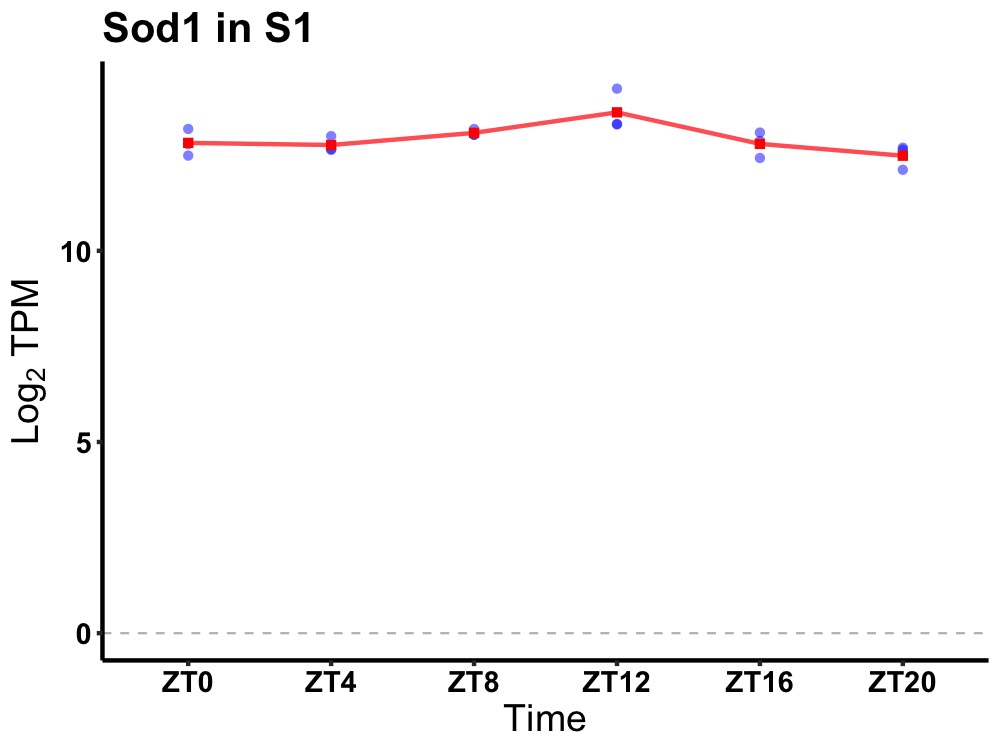 |
superoxide dismutase 1, soluble | 0.019 | 20 | 12 | 0.25 |
| ENSMUSG00000033308 | Mrps18a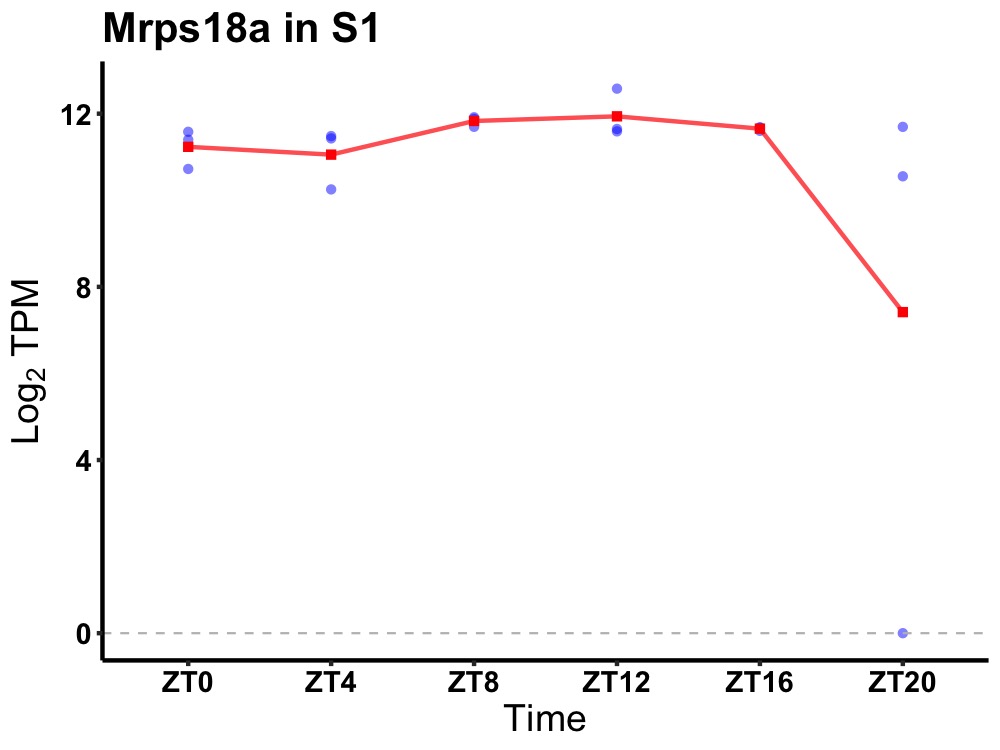 |
mitochondrial ribosomal protein S18A | 0.036 | 20 | 12 | 0.25 |
| ENSMUSG00000019433 | Arpc3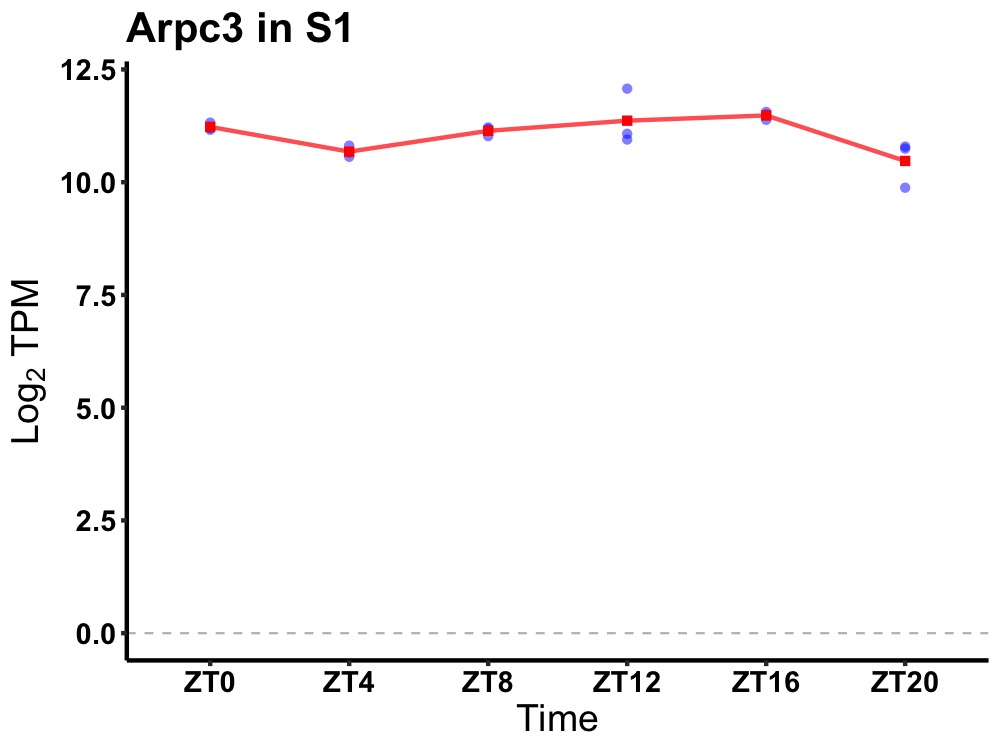 |
actin related protein 2/3 complex, subunit 3 | 0.036 | 20 | 14 | 0.25 |
| ENSMUSG00000021669 | Ndufc1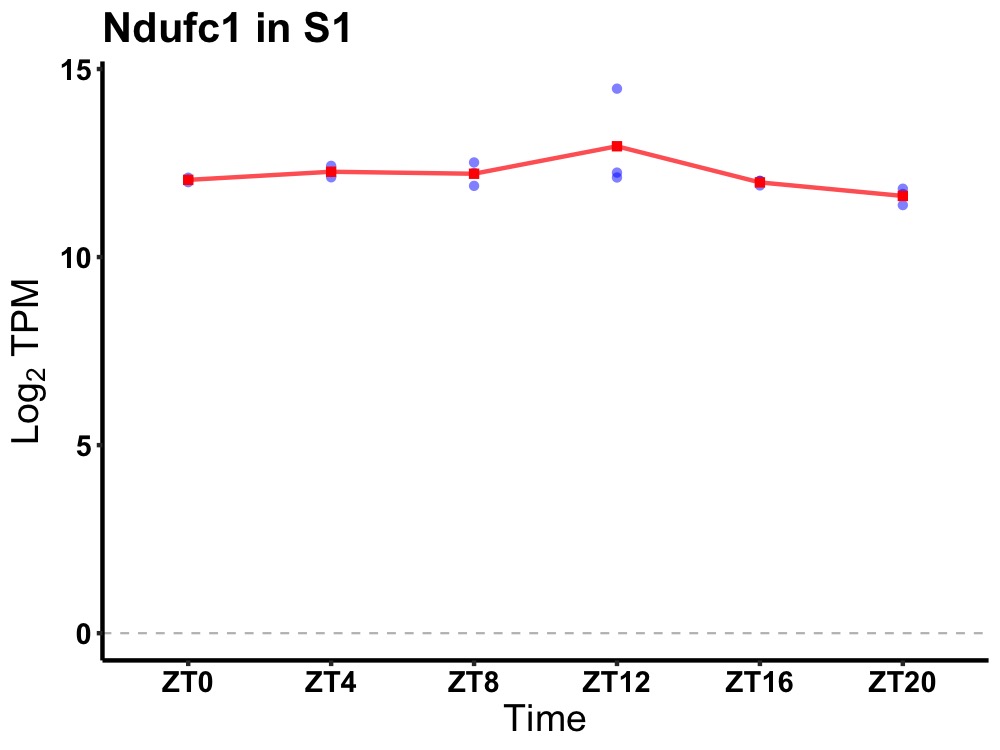 |
NADH:ubiquinone oxidoreductase subunit C1 | 0.007 | 24 | 8 | 0.24 |
| ENSMUSG00000021681 | Tmem174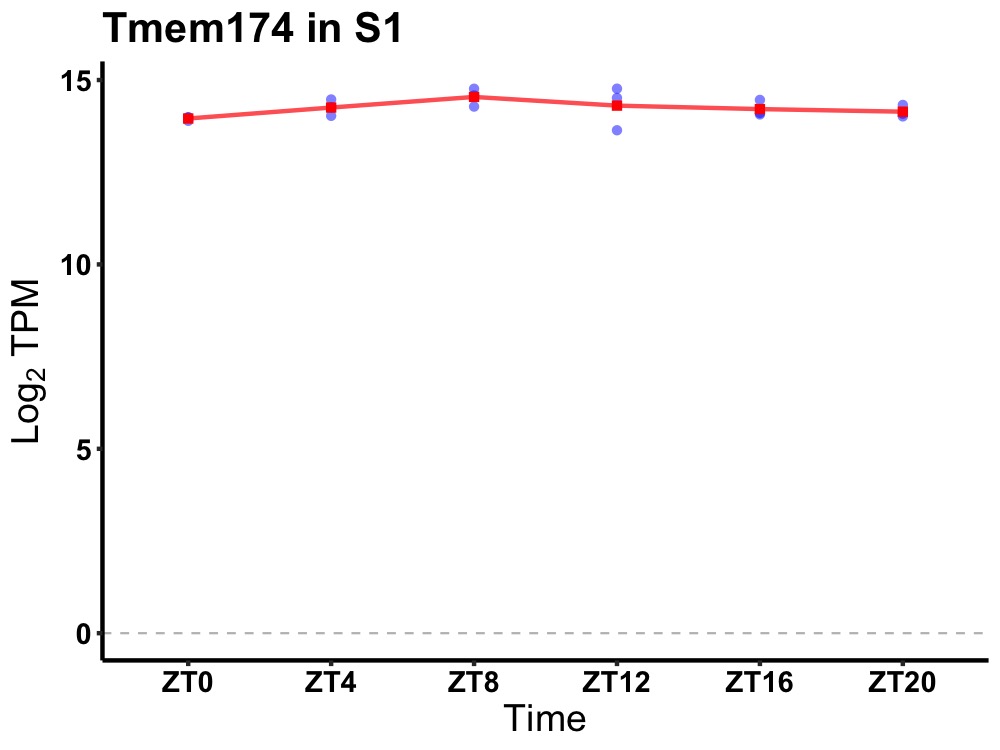 |
transmembrane protein 174 | 0.049 | 20 | 10 | 0.24 |
| ENSMUSG00000115160 | Mfn1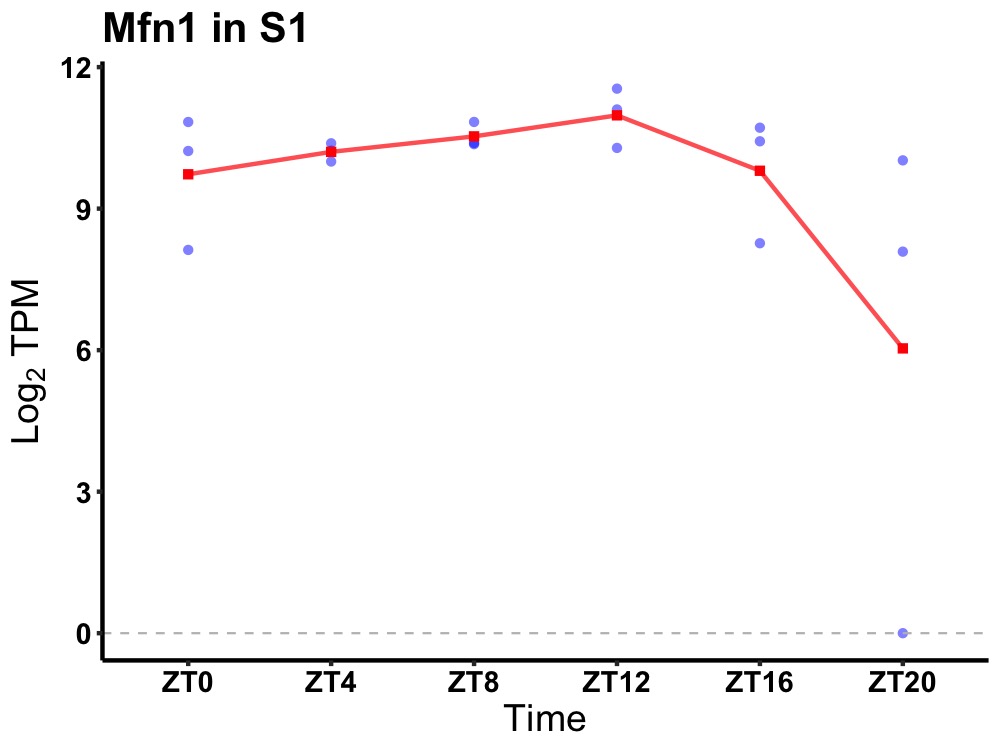 |
mitofusin 1 | 0.036 | 20 | 12 | 0.24 |
| ENSMUSG00000032560 | Hyi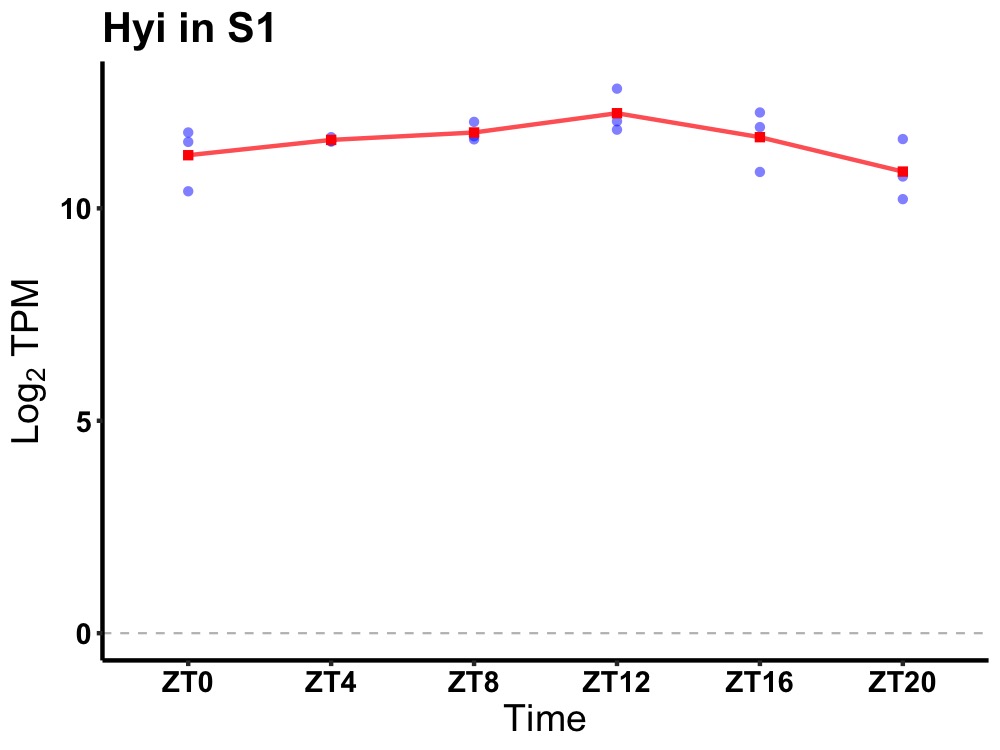 |
hydroxypyruvate isomerase (putative) | 0.027 | 20 | 12 | 0.24 |
| ENSMUSG00000100078 | Gm6170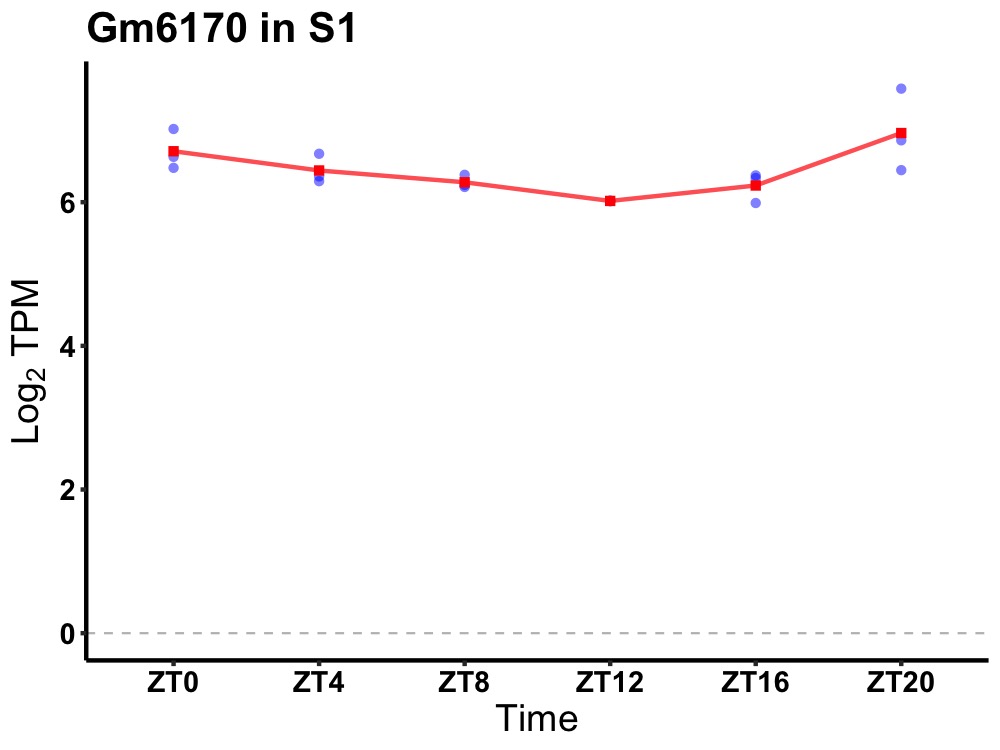 |
predicted gene 6170 | 0.002 | 20 | 2 | 0.24 |
| ENSMUSG00000117875 | AC124512.1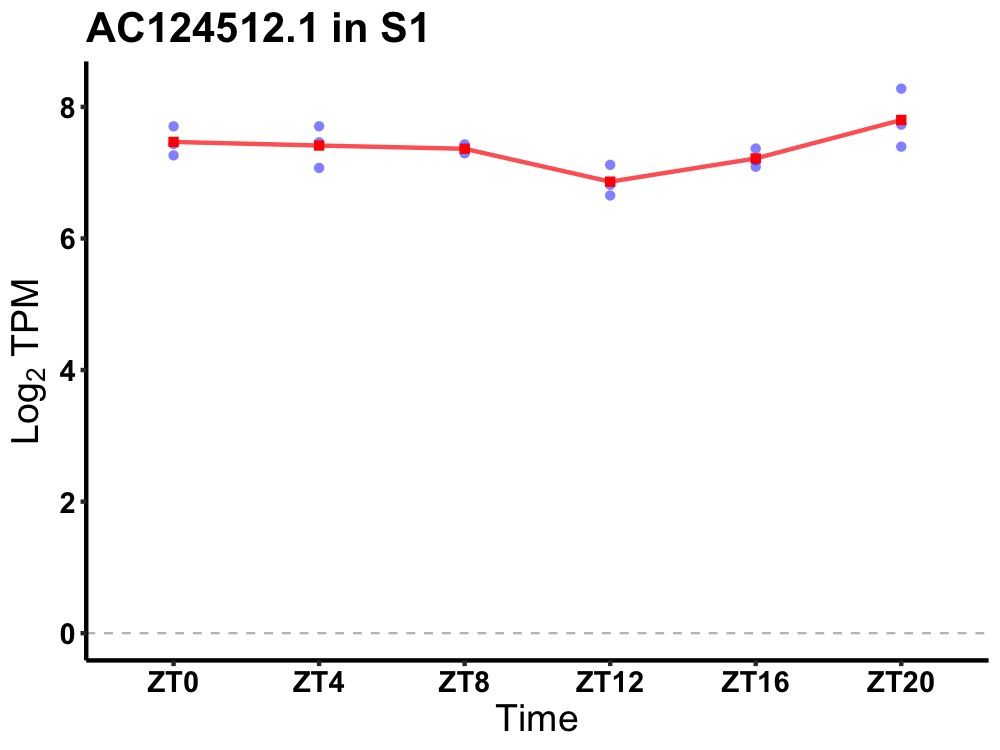 |
none | 0.036 | 20 | 4 | 0.23 |
| ENSMUSG00000037149 | Bckdk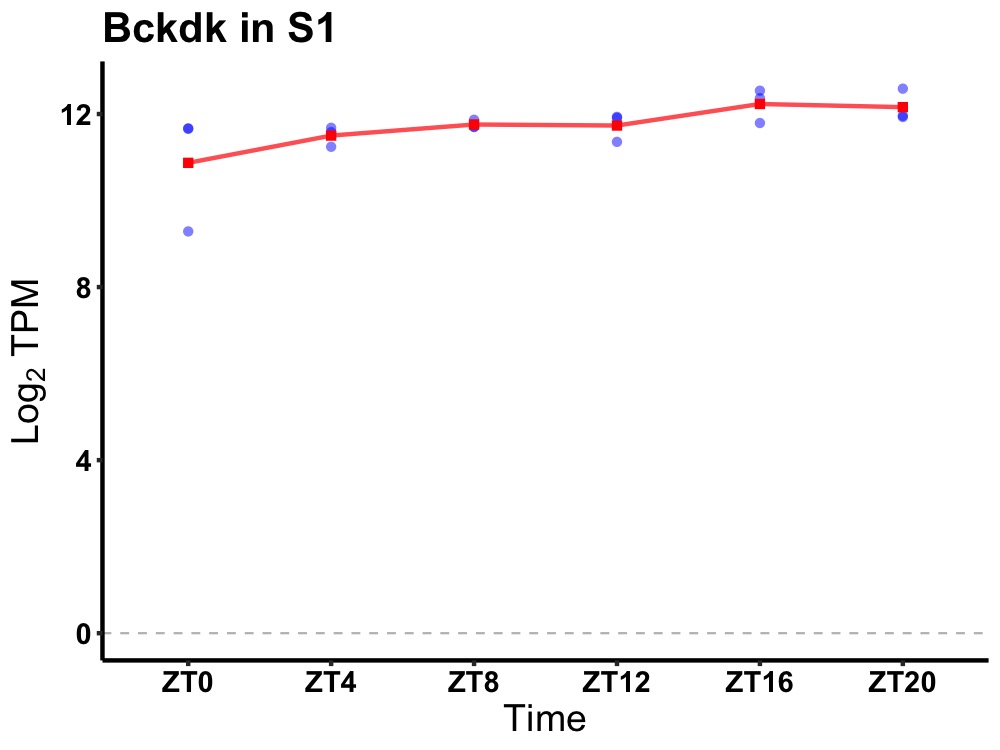 |
branched chain ketoacid dehydrogenase kinase | 0.036 | 24 | 16 | 0.23 |
| ENSMUSG00000084957 | Mpp1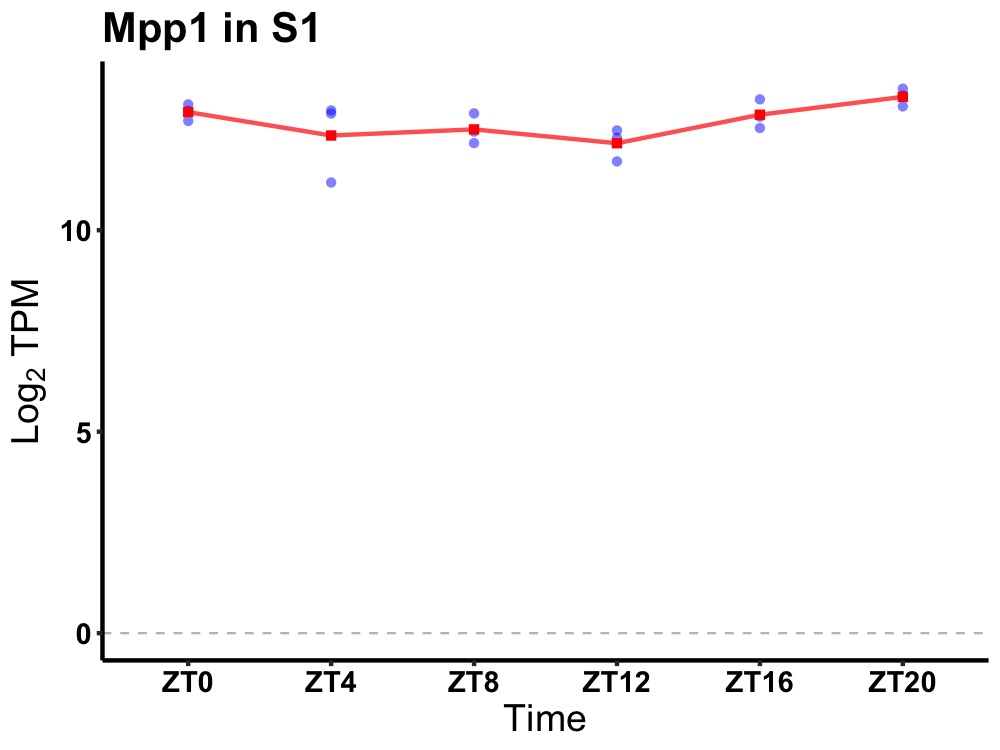 |
membrane protein, palmitoylated | 0.027 | 20 | 2 | 0.23 |
| ENSMUSG00000115070 | Bcas2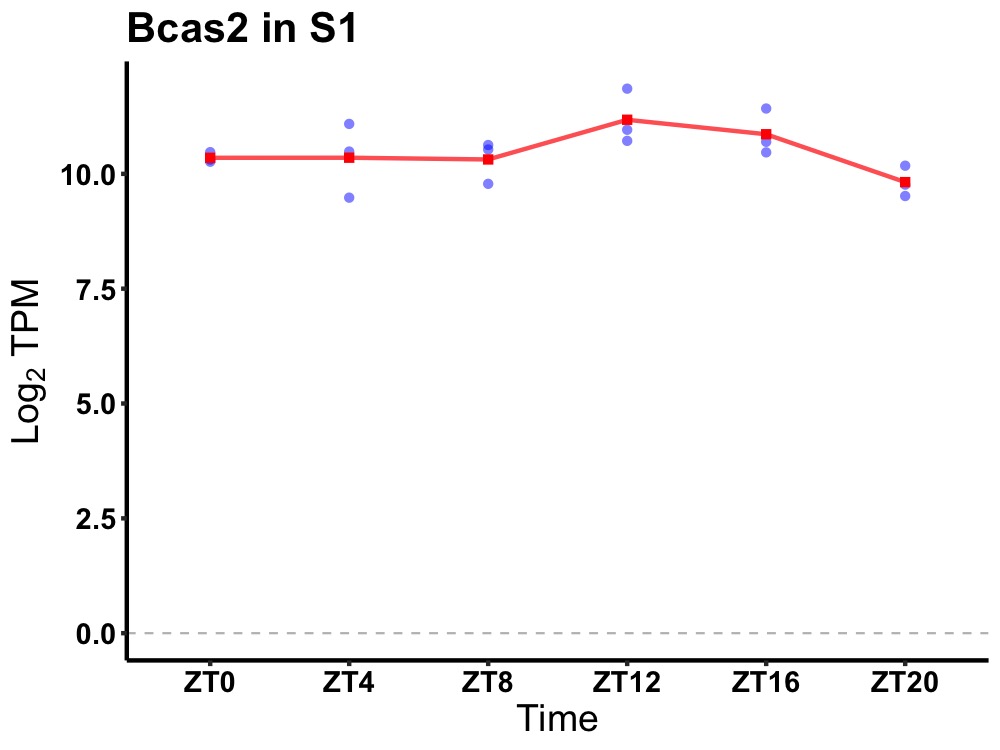 |
breast carcinoma amplified sequence 2 | 0.049 | 20 | 12 | 0.23 |
| ENSMUSG00000021226 | Slc47a1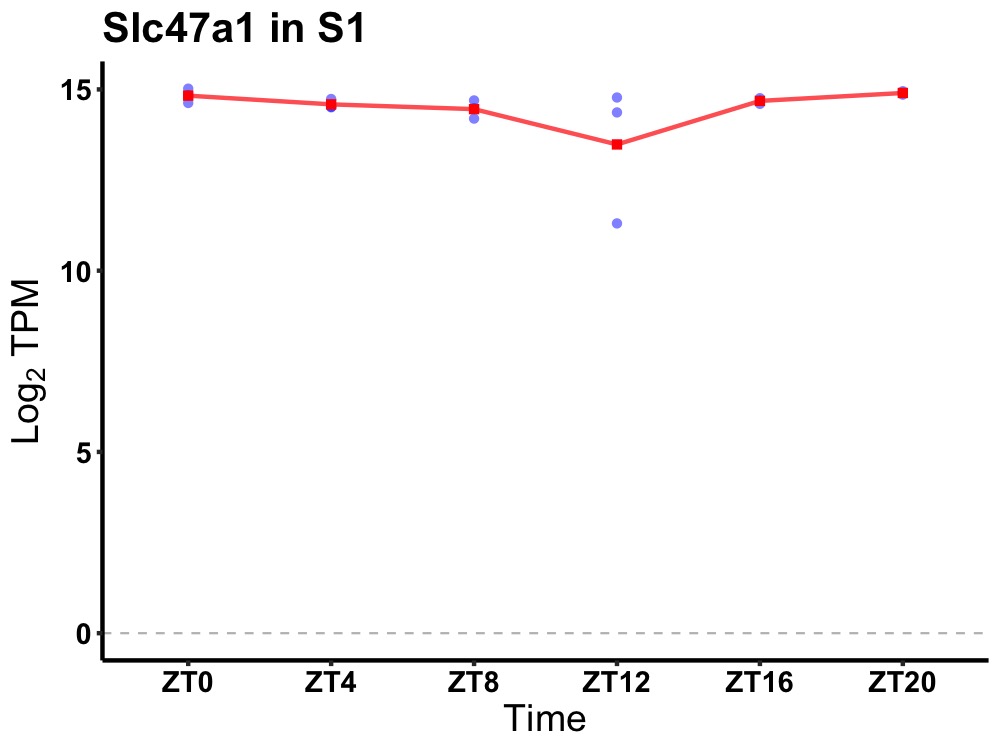 |
solute carrier family 47, member 1 | 0.014 | 24 | 22 | 0.22 |
| ENSMUSG00000107596 | Hmgn2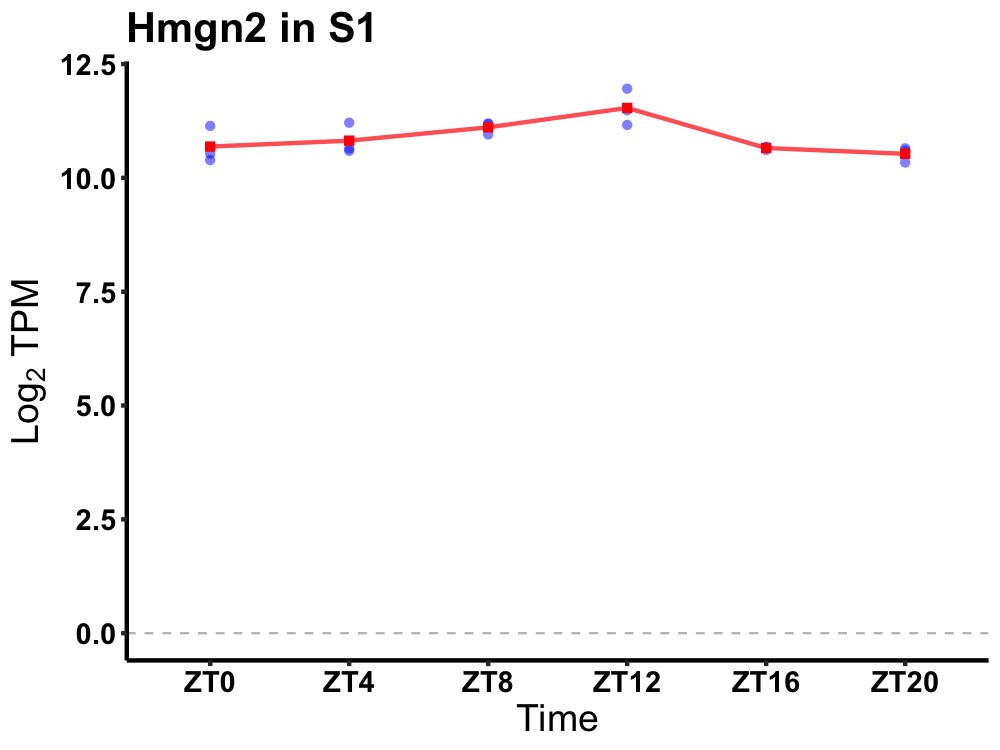 |
high mobility group nucleosomal binding domain 2 | 0.010 | 20 | 12 | 0.22 |
| ENSMUSG00000028973 | Acy3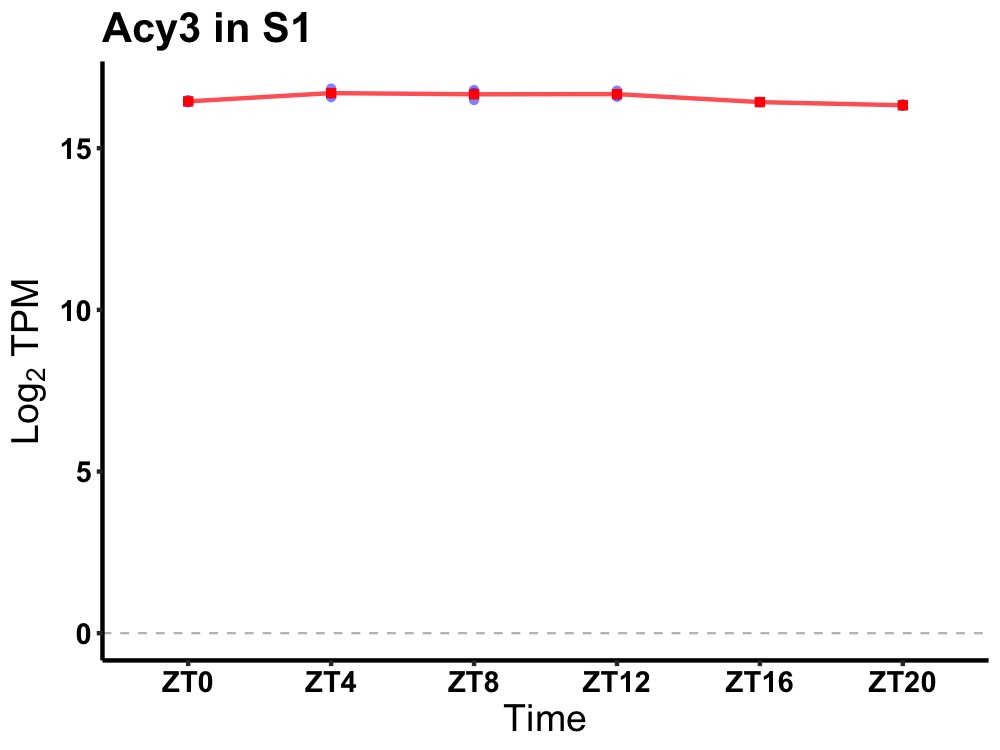 |
aspartoacylase (aminoacylase) 3 | 0.000 | 24 | 8 | 0.20 |
| ENSMUSG00000066809 | Coa6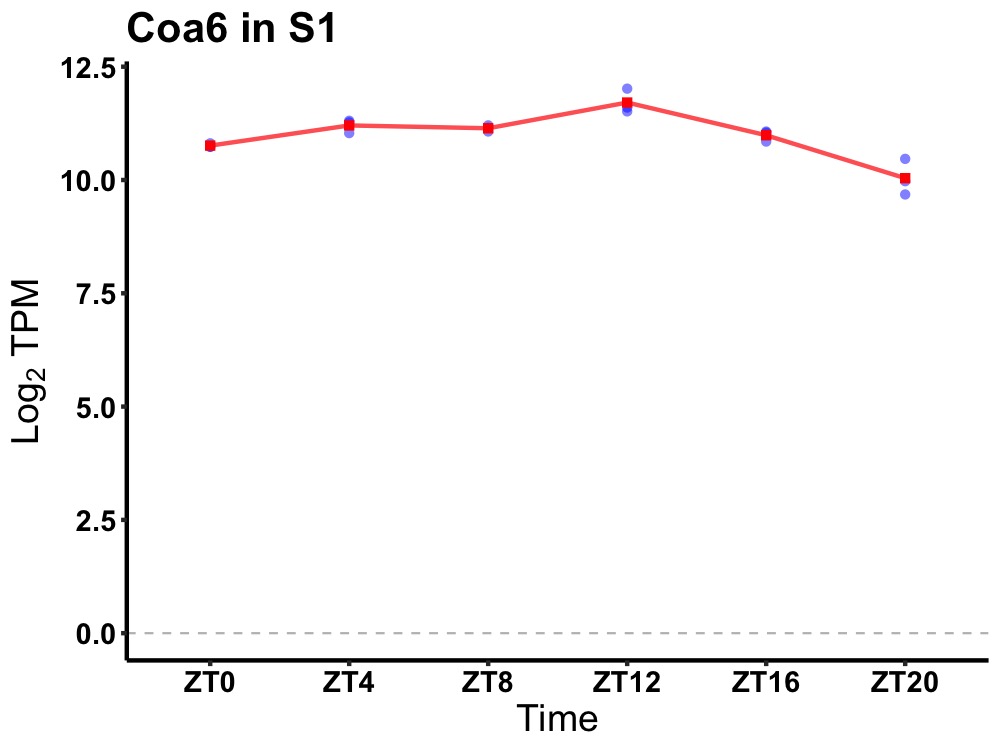 |
cytochrome c oxidase assembly factor 6 | 0.000 | 20 | 12 | 0.18 |
| ENSMUSG00000032437 | Gm13456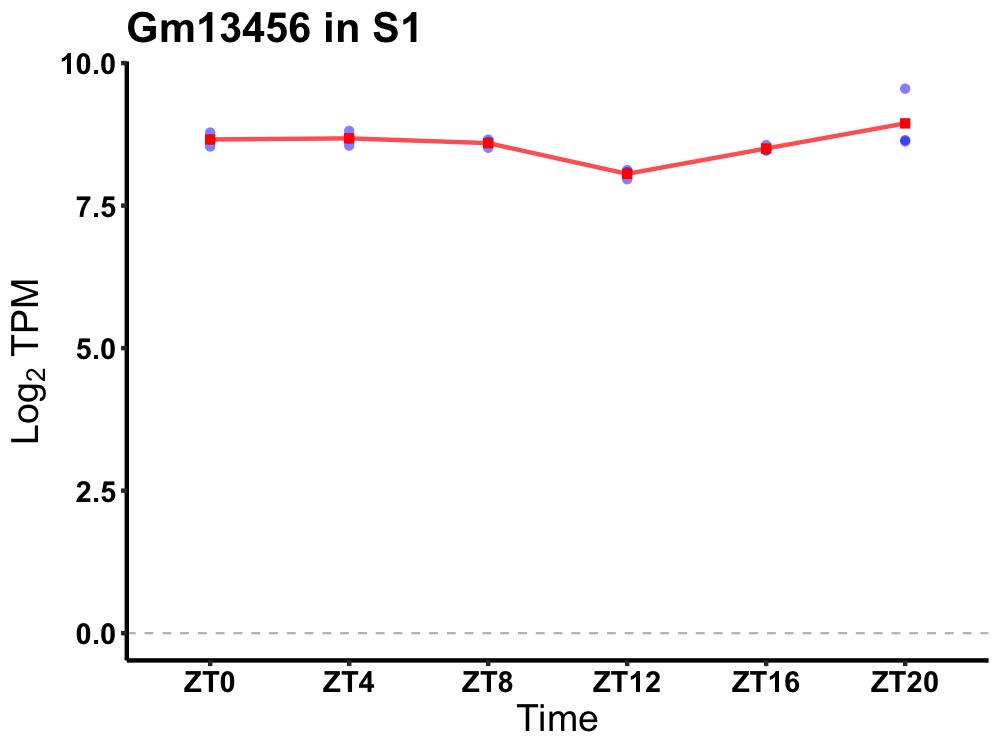 |
predicted gene 13456 | 0.003 | 20 | 4 | 0.18 |
| ENSMUSG00000086507 | Mpc2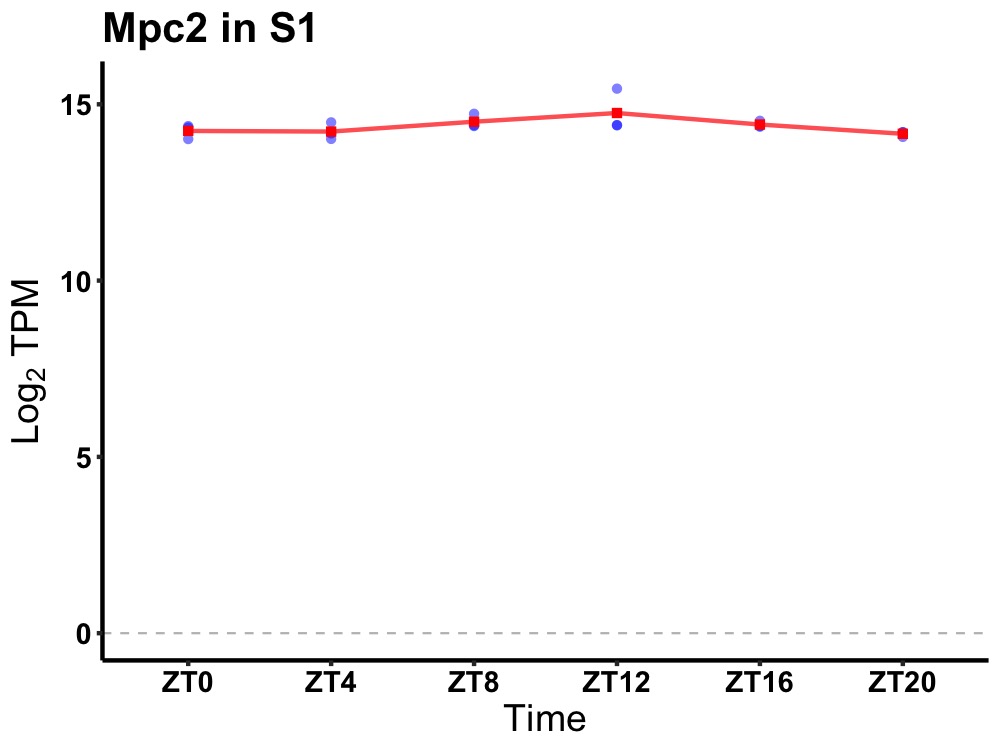 |
mitochondrial pyruvate carrier 2 | 0.014 | 20 | 12 | 0.16 |
| ENSMUSG00000026687 | Calml4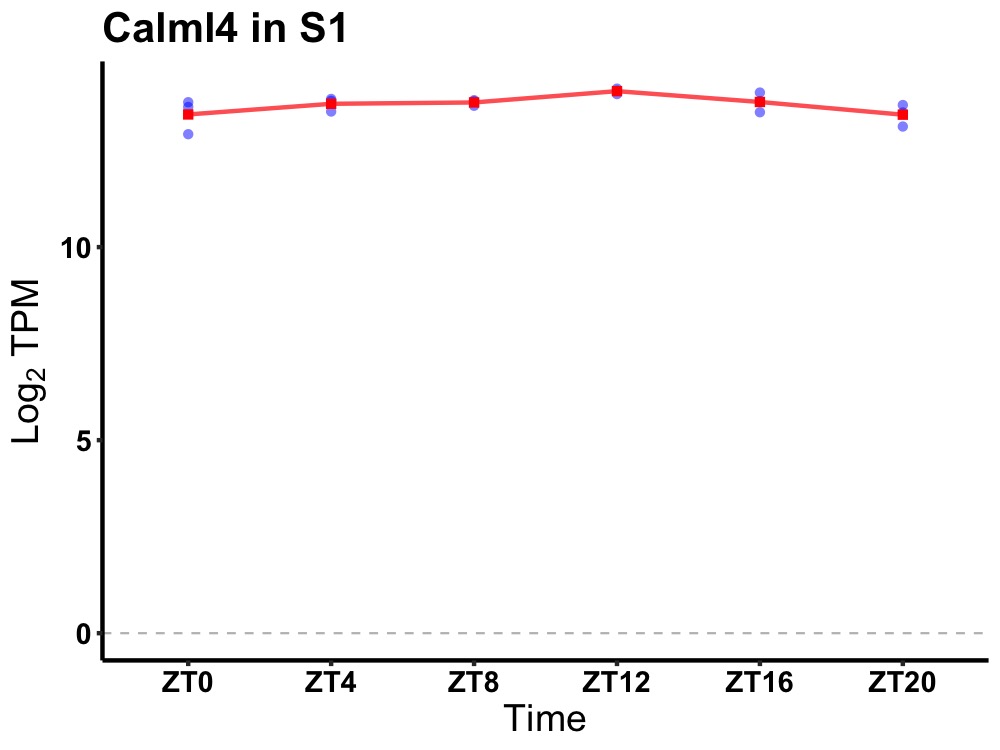 |
calmodulin-like 4 | 0.036 | 20 | 12 | 0.15 |
| ENSMUSG00000048701 | Mcrip2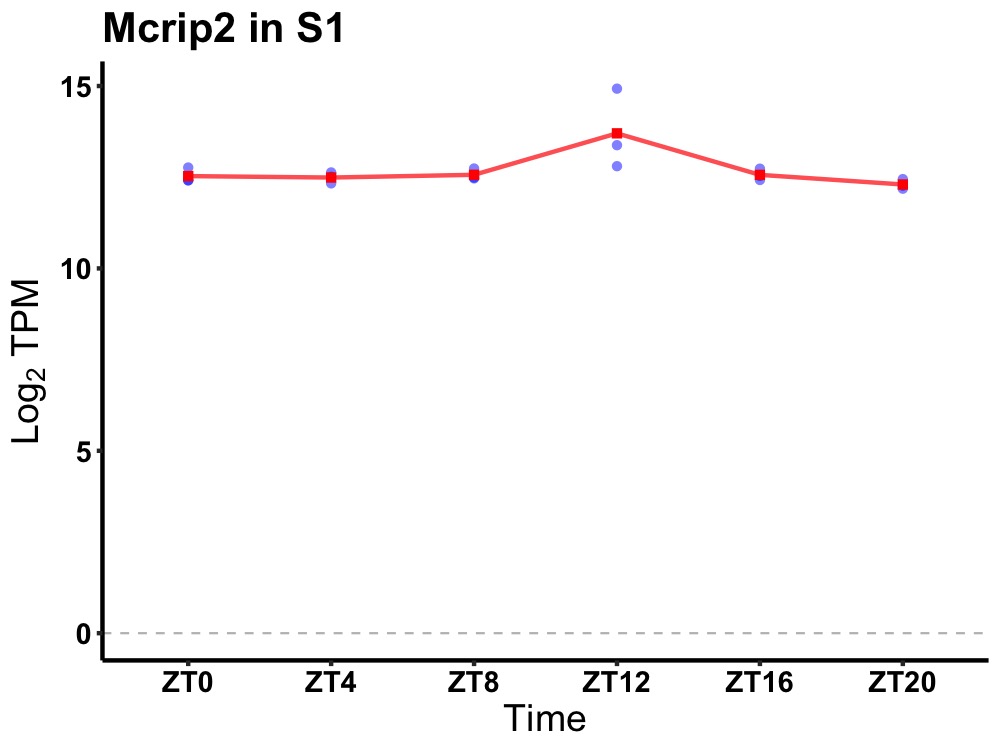 |
MAPK regulated corepressor interacting protein 2 | 0.019 | 20 | 12 | 0.12 |
| Ensembl ID | Gene Symbol | Annotation | PCT | |||
| P-value | Period, hr | Phase, hr | Amplitude (Log2[TPM]) | |||
