Circadian mRNA Abundance Changes in Microdissected Proximal Convoluted and Straight Tubules from Mouse Kidney
Method: Proximal convoluted tubules (PCTs [S1]) and proximal straight tubules (PSTs [S2]) were microdissected from kidneys of mice (male C57BL/6, 4-6 weeks) every 4 hours. Each time point was represented by 3 samples, each from a different mouse. RNA-Seq used the NEBNext(TM) Low Input RNA library preparation kit and a NovaSeq6000 DNA sequencer. The data were analyzed using the JTK_CYCLE algorithm to identify transcripts that vary with time of day with a 20 or 24 hour period (Hughes et al. PMID: 20876817). To see only those transcripts that significantly vary in a circadian pattern, select a sort option in the drop-down menu below.
This website was created by Molly Bingham, Chin-Rang Yang, Hyun Jun Jung, Joe Chou, Viswanathan Raghuram, Mark Knepper and Margo Dona (July 2021). Download data. Contact us with questions or comments: knep@helix.nih.gov
Sort transcripts by:
"Transcripts that significantly vary" was defined as having P<0.05 and a period of either 20 or 24 hours and are highlighted in gray.
To search, use browser's Find command (Ctrl-F)
| Ensembl ID | Gene Symbol | Annotation | PST | |||
| P-value | Period, hr | Phase, hr | Amplitude (Log2[TPM]) | |||
| ENSMUSG00000018427 | Ypel2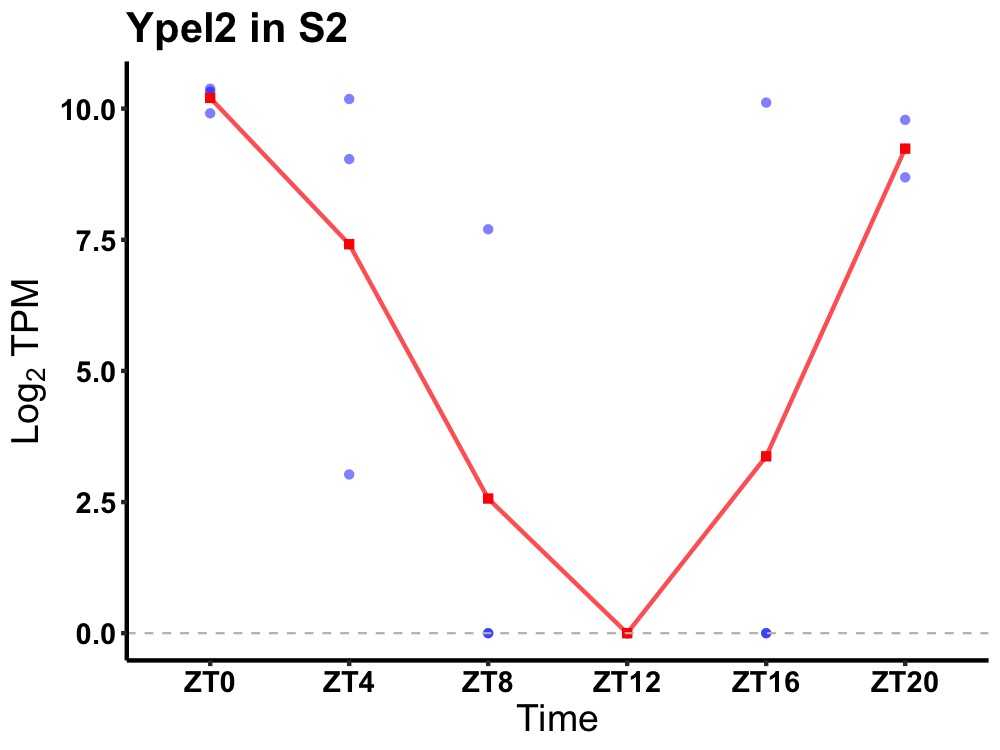 |
protein yippee-like 2 | 0.005 | 24 | 0 | 6.53 |
| ENSMUSG00000038806 | Sde2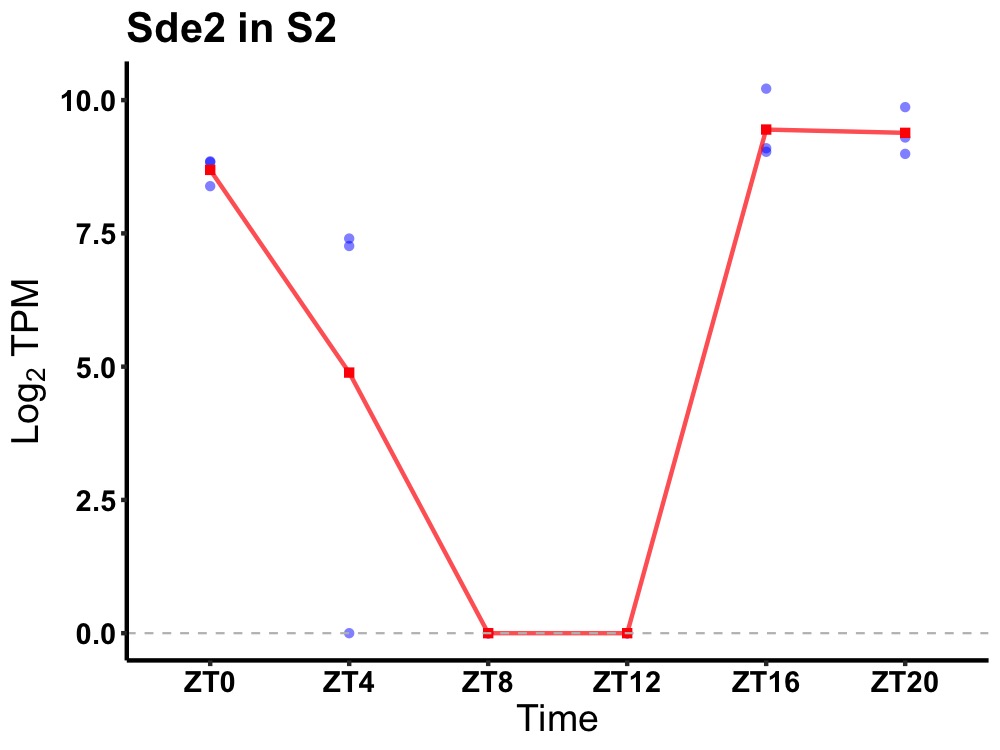 |
replication stress response regulator SDE2 | 0.001 | 24 | 20 | 6.36 |
| ENSMUSG00000041052 | Dcbld1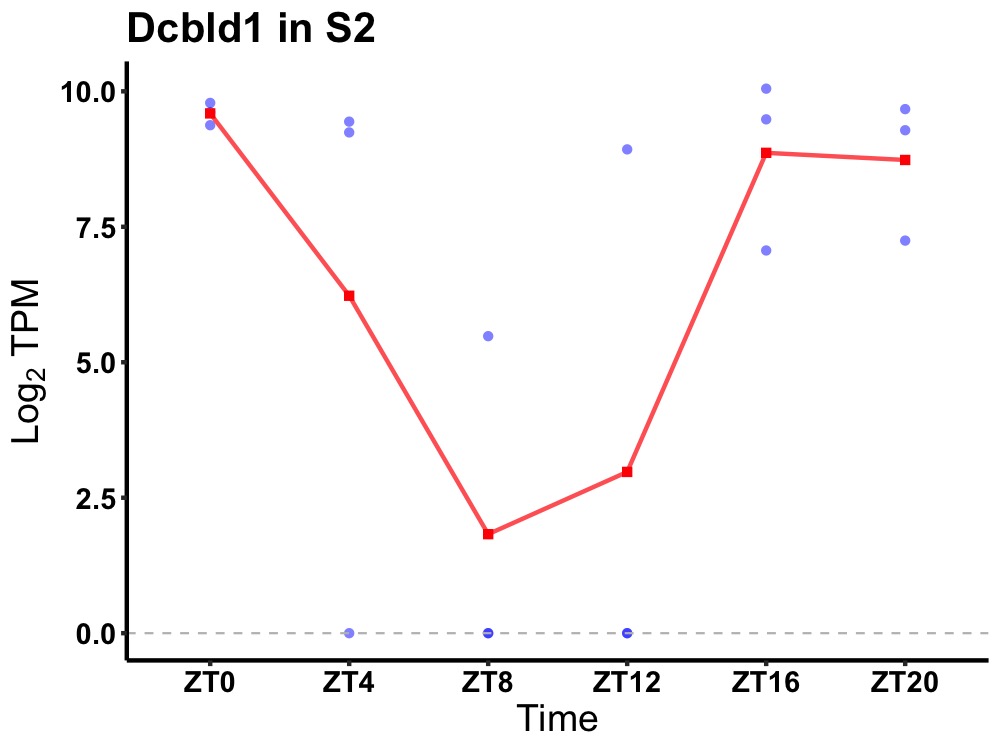 |
discoidin, CUB and LCCL domain-containing protein 1 | 0.027 | 20 | 0 | 6.31 |
| ENSMUSG00000026816 | Gtf3c5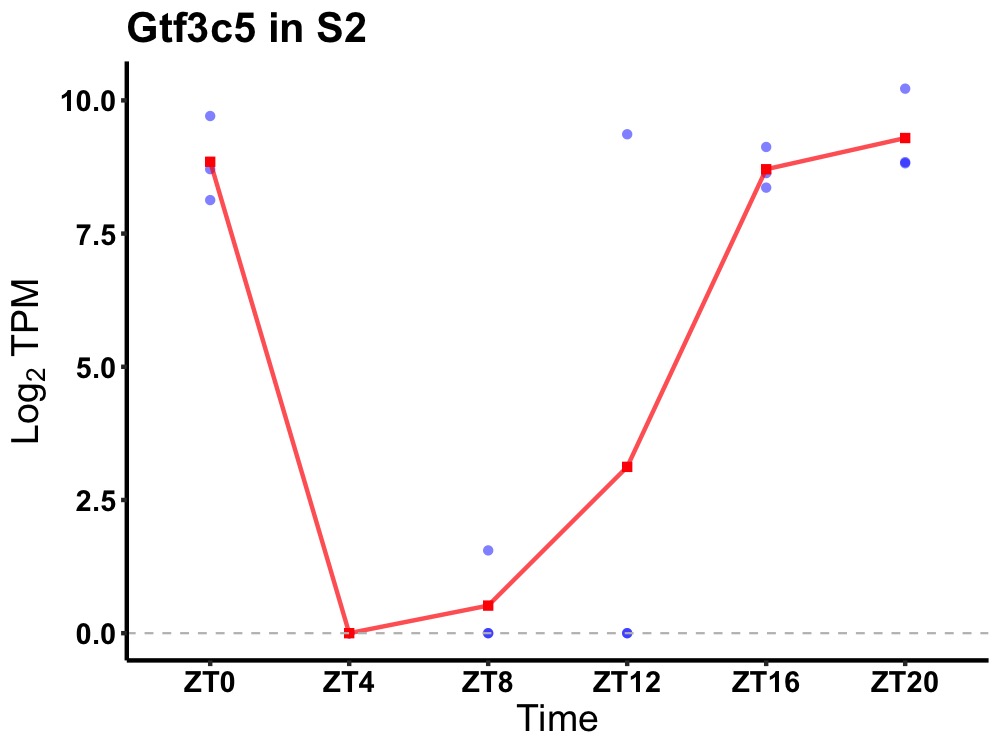 |
general transcription factor 3C polypeptide 5 | 0.049 | 24 | 20 | 6.16 |
| ENSMUSG00000019866 | Crybg1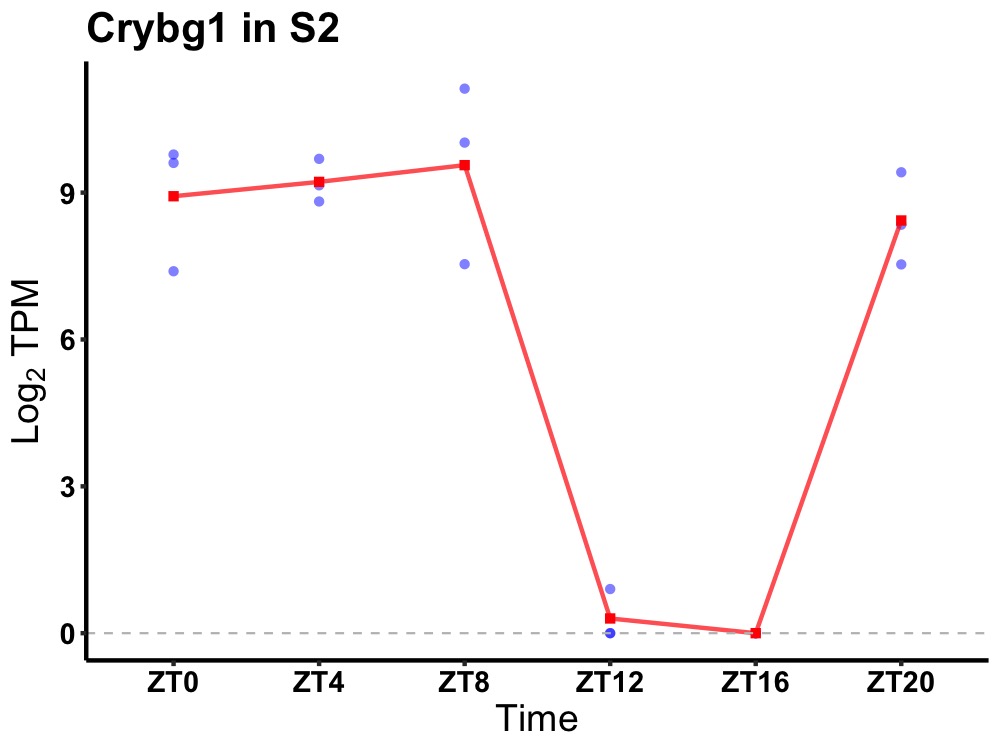 |
beta/gamma crystallin domain-containing protein 1 | 0.019 | 20 | 6 | 6.06 |
| ENSMUSG00000028229 | Pdk4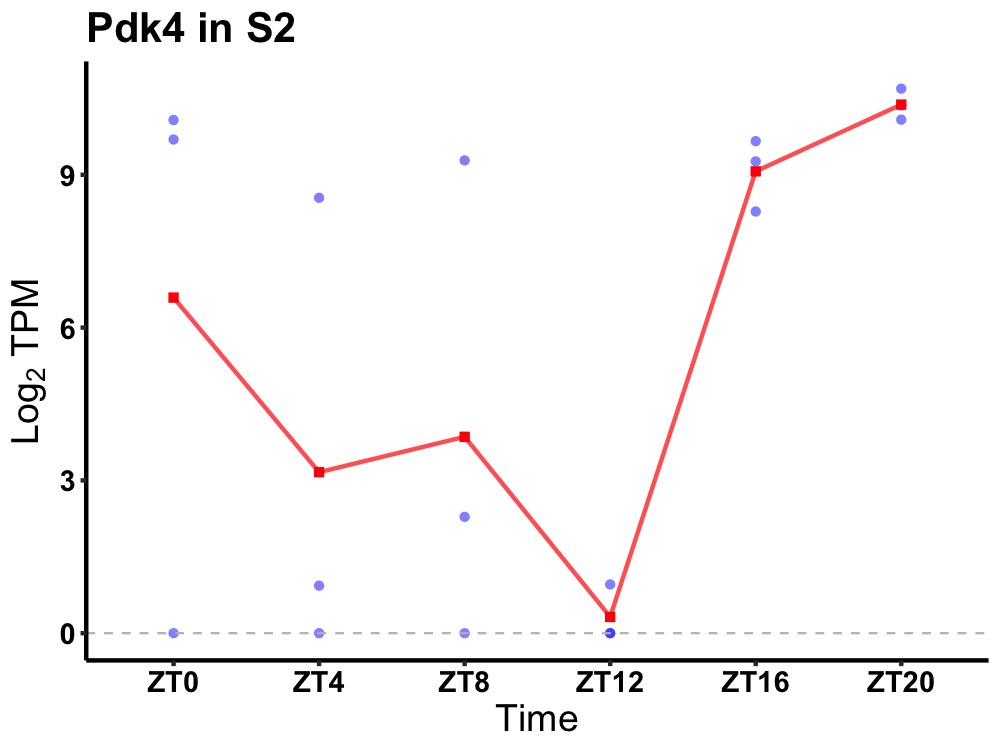 |
[Pyruvate dehydrogenase (acetyl-transferring)] kinase isozyme 4, mitochondrial | 0.043 | 24 | 22 | 6.05 |
| ENSMUSG00000035235 | Trim13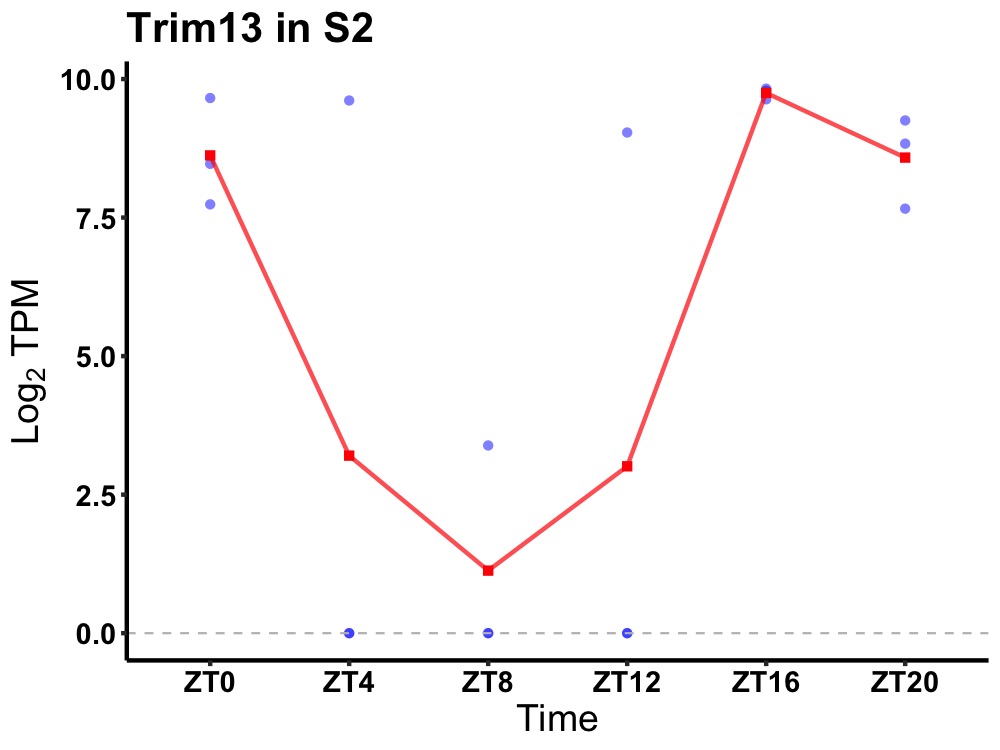 |
E3 ubiquitin-protein ligase TRIM13 | 0.027 | 20 | 18 | 5.99 |
| ENSMUSG00000032477 | Cdc25a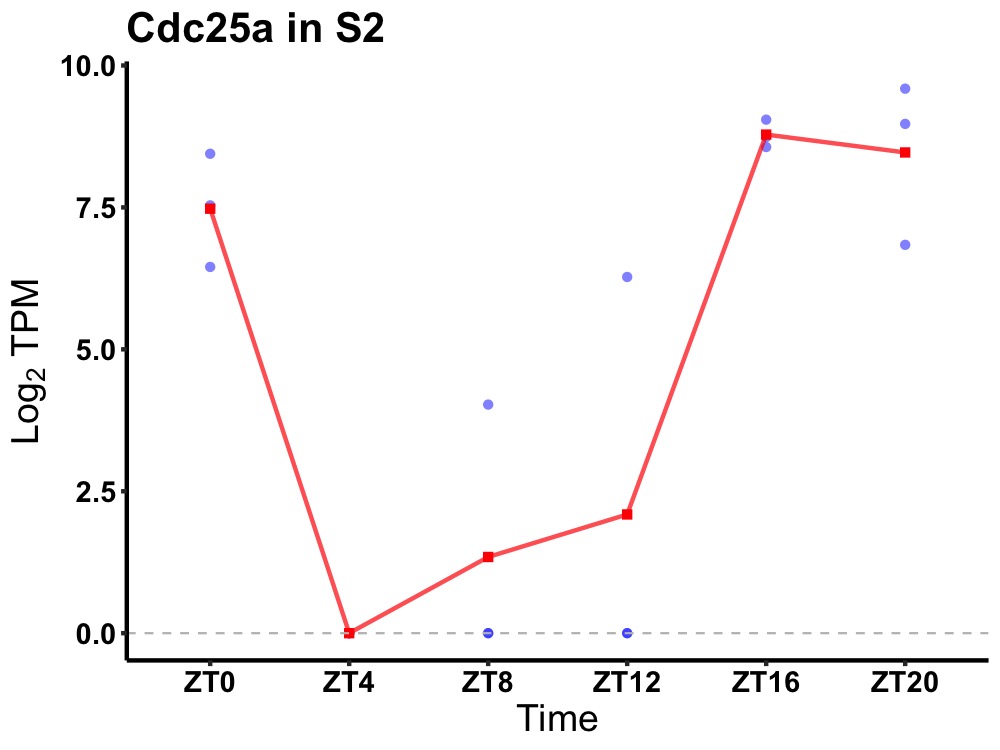 |
M-phase inducer phosphatase 1 | 0.001 | 24 | 20 | 5.97 |
| ENSMUSG00000039701 | Usp53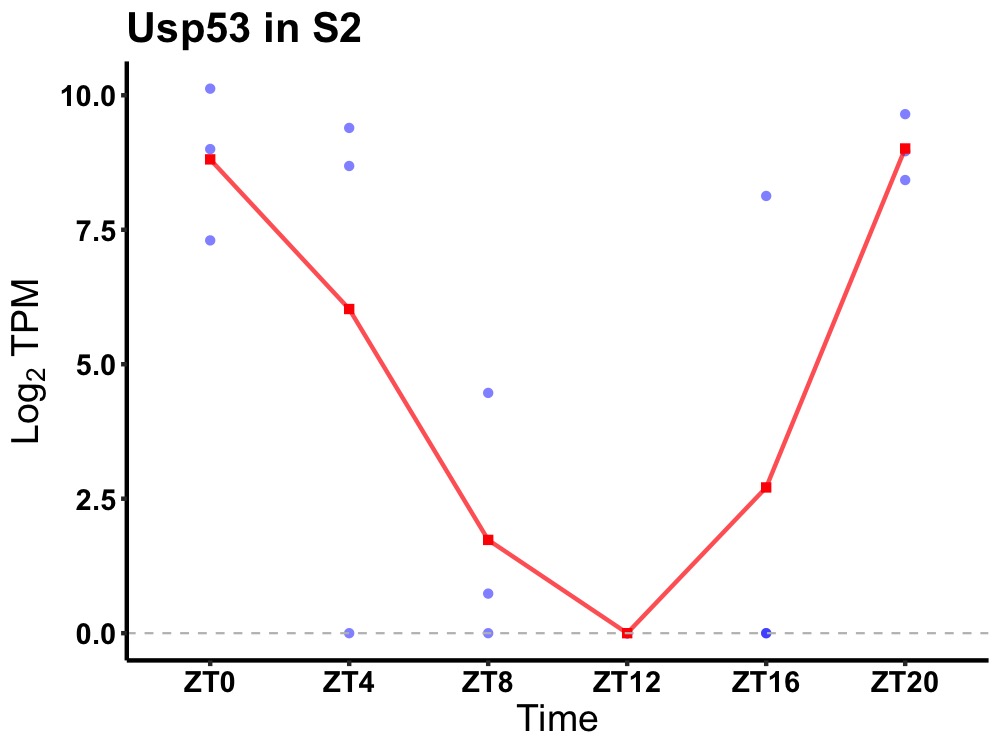 |
inactive ubiquitin carboxyl-terminal hydrolase 53 | 0.023 | 24 | 0 | 5.96 |
| ENSMUSG00000031355 | Arhgap6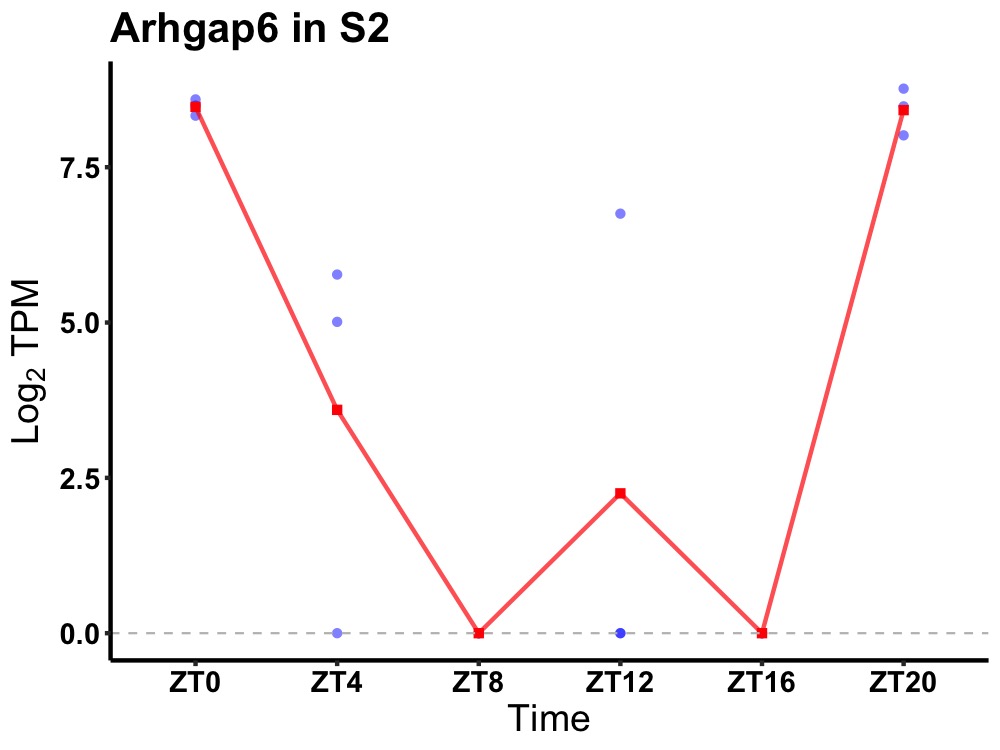 |
rho GTPase-activating protein 6 | 0.031 | 24 | 0 | 5.89 |
| ENSMUSG00000050973 | Gdpgp1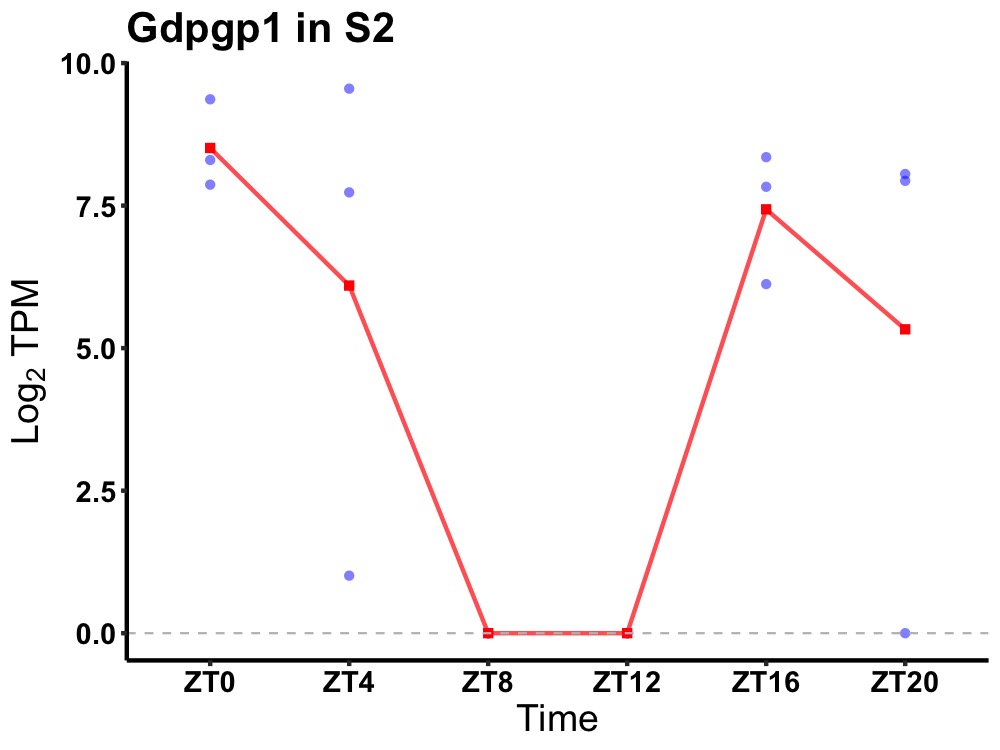 |
GDP-D-glucose phosphorylase 1 | 0.043 | 20 | 0 | 5.85 |
| ENSMUSG00000033985 | Tesk2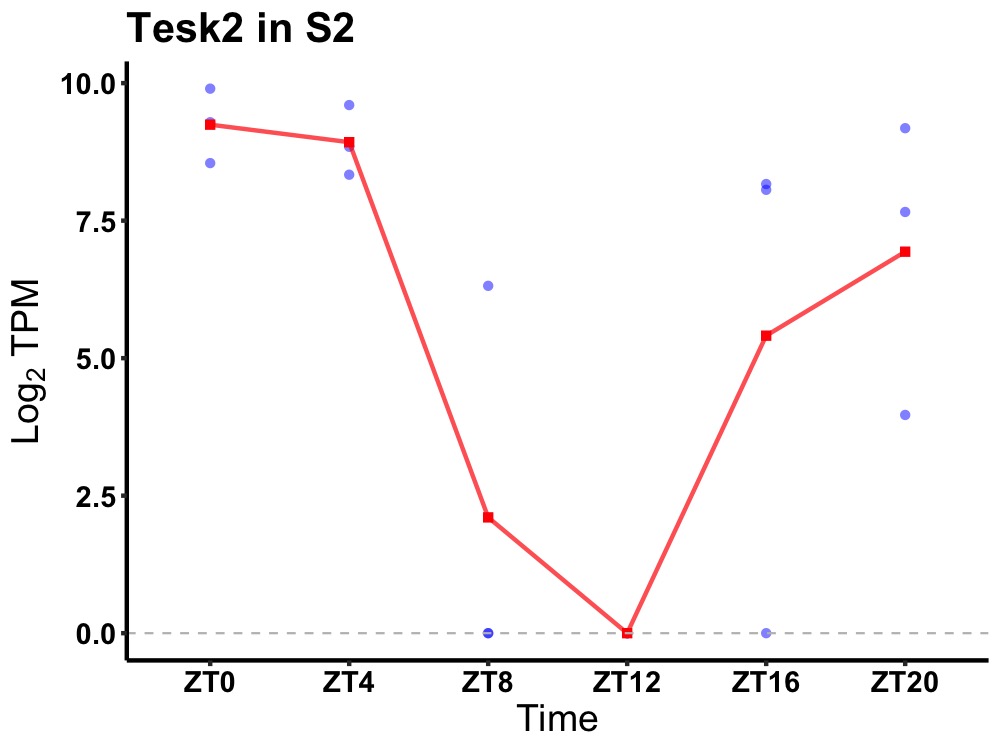 |
dual specificity testis-specific protein kinase 2 | 0.006 | 24 | 0 | 5.63 |
| ENSMUSG00000035852 | Misp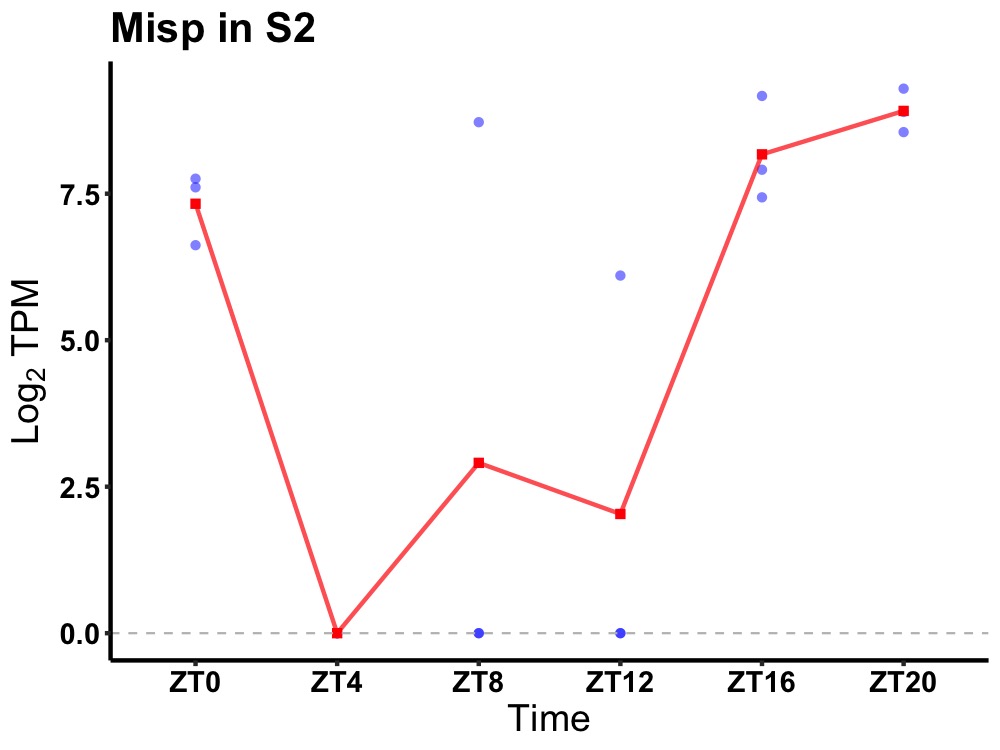 |
mitotic interactor and substrate of PLK1 | 0.010 | 24 | 20 | 5.48 |
| ENSMUSG00000044991 | Shld1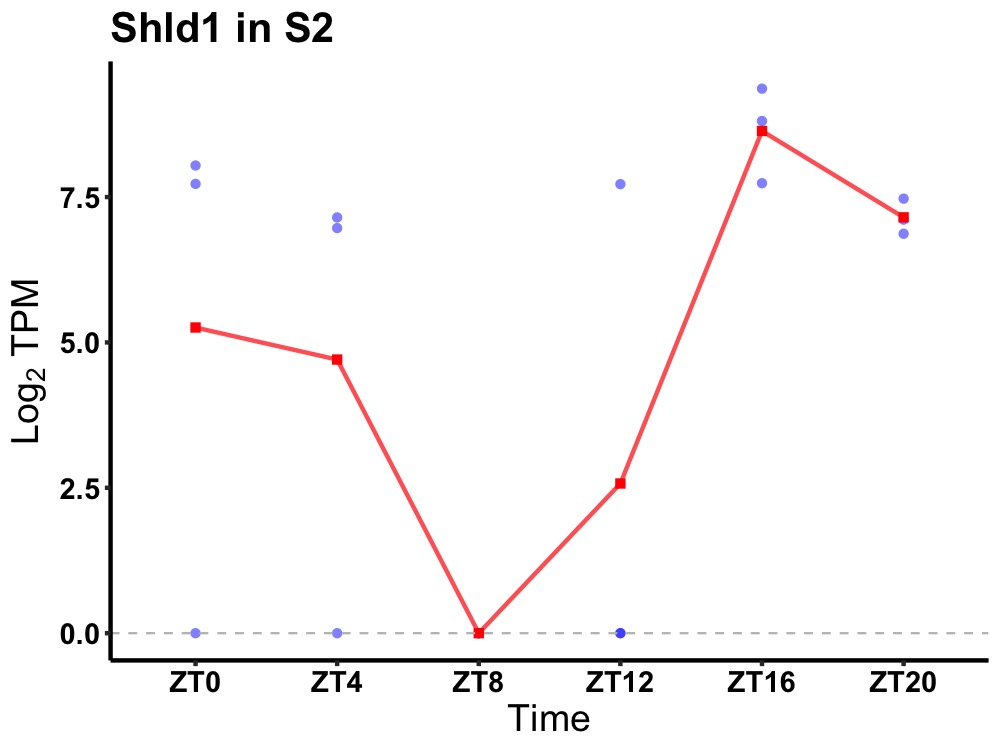 |
shieldin complex subunit 1 | 0.016 | 20 | 18 | 5.46 |
| ENSMUSG00000034889 | Cactin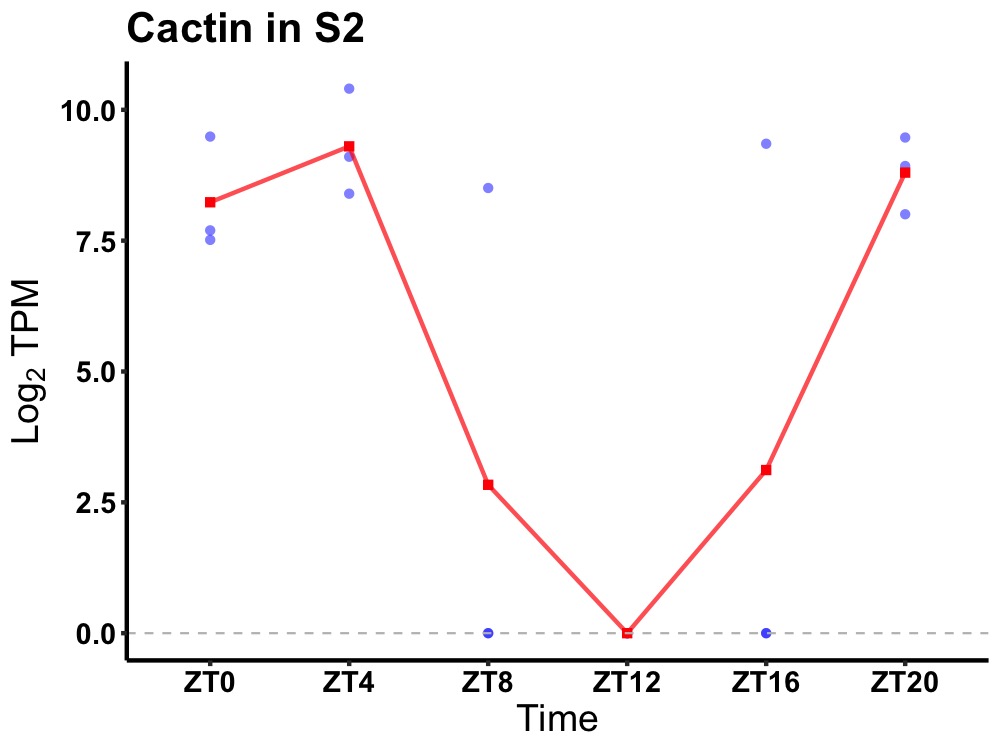 |
cactin | 0.049 | 20 | 4 | 5.44 |
| ENSMUSG00000024386 | Ascc2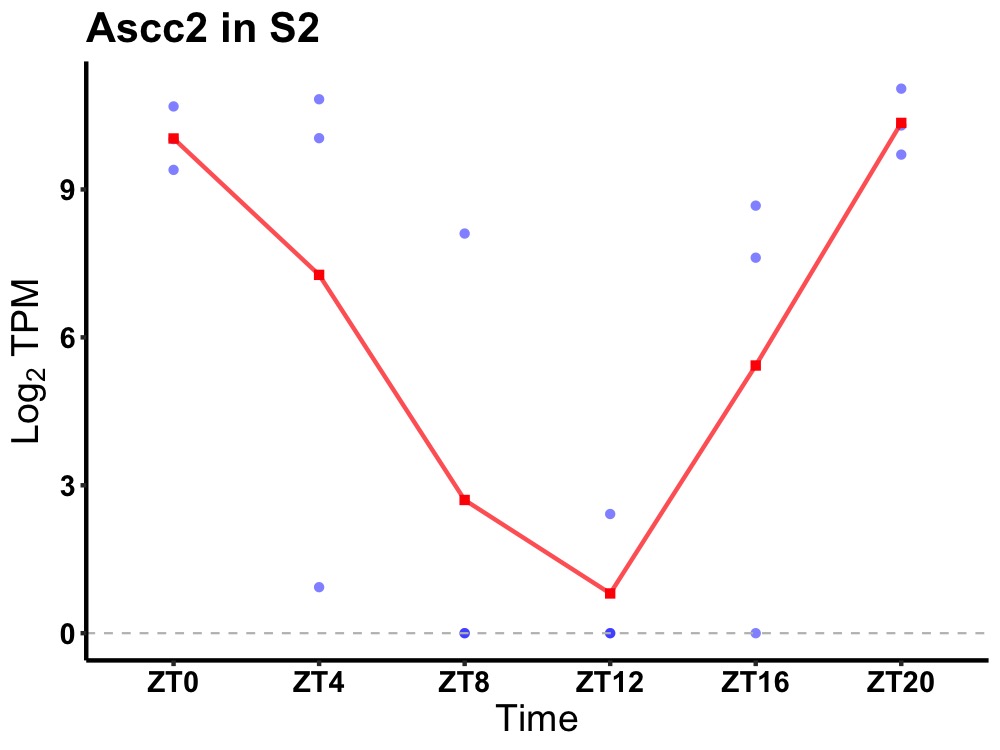 |
activating signal cointegrator 1 complex subunit 2 | 0.010 | 20 | 2 | 5.39 |
| ENSMUSG00000020823 | Sec14l1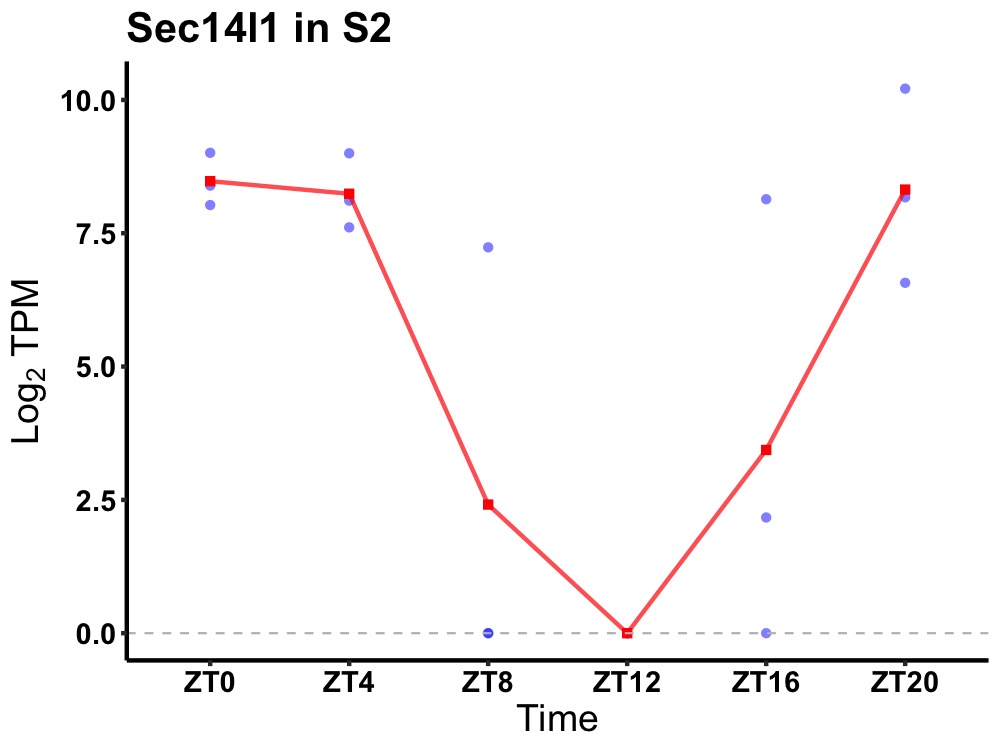 |
SEC14-like protein 1 | 0.008 | 24 | 0 | 5.38 |
| ENSMUSG00000011958 | Bnip2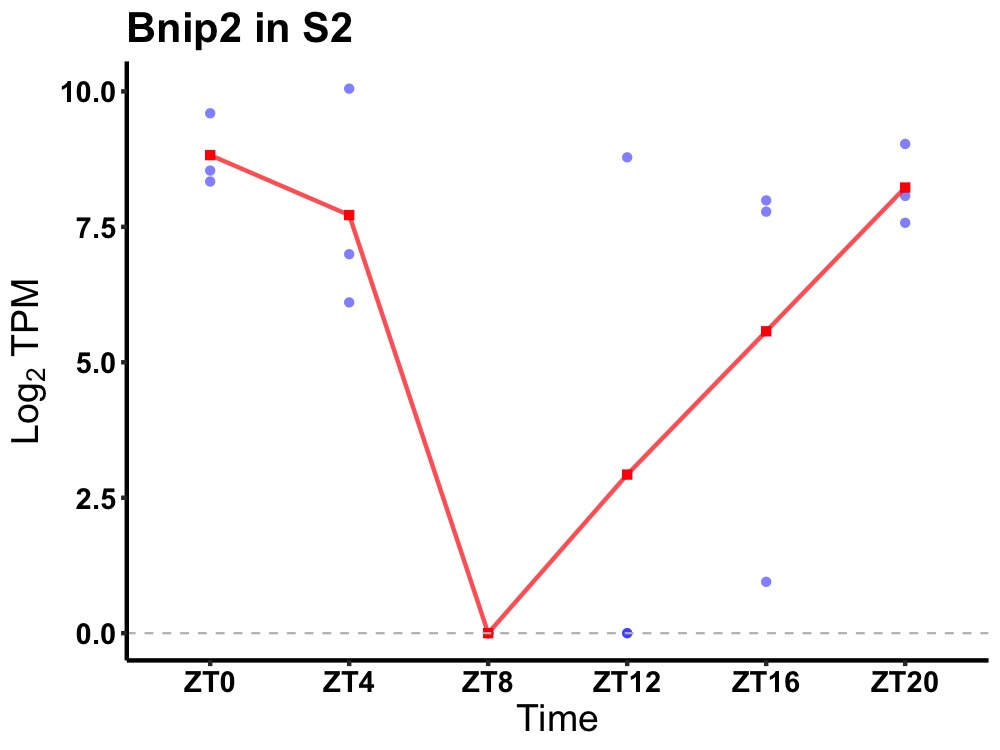 |
BCL2/adenovirus E1B 19 kDa protein-interacting protein 2 | 0.027 | 20 | 0 | 5.32 |
| ENSMUSG00000071632 | Mettl15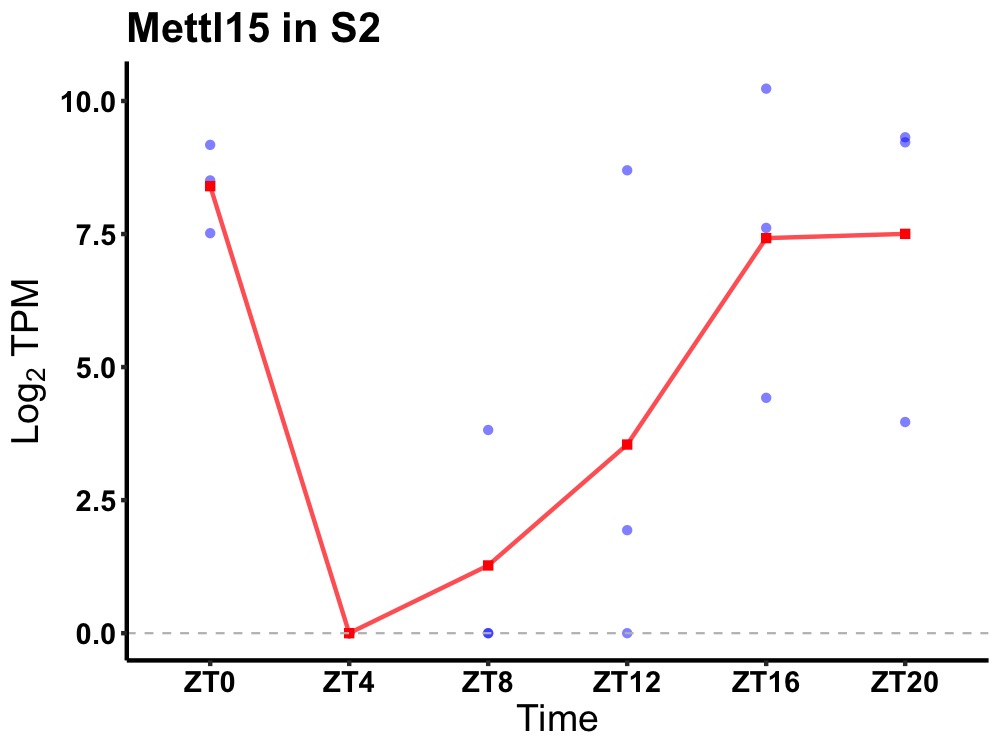 |
12S rRNA N4-methylcytidine methyltransferase | 0.043 | 24 | 20 | 5.31 |
| ENSMUSG00000048489 | Depp1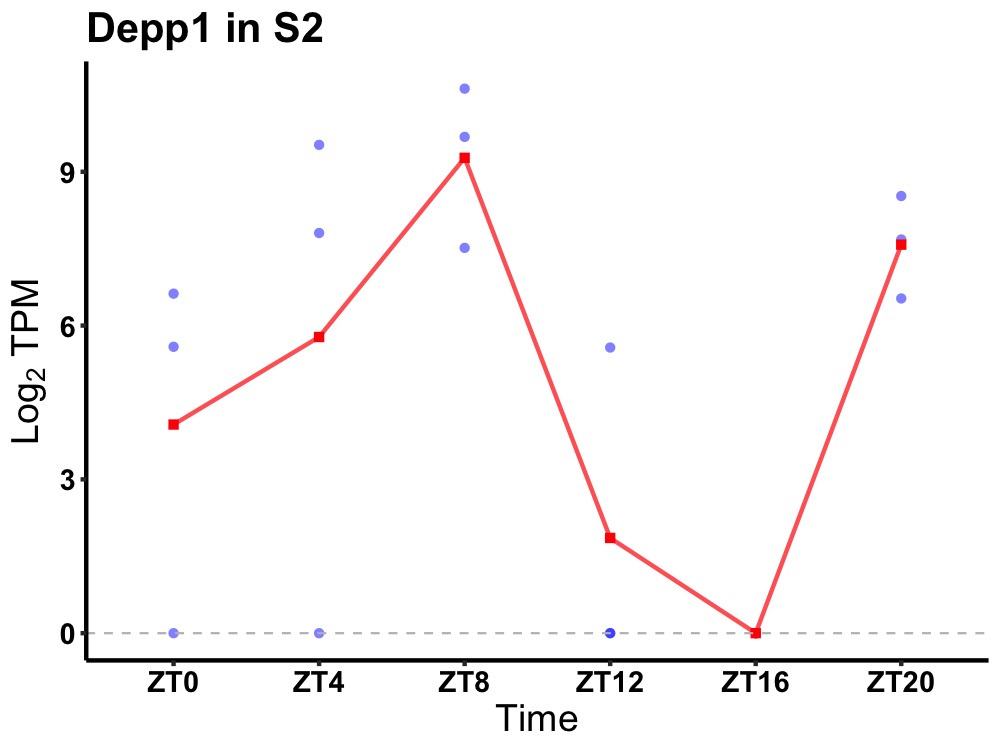 |
protein DEPP1 | 0.023 | 20 | 6 | 5.31 |
| ENSMUSG00000027423 | Pwp1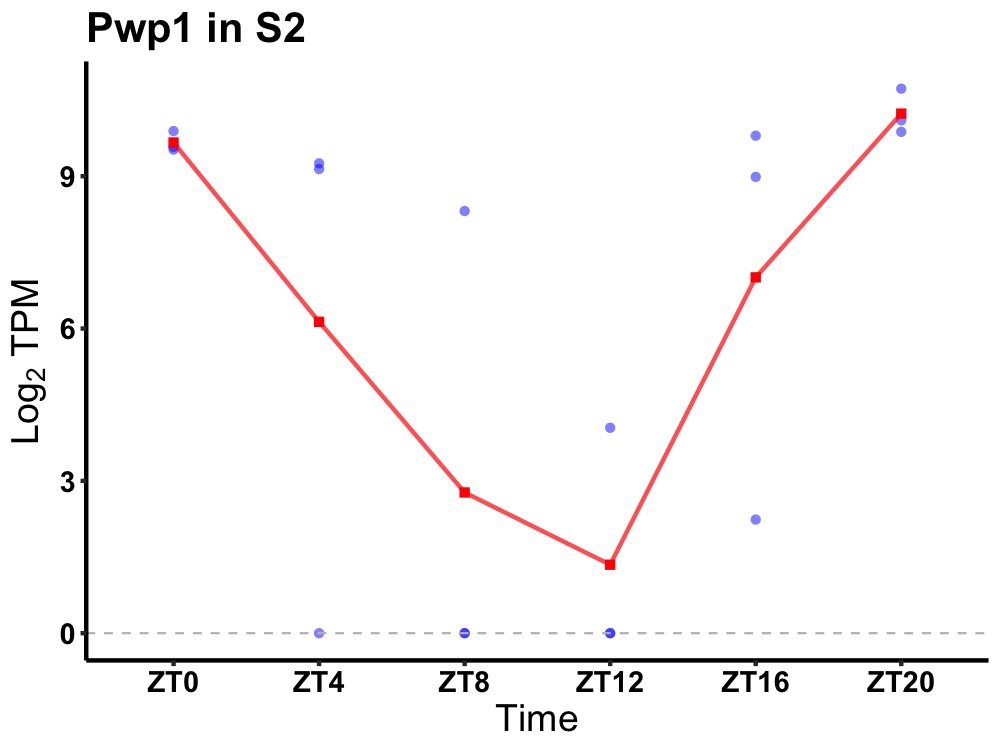 |
periodic tryptophan protein 1 homolog | 0.001 | 24 | 22 | 5.30 |
| ENSMUSG00000025332 | Kdm5c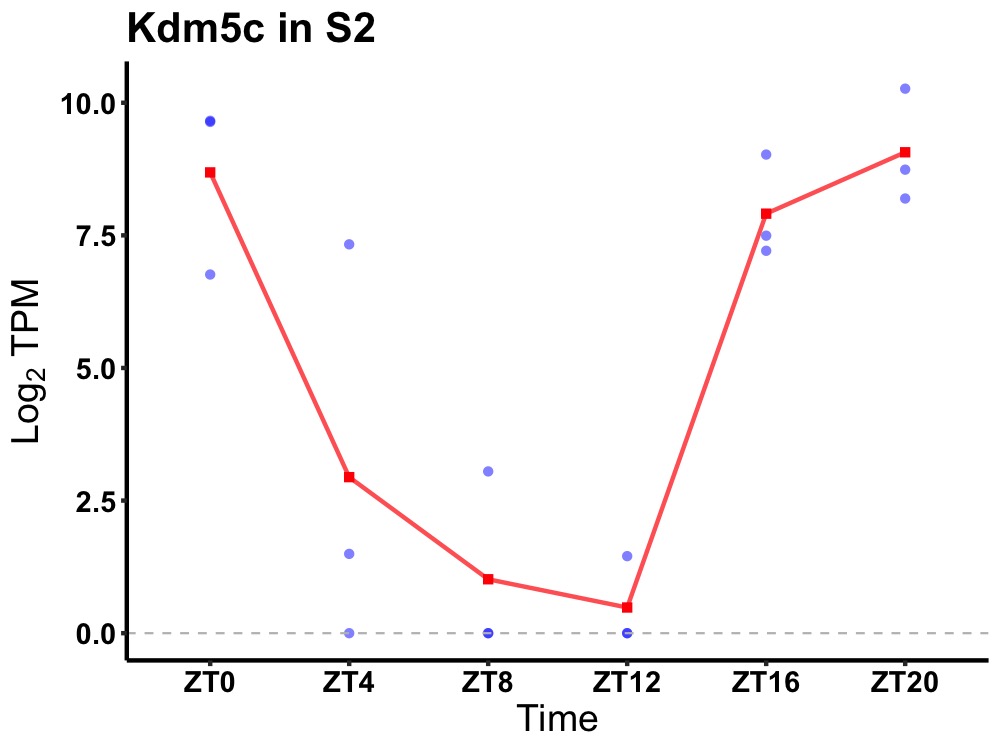 |
lysine-specific demethylase 5C | 0.003 | 24 | 22 | 5.30 |
| ENSMUSG00000108442 | Ddx20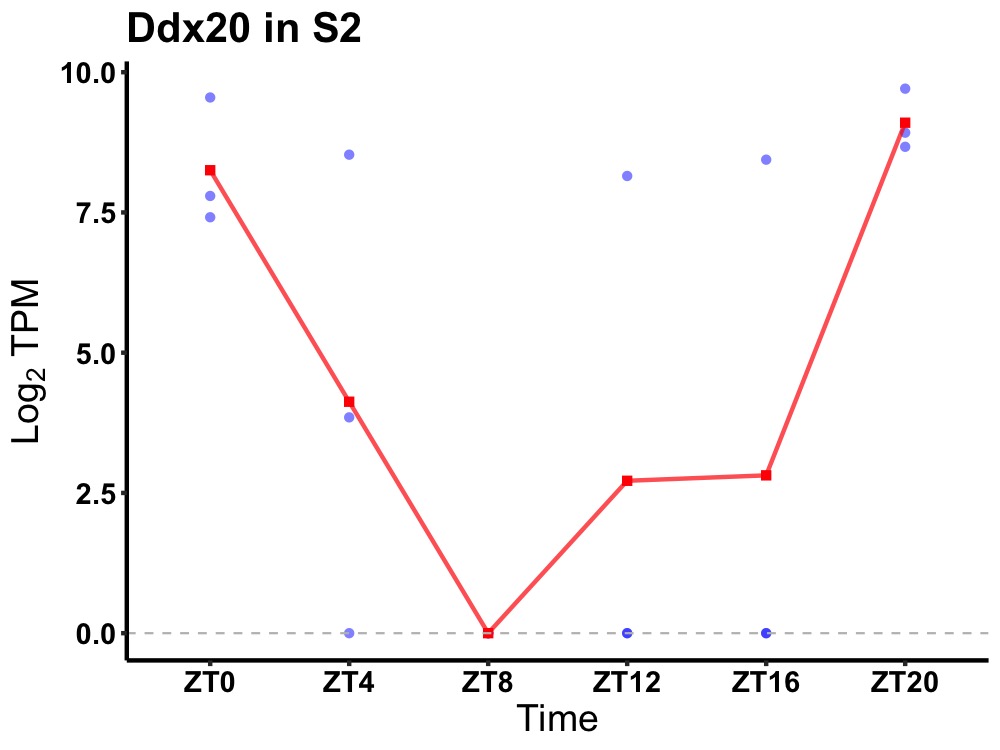 |
probable ATP-dependent RNA helicase DDX20 | 0.043 | 20 | 2 | 5.24 |
| ENSMUSG00000045252 | Zfp574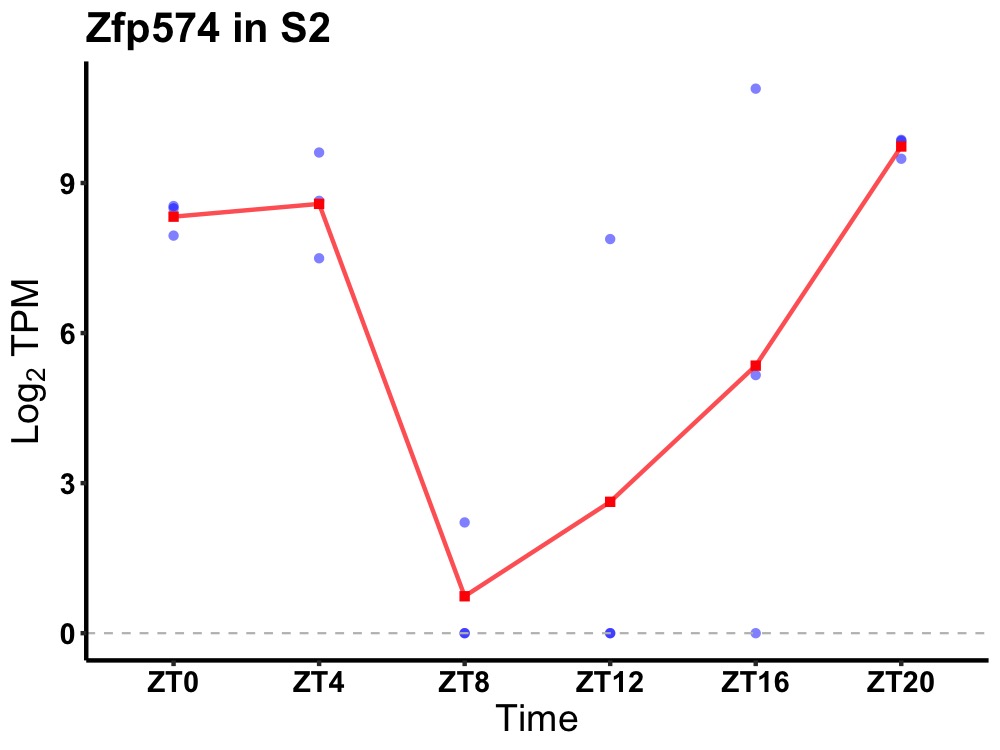 |
zinc finger protein 574 | 0.027 | 20 | 2 | 5.23 |
| ENSMUSG00000021254 | Gpatch2l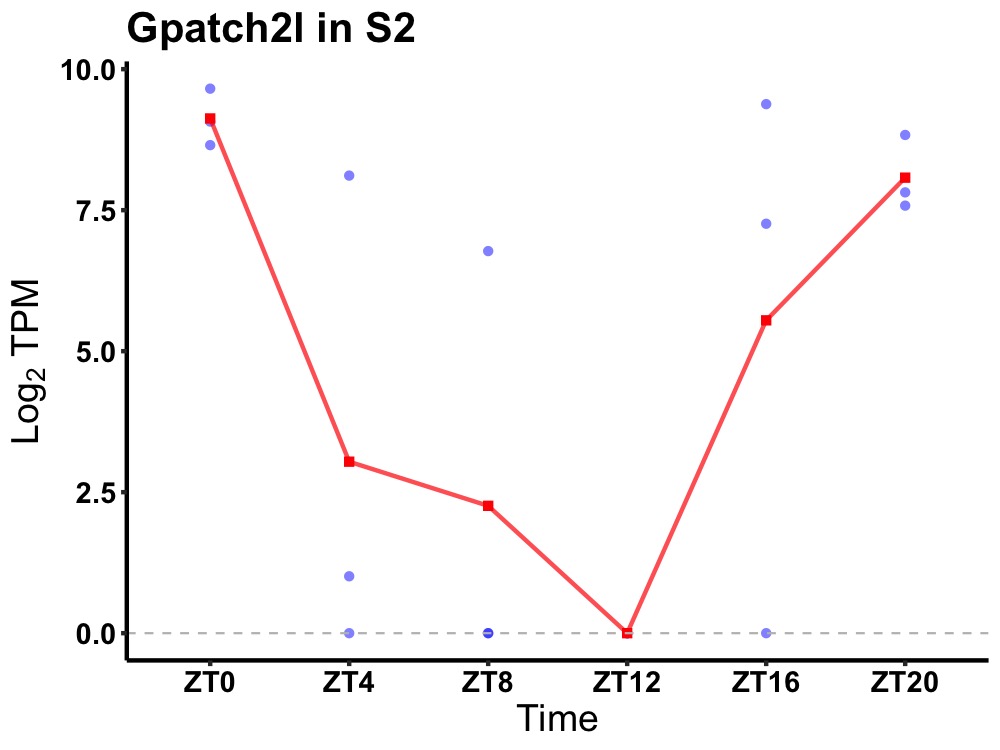 |
G patch domain-containing protein 2-like | 0.012 | 24 | 0 | 5.23 |
| ENSMUSG00000052512 | Nav2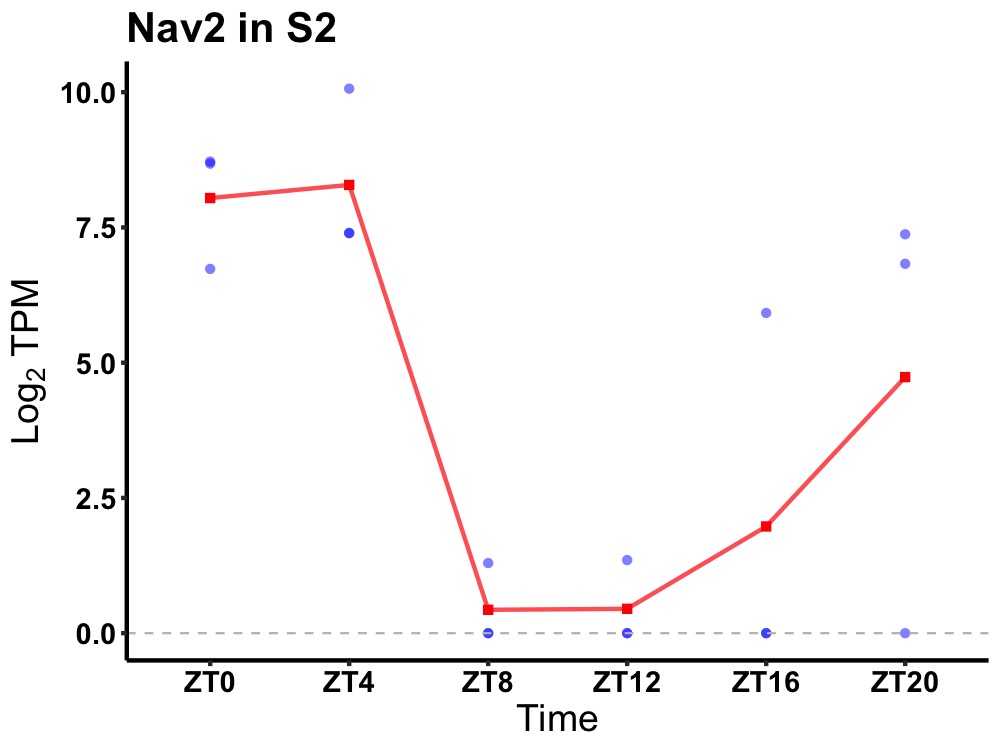 |
neuron navigator 2 | 0.027 | 24 | 2 | 5.21 |
| ENSMUSG00000024098 | Twsg1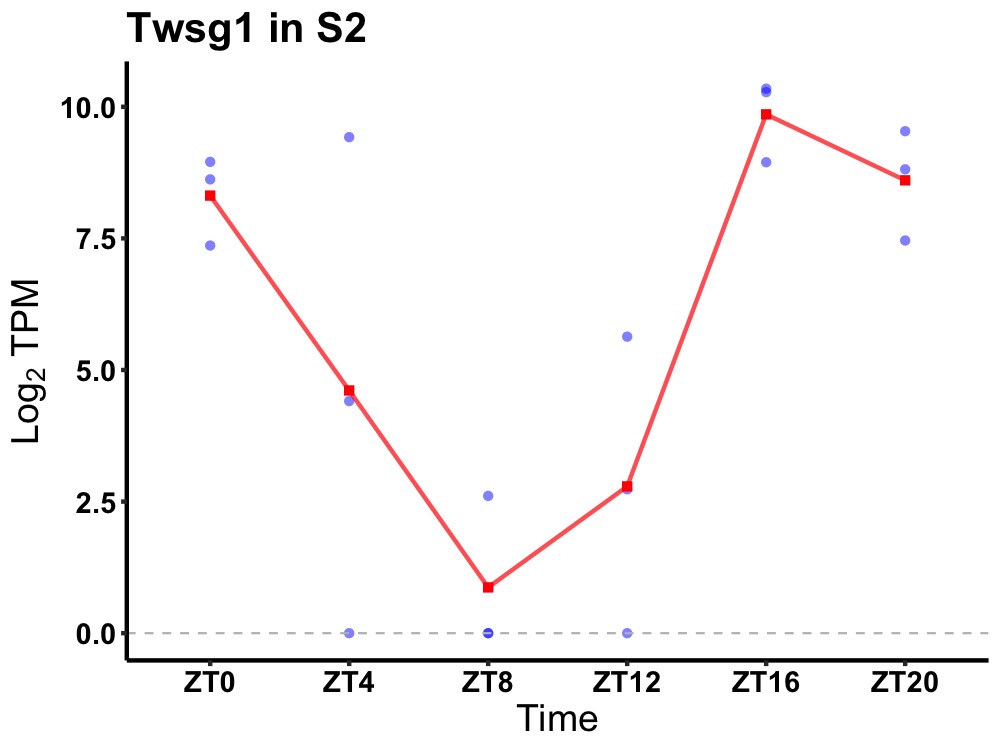 |
twisted gastrulation protein homolog 1 | 0.008 | 20 | 18 | 5.18 |
| ENSMUSG00000020108 | Ddit4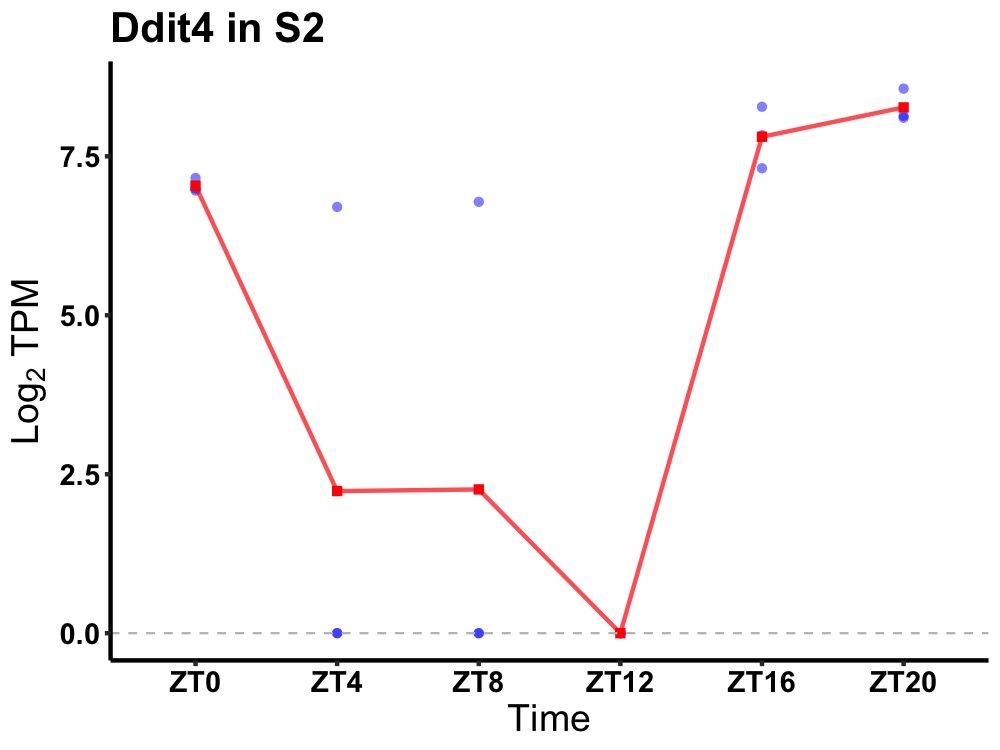 |
DNA damage-inducible transcript 4 protein | 0.001 | 24 | 20 | 5.17 |
| ENSMUSG00000005397 | Nid1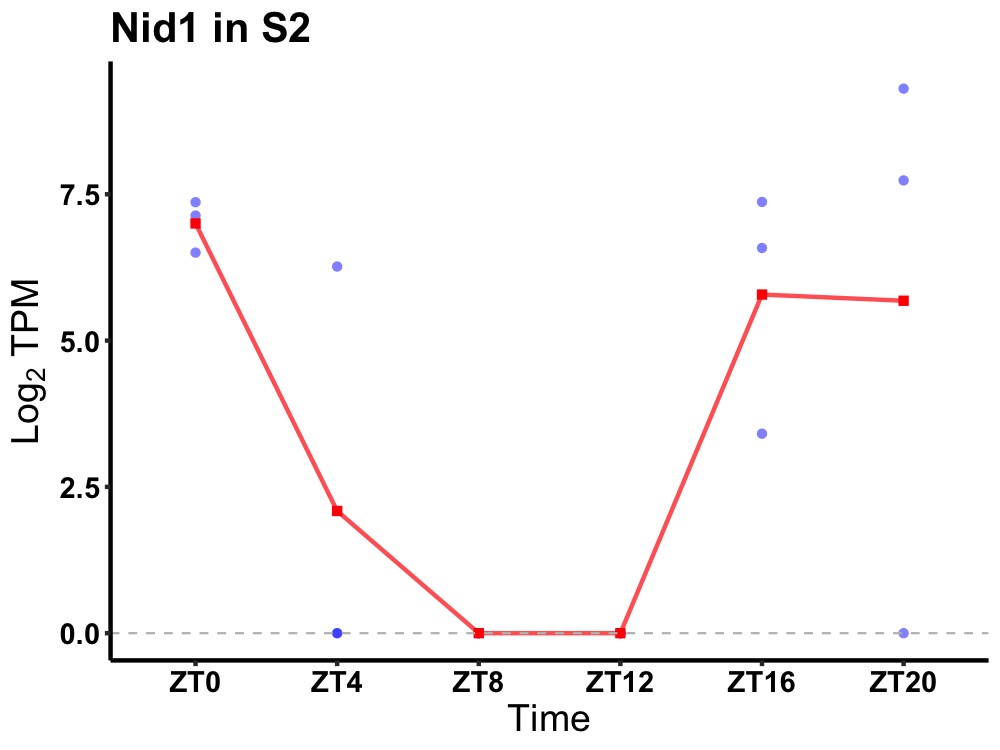 |
nidogen-1 | 0.023 | 24 | 22 | 5.05 |
| ENSMUSG00000029135 | Fosl2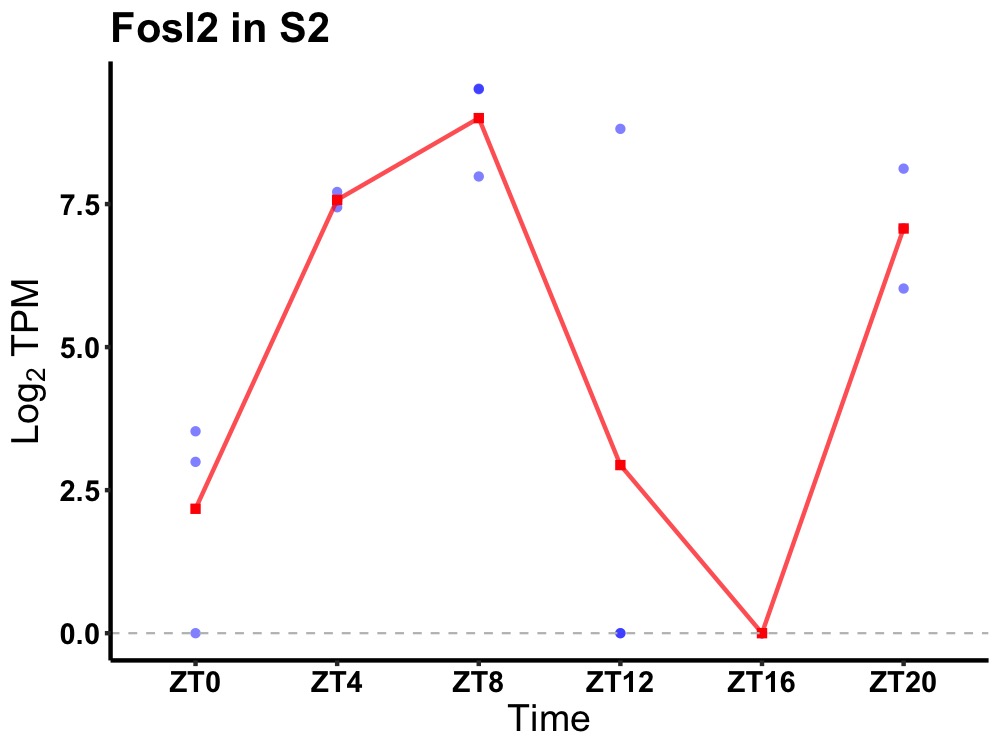 |
fos-related antigen 2 | 0.008 | 20 | 8 | 4.97 |
| ENSMUSG00000032041 | Tirap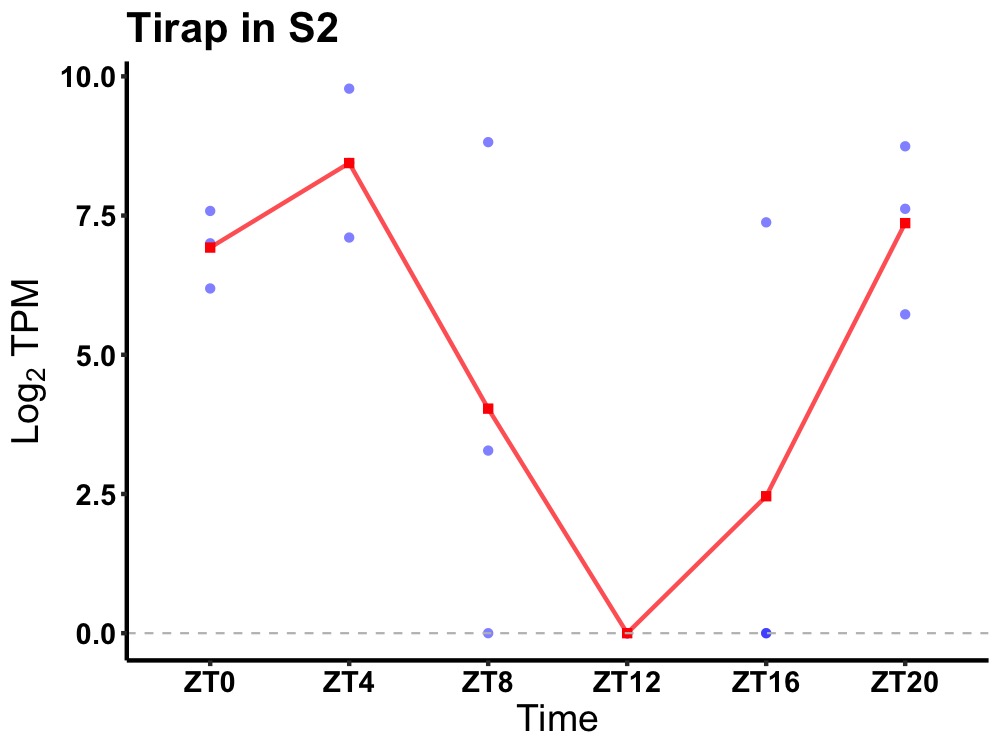 |
toll/interleukin-1 receptor domain-containing adapter protein | 0.023 | 20 | 4 | 4.95 |
| ENSMUSG00000035898 | Uba6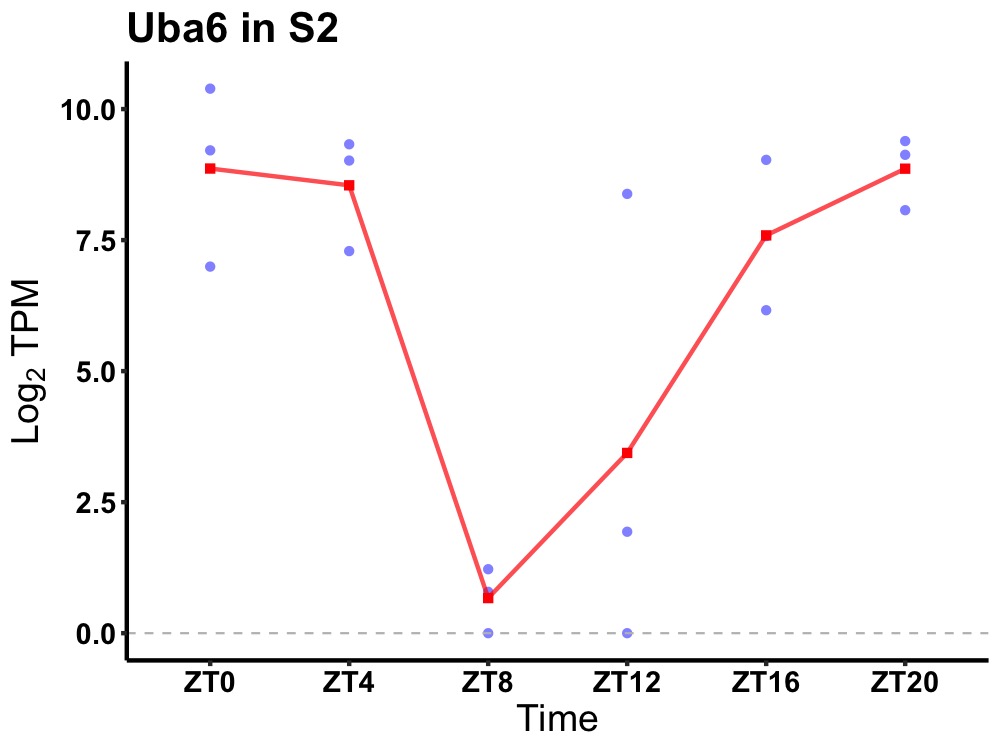 |
ubiquitin-like modifier-activating enzyme 6 | 0.031 | 20 | 0 | 4.94 |
| ENSMUSG00000051351 | Zfp46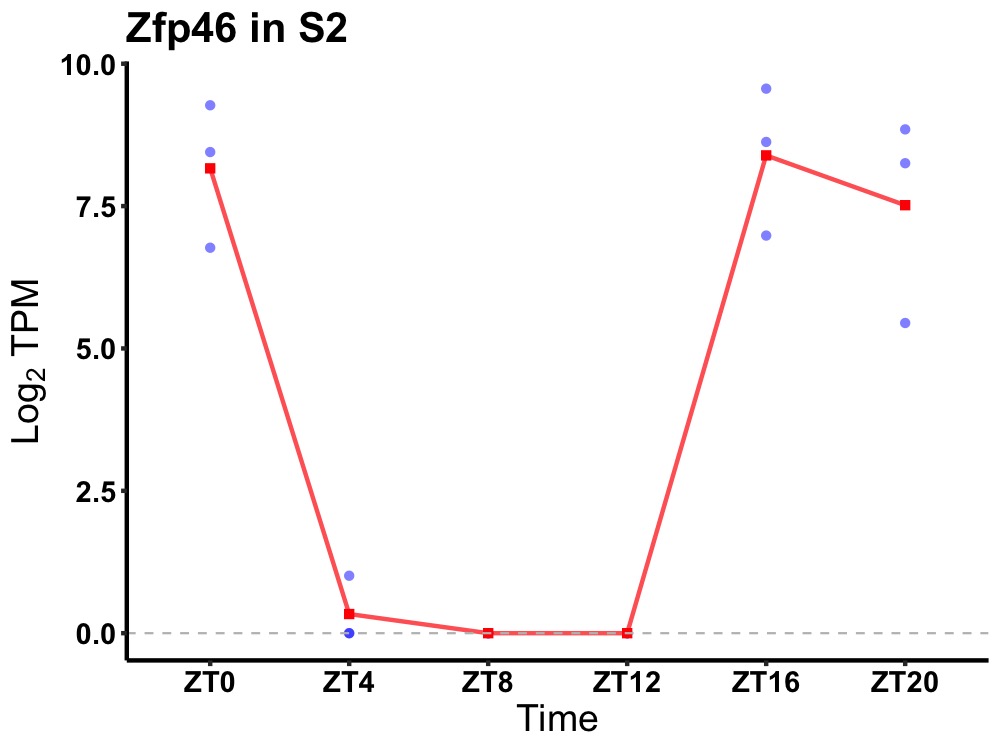 |
zinc finger protein 436 | 0.006 | 20 | 18 | 4.94 |
| ENSMUSG00000032193 | Ldlr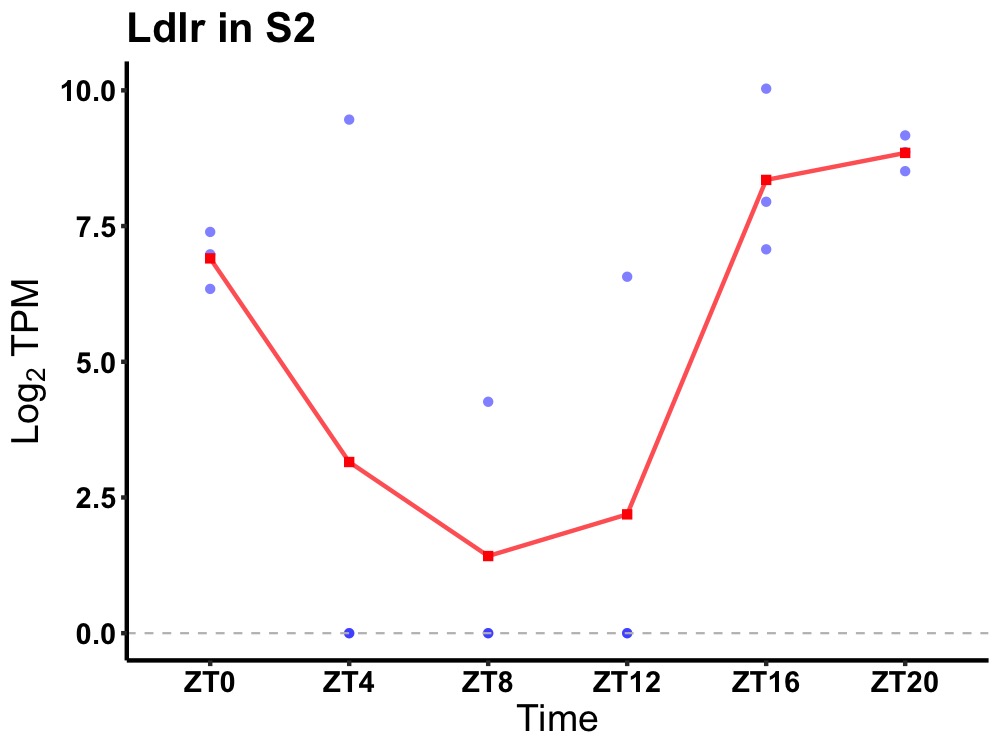 |
low-density lipoprotein receptor presursor | 0.019 | 24 | 20 | 4.93 |
| ENSMUSG00000004508 | Gab2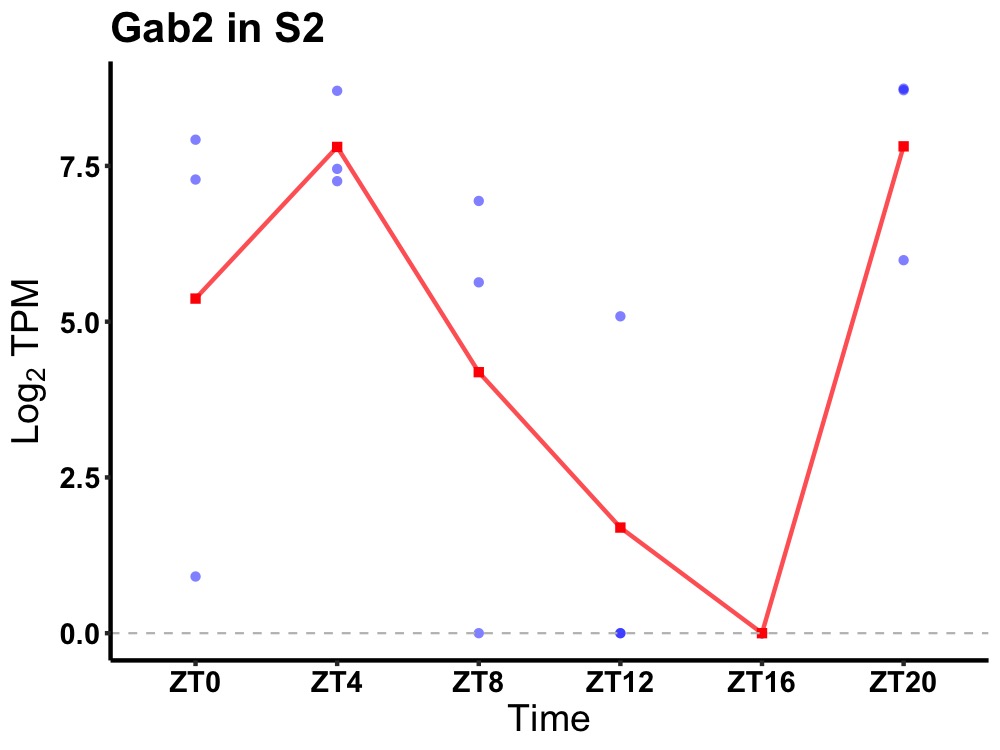 |
GRB2-associated-binding protein 2 | 0.012 | 20 | 4 | 4.91 |
| ENSMUSG00000042579 | 4632404H12Rik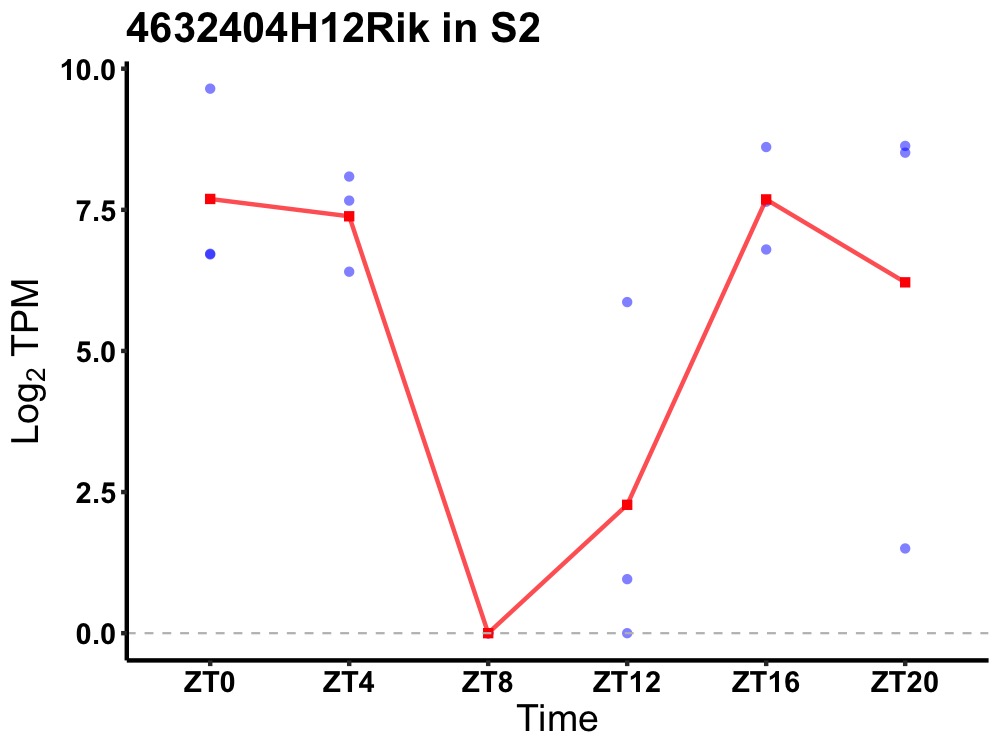 |
none | 0.031 | 20 | 0 | 4.74 |
| ENSMUSG00000019856 | Fam184a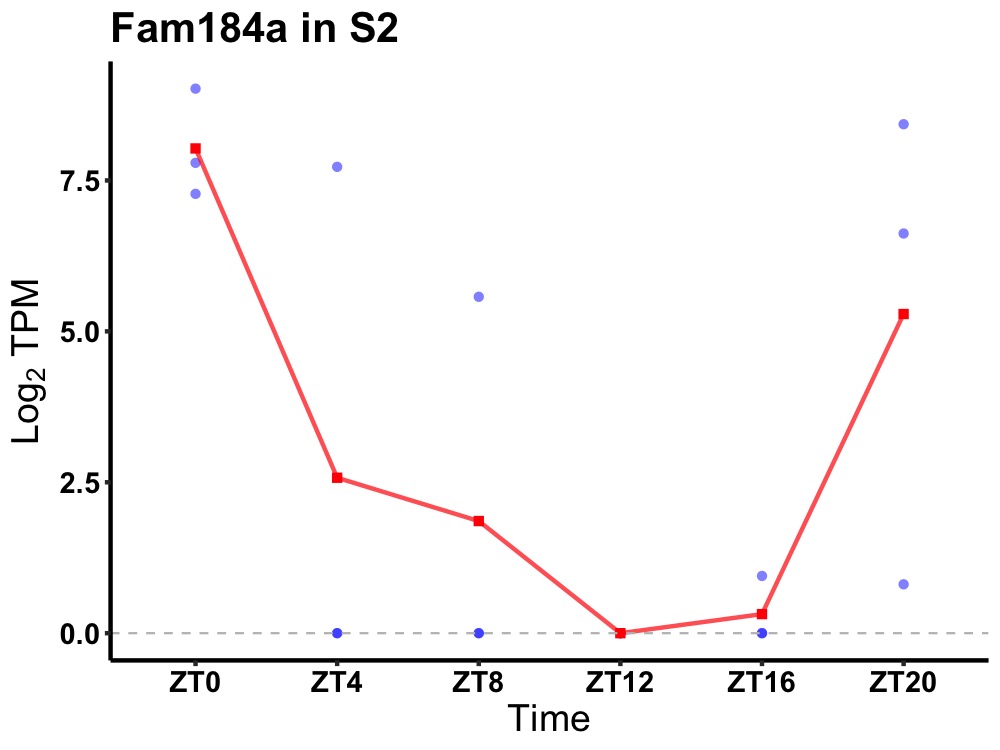 |
protein FAM184A | 0.049 | 24 | 0 | 4.68 |
| ENSMUSG00000034751 | Mast4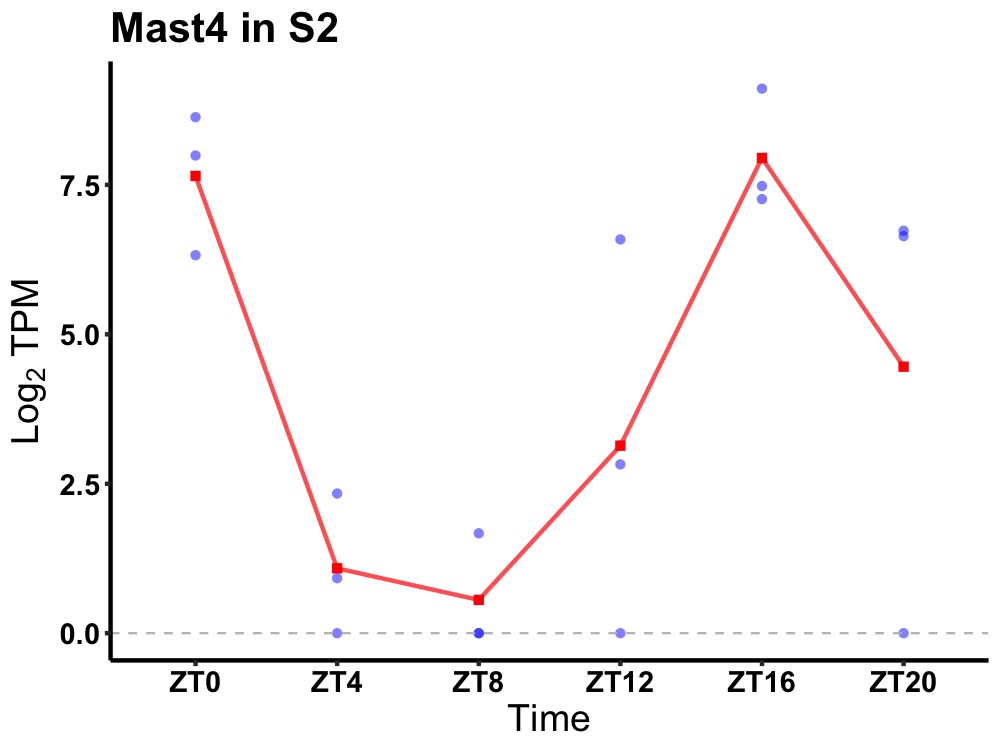 |
microtubule-associated serine/threonine-protein kinase 4 | 0.004 | 20 | 18 | 4.62 |
| ENSMUSG00000001065 | Zfp276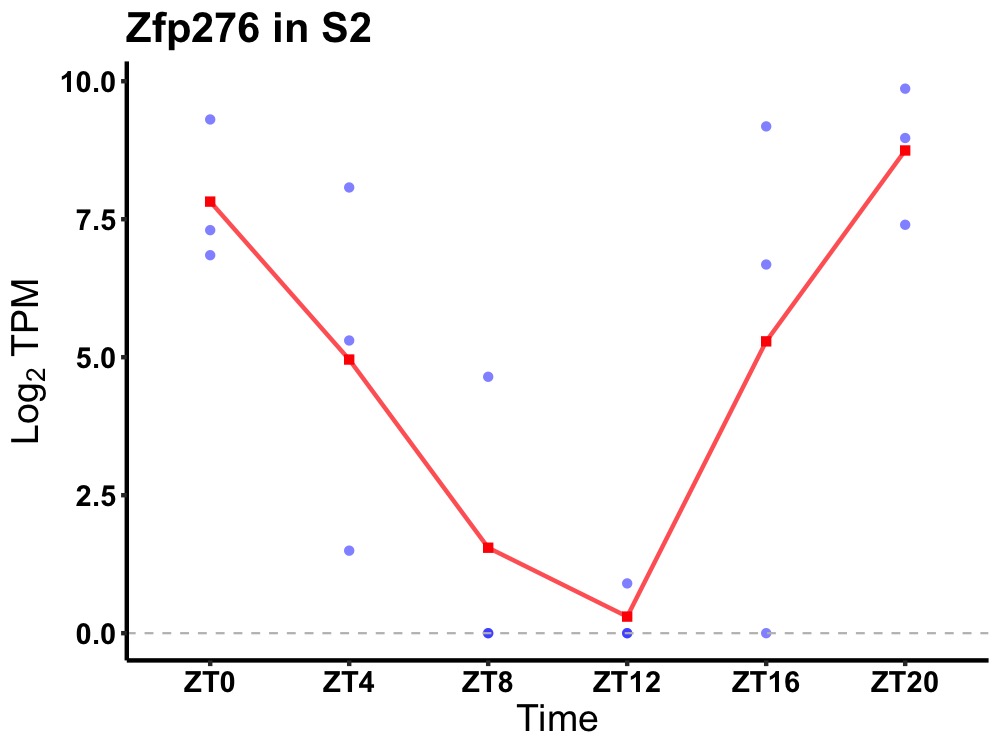 |
zinc finger protein 276 | 0.007 | 24 | 22 | 4.59 |
| ENSMUSG00000020740 | Gga3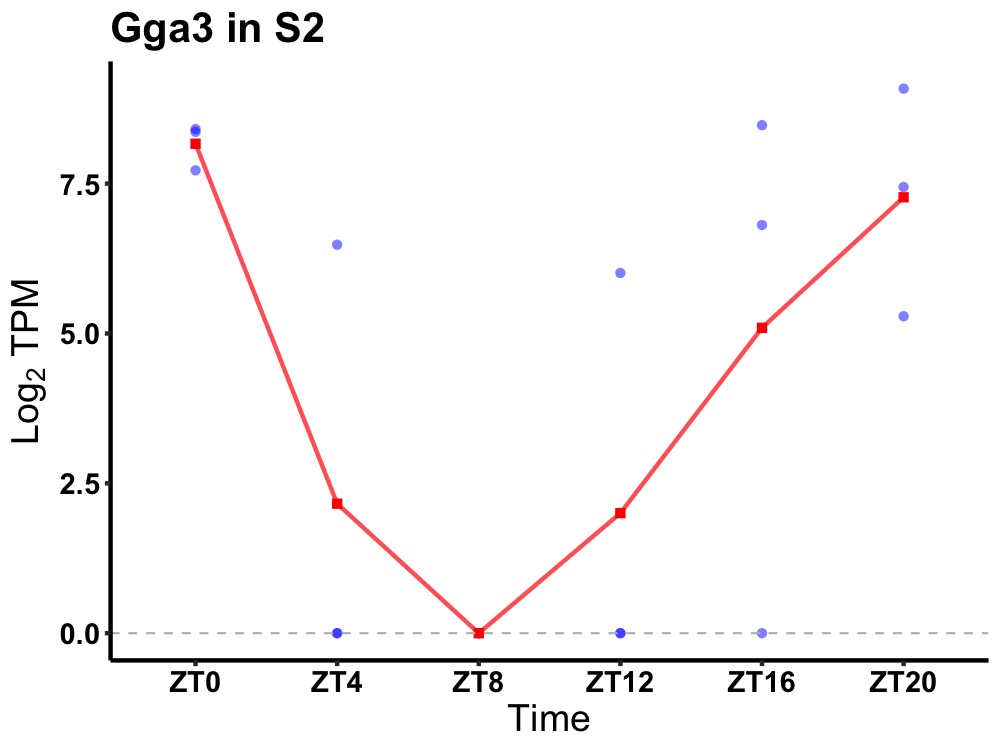 |
ADP-ribosylation factor-binding protein GGA3 | 0.023 | 20 | 0 | 4.58 |
| ENSMUSG00000043432 | Leng9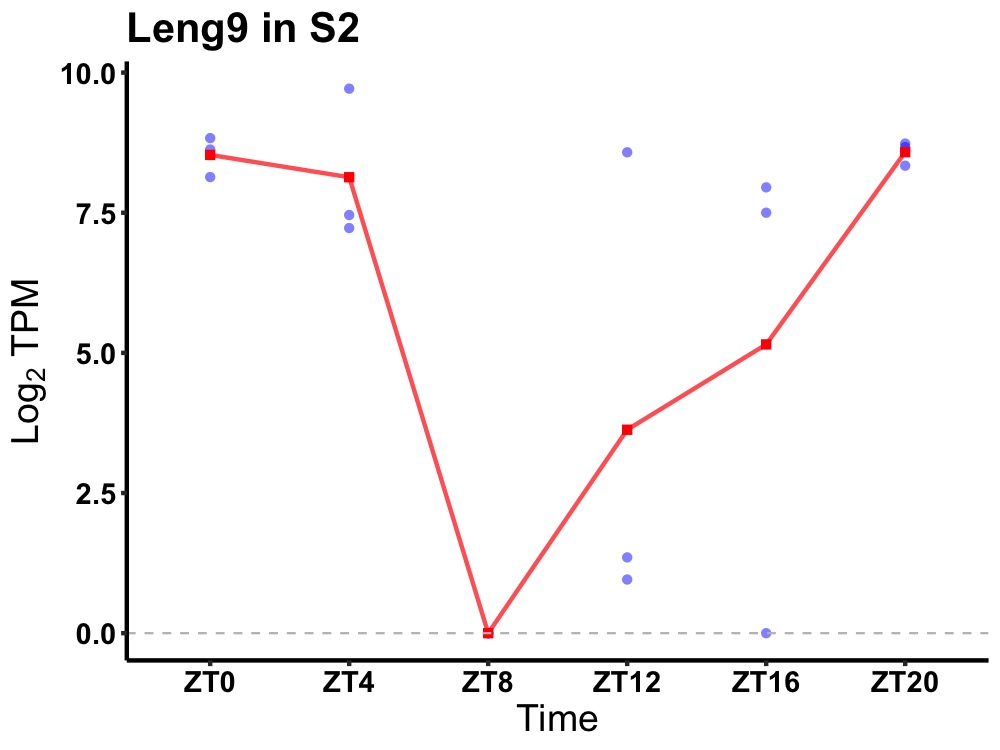 |
leukocyte receptor cluster member 9 | 0.016 | 24 | 22 | 4.35 |
| ENSMUSG00000034218 | Atm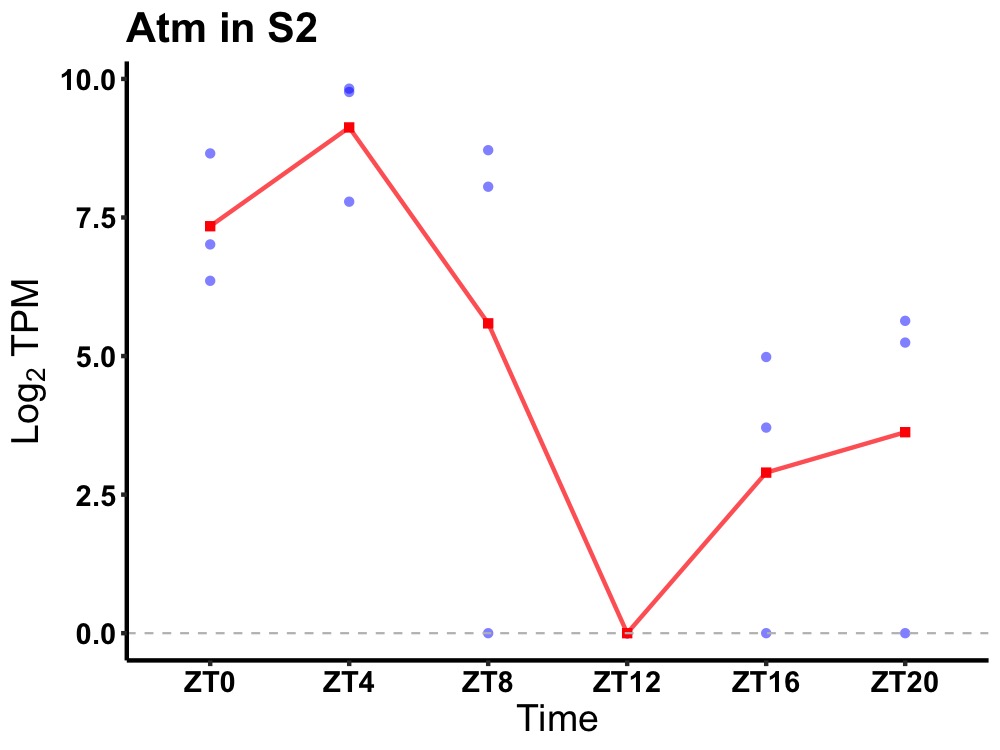 |
serine-protein kinase ATM | 0.019 | 24 | 4 | 4.28 |
| ENSMUSG00000036898 | Zfp157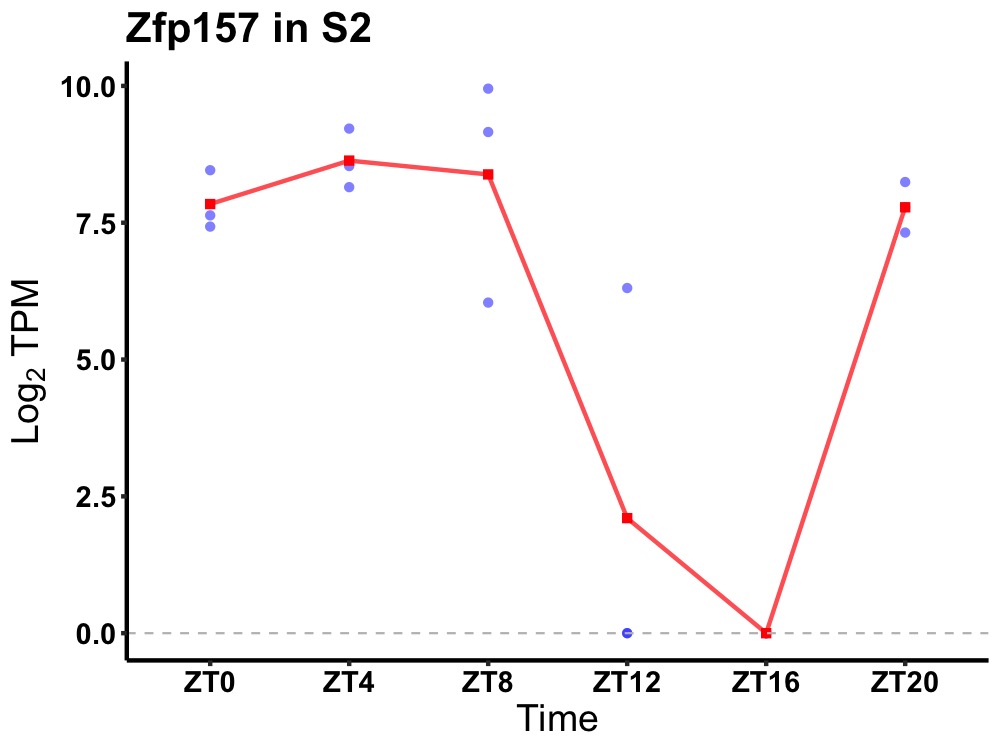 |
zinc finger protein 157 | 0.005 | 20 | 6 | 4.27 |
| ENSMUSG00000024871 | Doc2g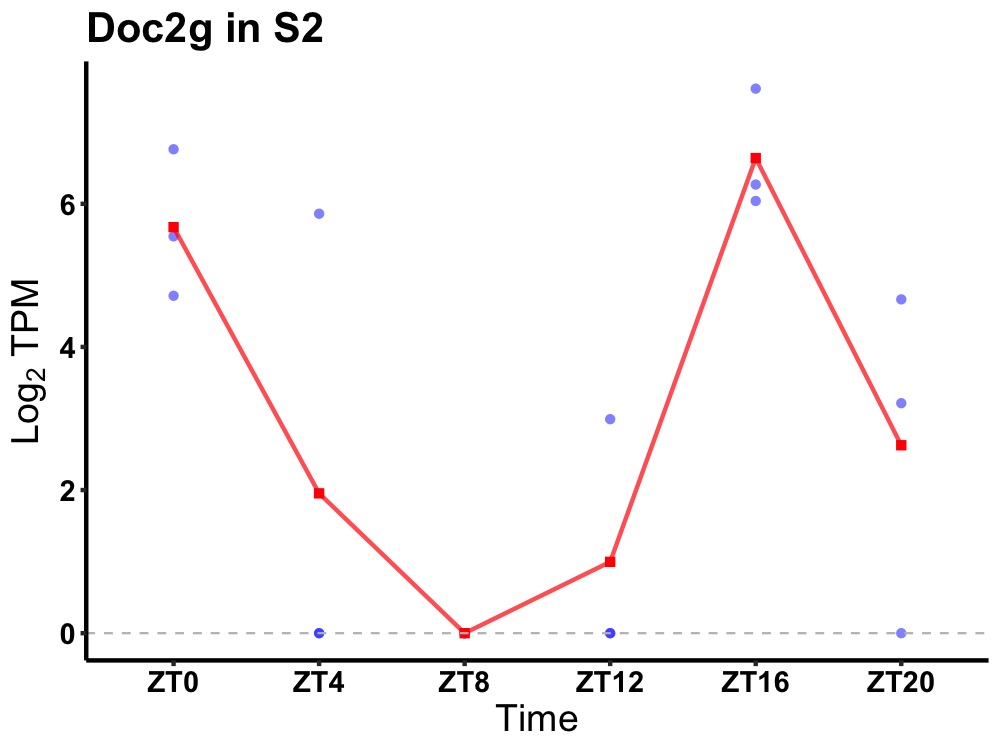 |
double C2-like domain-containing protein gamma | 0.006 | 20 | 18 | 4.27 |
| ENSMUSG00000092416 | Zfp141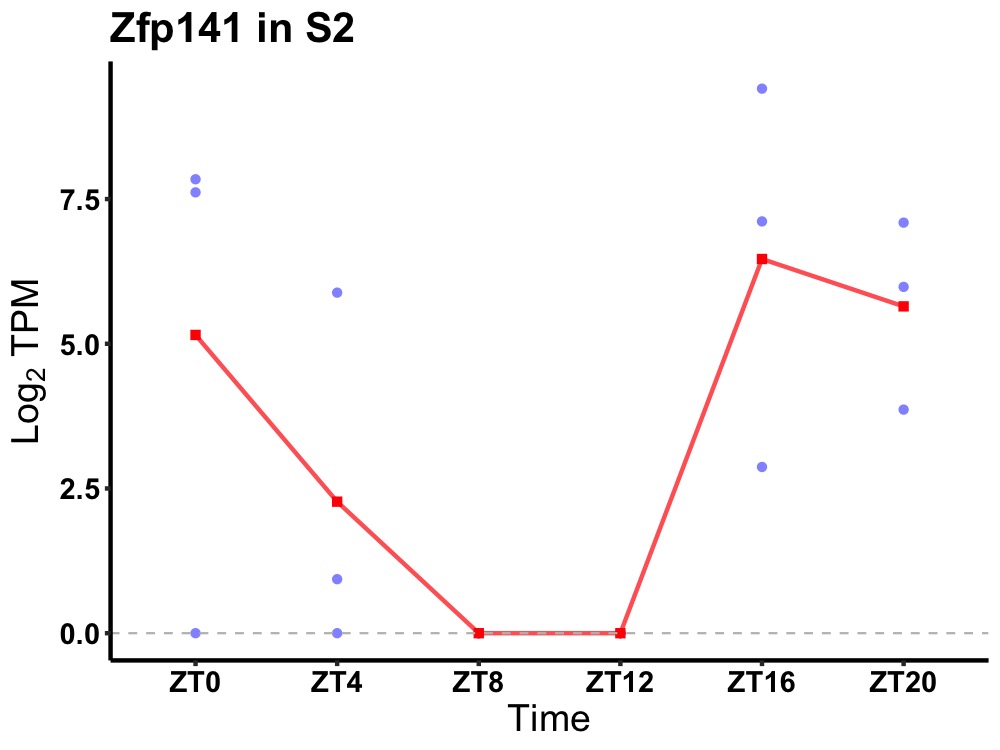 |
zinc finger protein 141 | 0.049 | 20 | 0 | 4.16 |
| ENSMUSG00000009681 | Bcr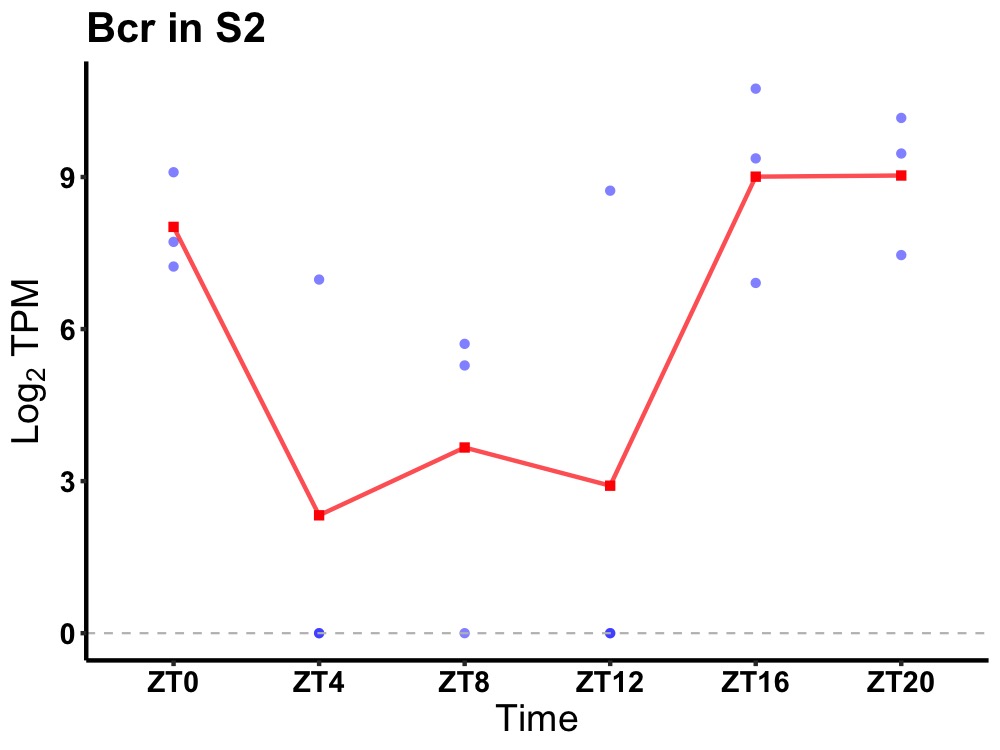 |
breakpoint cluster region protein | 0.036 | 24 | 20 | 4.02 |
| ENSMUSG00000099966 | 2810402E24Rik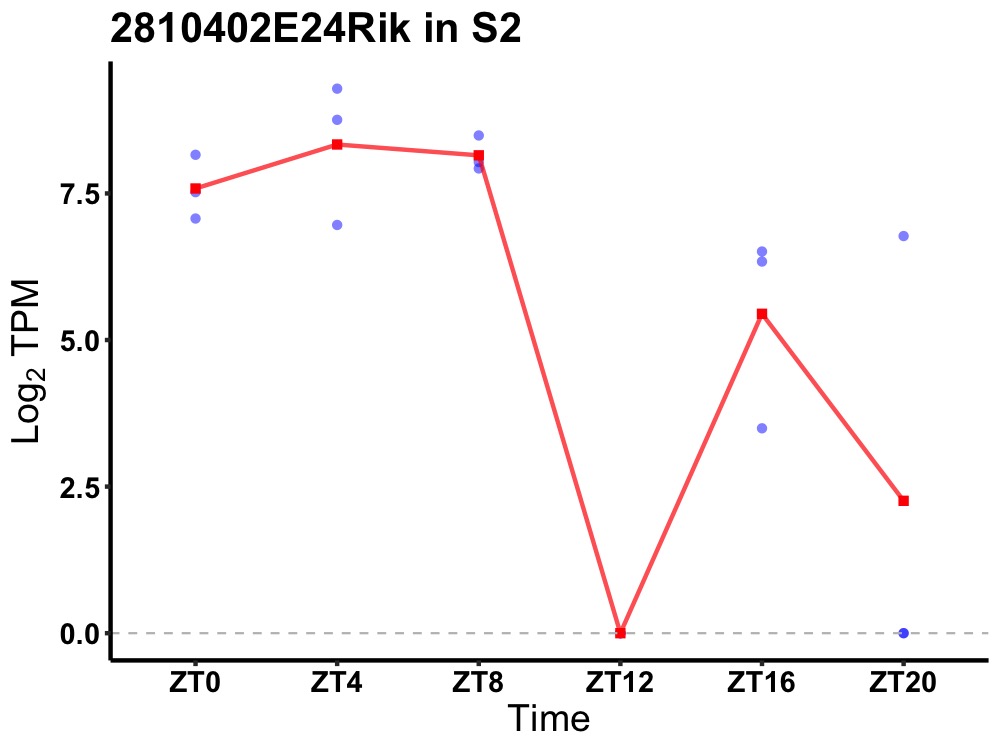 |
none | 0.049 | 24 | 6 | 4.02 |
| ENSMUSG00000056310 | Gtf2h4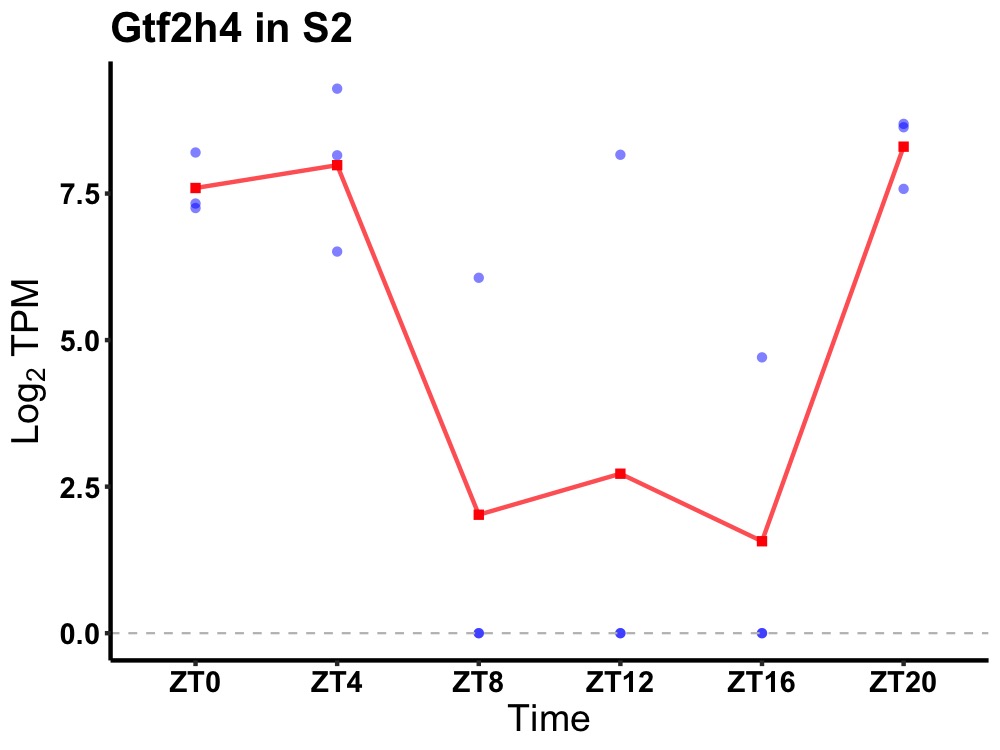 |
general transcription factor IIH subunit 4 | 0.049 | 20 | 4 | 3.99 |
| ENSMUSG00000032109 | Nlrx1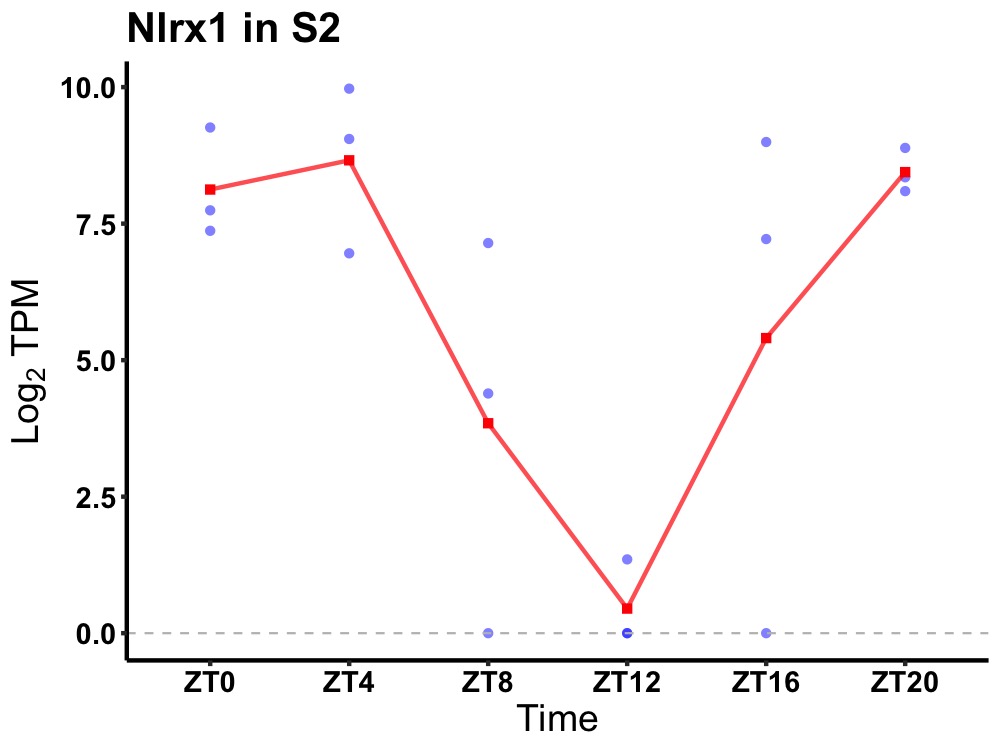 |
NLR family member X1 | 0.043 | 20 | 2 | 3.96 |
| ENSMUSG00000024769 | Cdc42bpg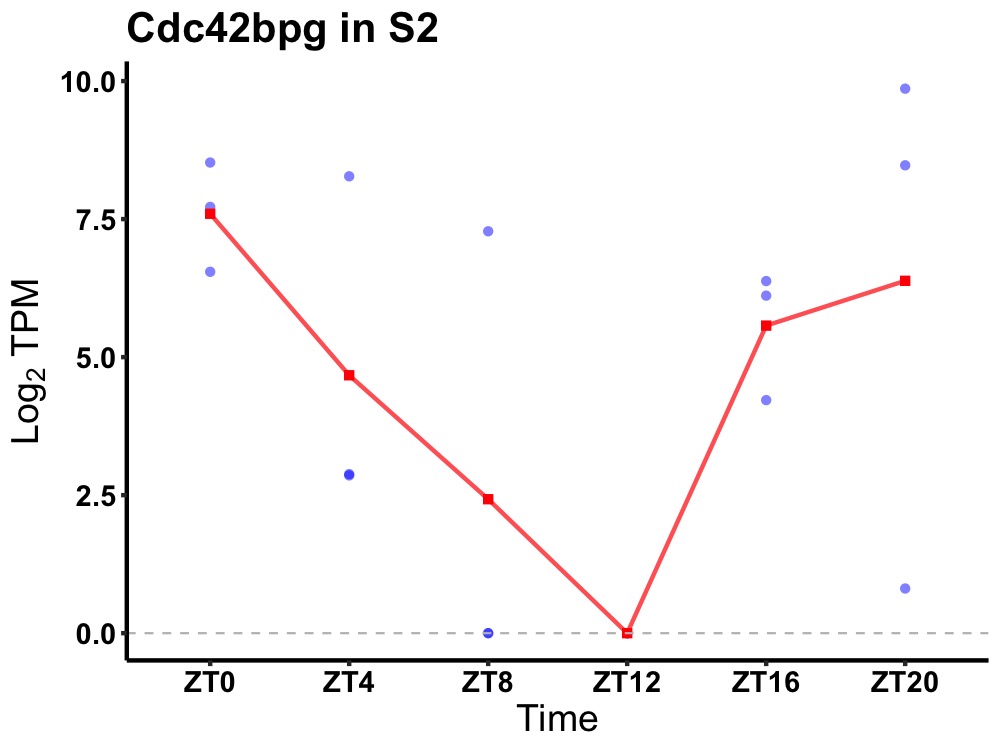 |
serine/threonine-protein kinase MRCK gamma | 0.036 | 24 | 22 | 3.96 |
| ENSMUSG00000018920 | Cxcl16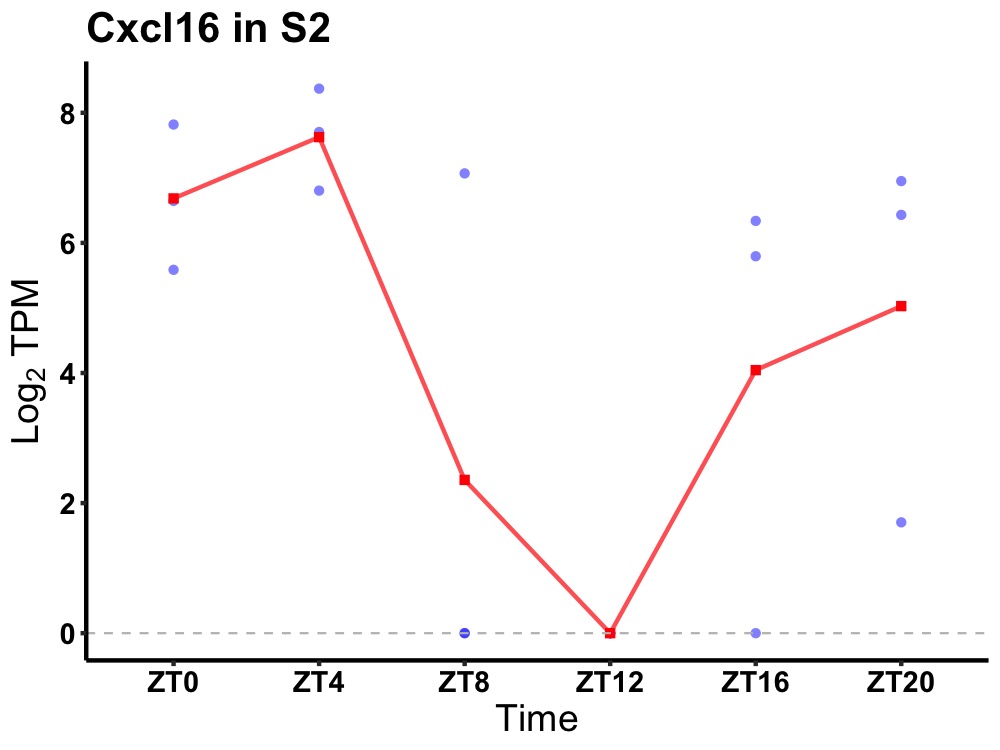 |
C-X-C motif chemokine 16 | 0.043 | 20 | 4 | 3.95 |
| ENSMUSG00000021770 | Sap130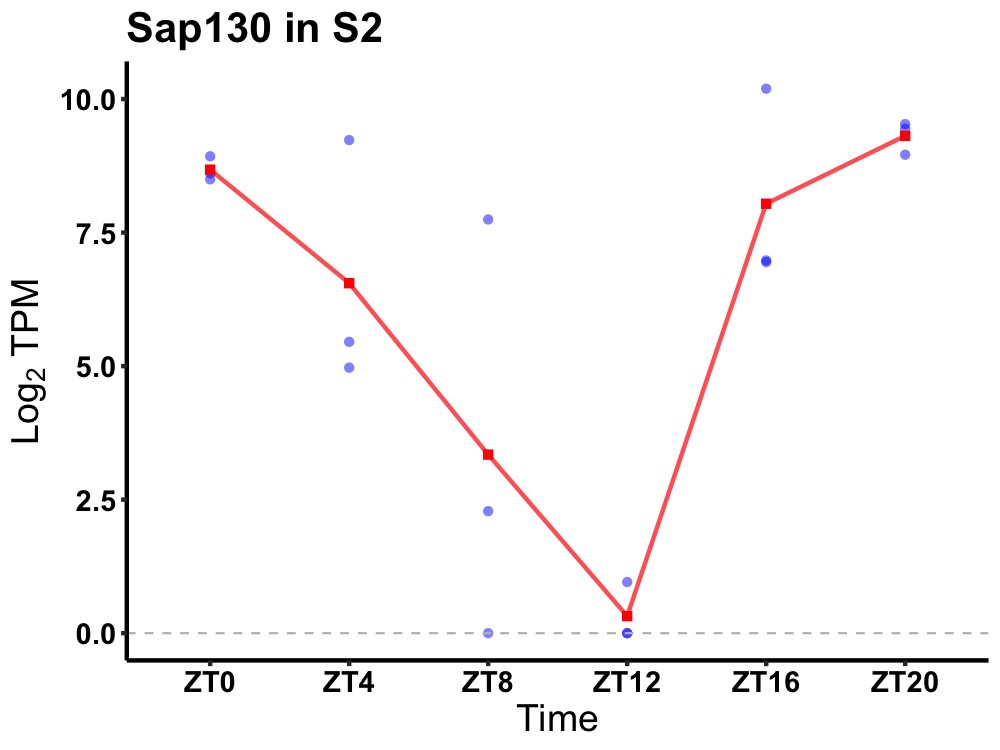 |
histone deacetylase complex subunit SAP130 | 0.007 | 24 | 22 | 3.86 |
| ENSMUSG00000024869 | Gm49405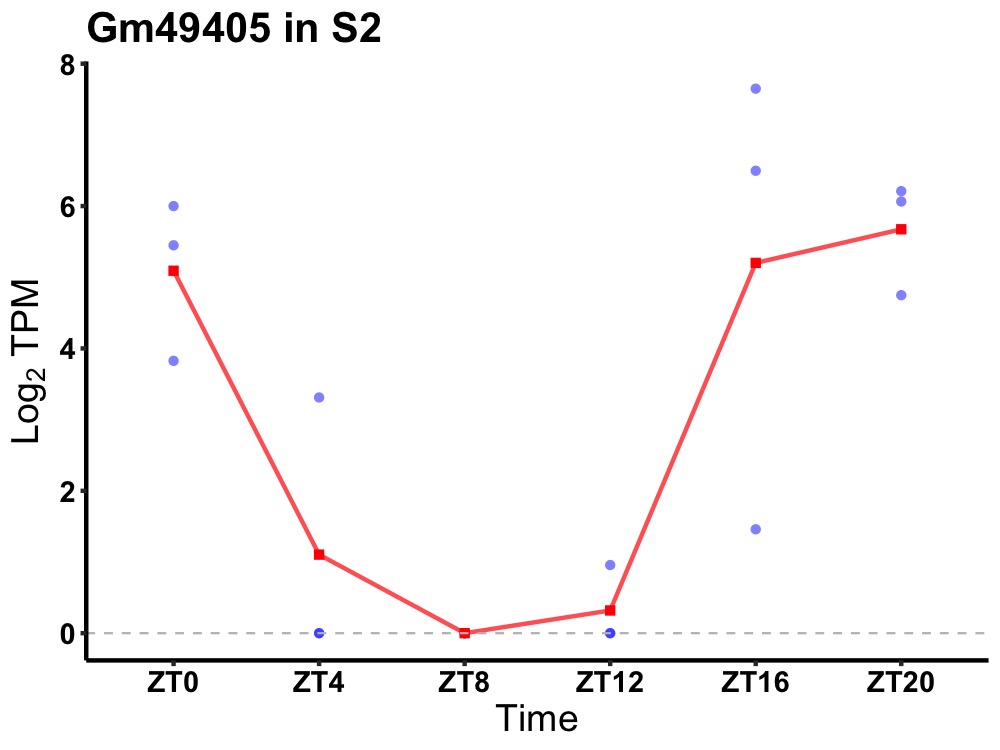 |
none | 0.007 | 24 | 20 | 3.85 |
| ENSMUSG00000007029 | Vars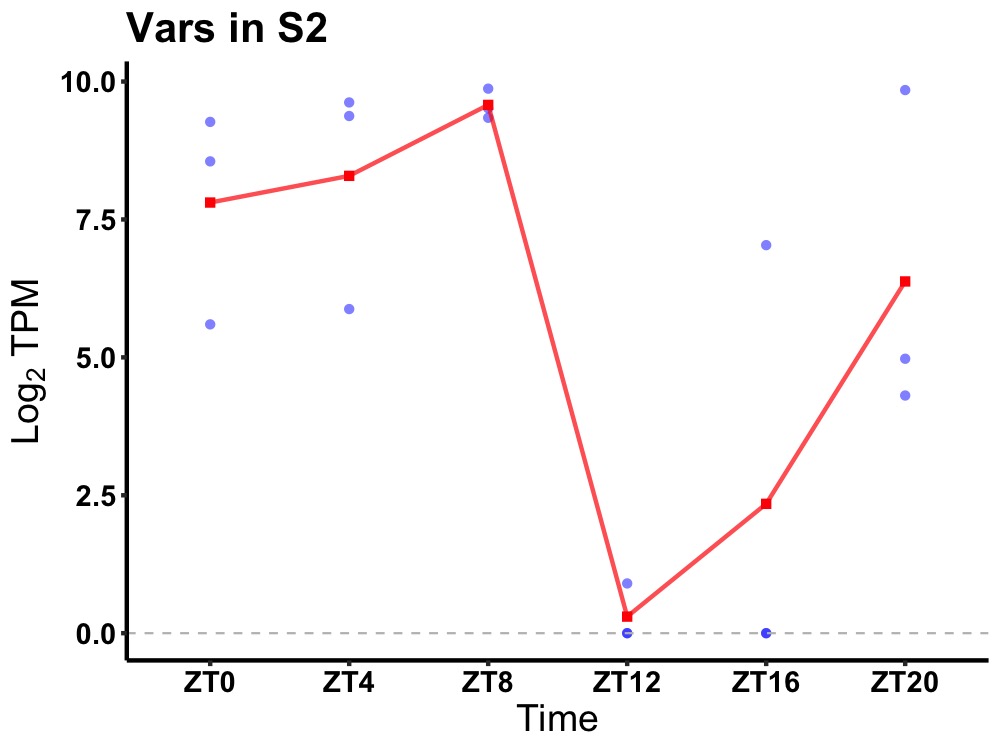 |
valine--tRNA ligase | 0.036 | 20 | 6 | 3.83 |
| ENSMUSG00000024891 | Slc29a2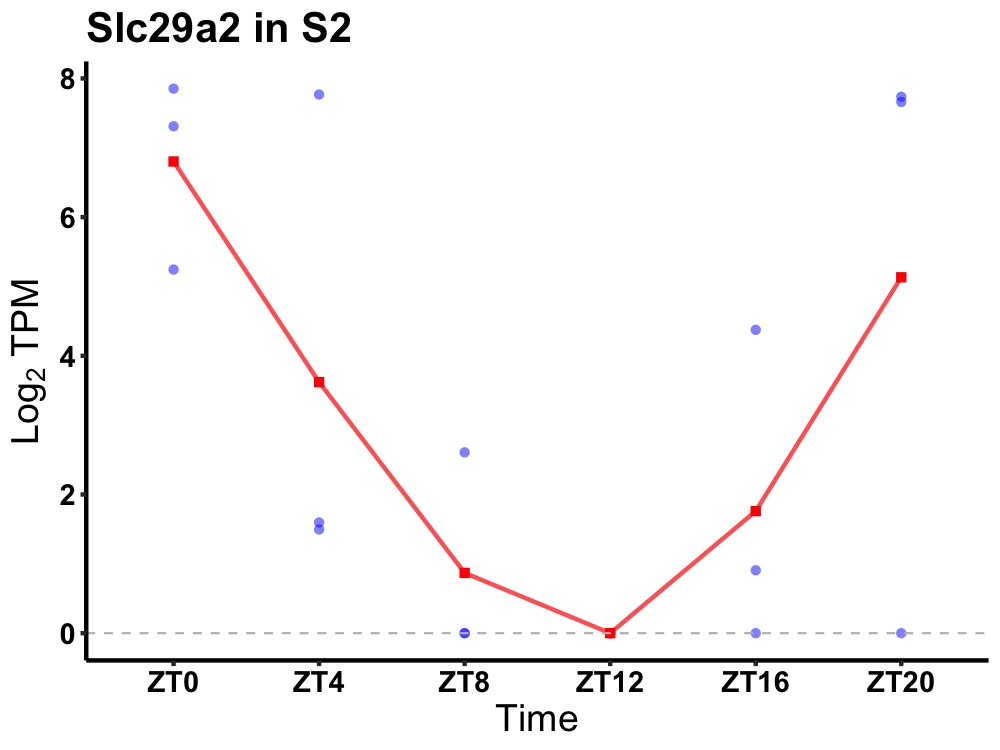 |
equilibrative nucleoside transporter 2 | 0.043 | 24 | 0 | 3.71 |
| ENSMUSG00000033790 | Tubgcp5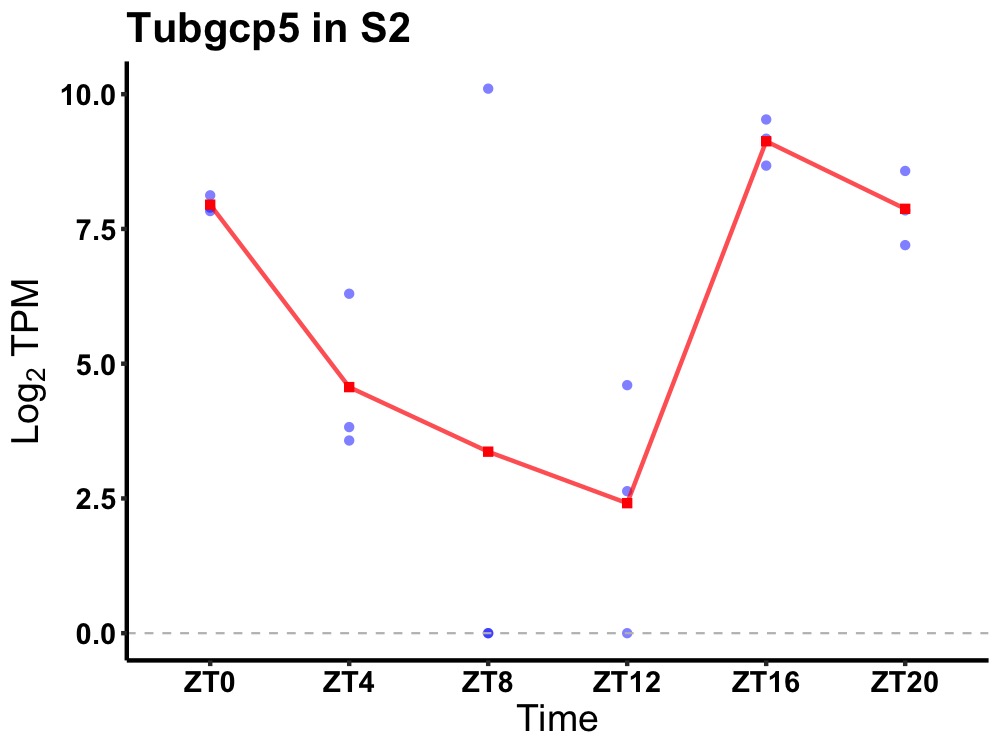 |
gamma-tubulin complex component 5 | 0.027 | 20 | 18 | 3.68 |
| ENSMUSG00000026960 | Arl6ip6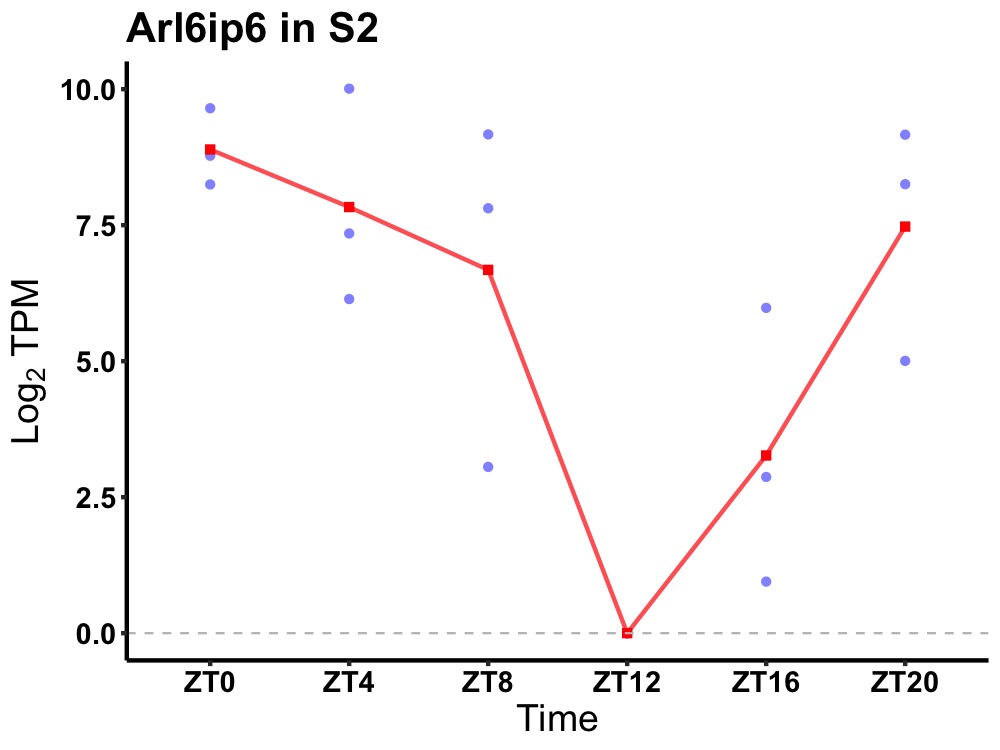 |
ADP-ribosylation factor-like protein 6-interacting protein 6 | 0.005 | 24 | 2 | 3.67 |
| ENSMUSG00000001415 | Fam222b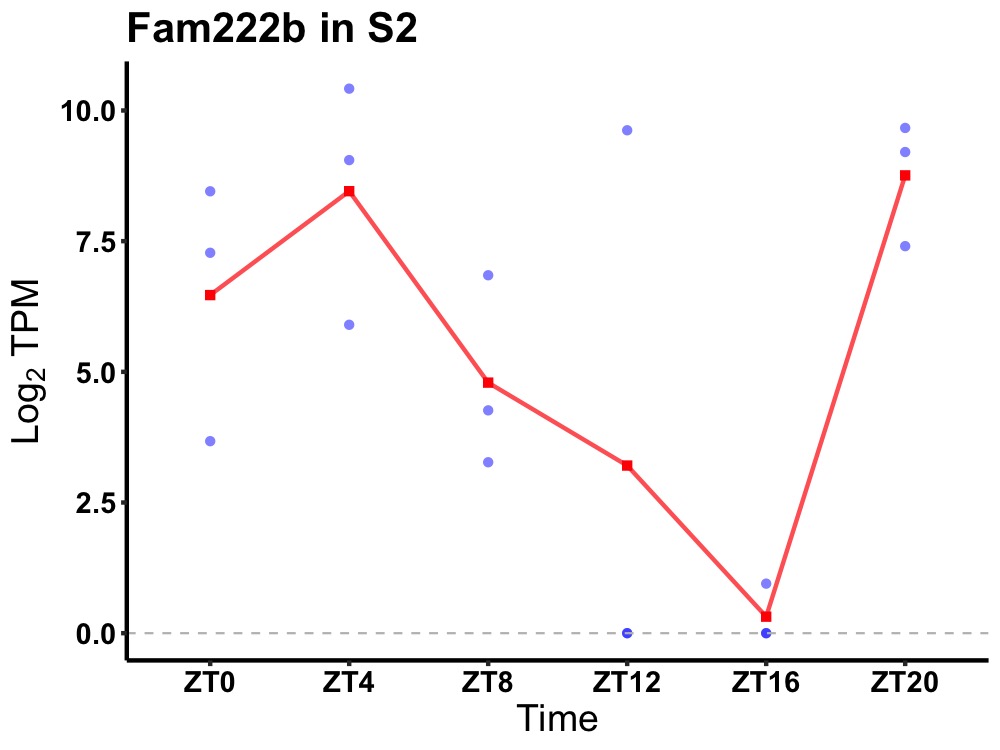 |
protein FAM222B | 0.019 | 20 | 4 | 3.65 |
| ENSMUSG00000002250 | Ppard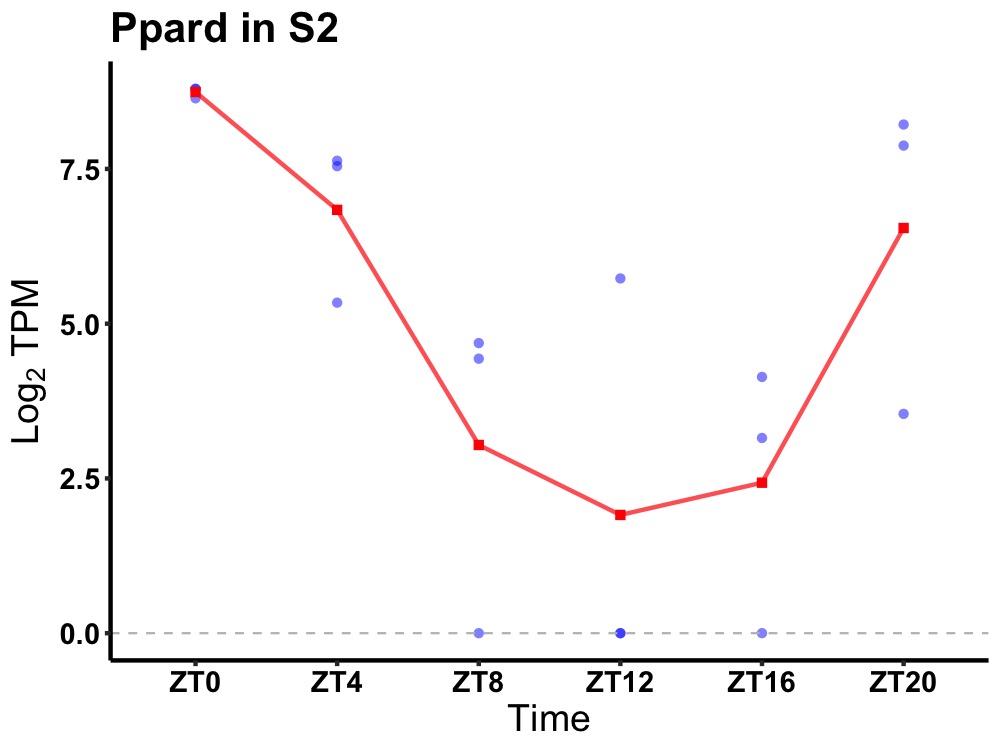 |
peroxisome proliferator-activated receptor delta | 0.004 | 24 | 2 | 3.57 |
| ENSMUSG00000074217 | 2210011C24Rik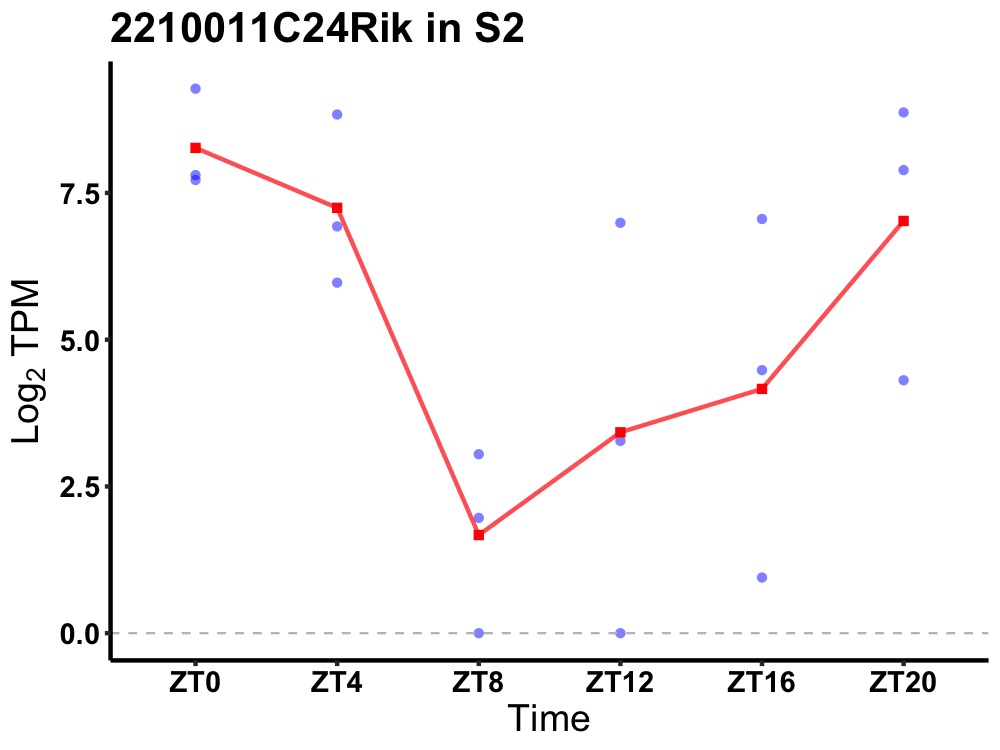 |
none | 0.043 | 24 | 0 | 3.30 |
| ENSMUSG00000027907 | Fzd1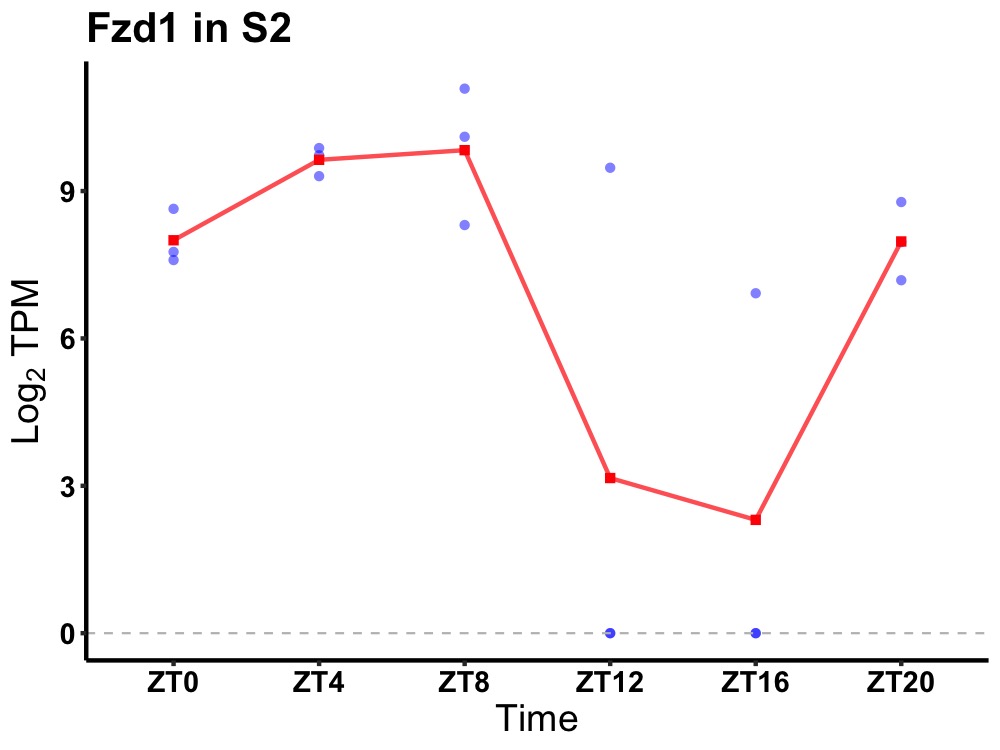 |
frizzled-1 | 0.010 | 20 | 6 | 3.29 |
| ENSMUSG00000043183 | Simc1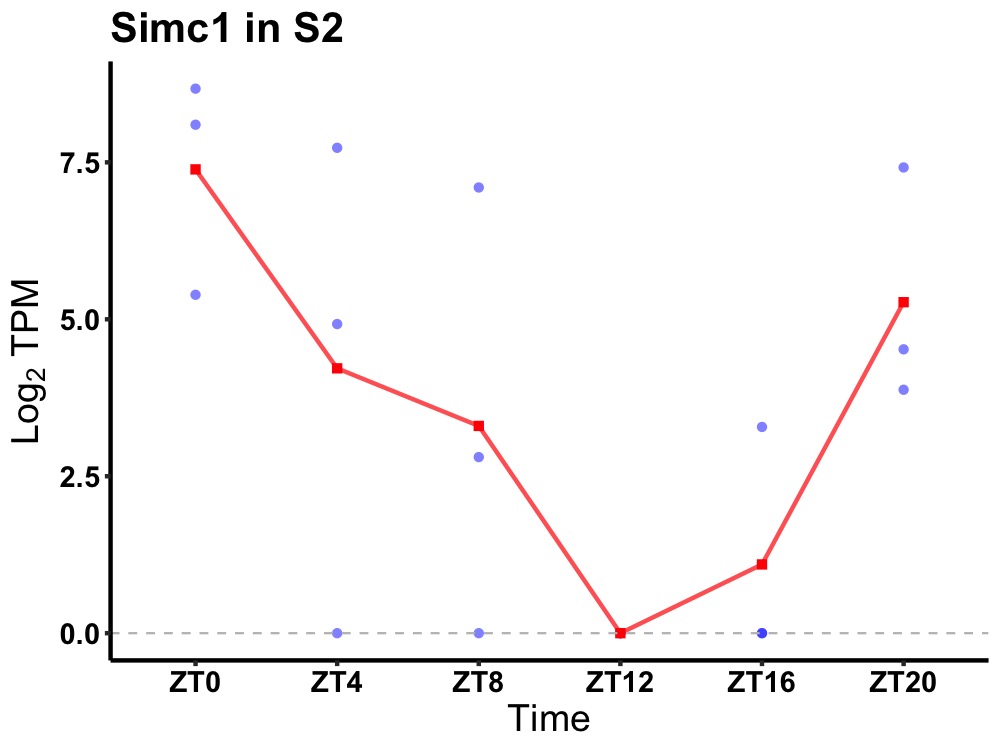 |
SUMO-interacting motif-containing protein 1 | 0.008 | 24 | 2 | 3.26 |
| ENSMUSG00000060935 | Tmem263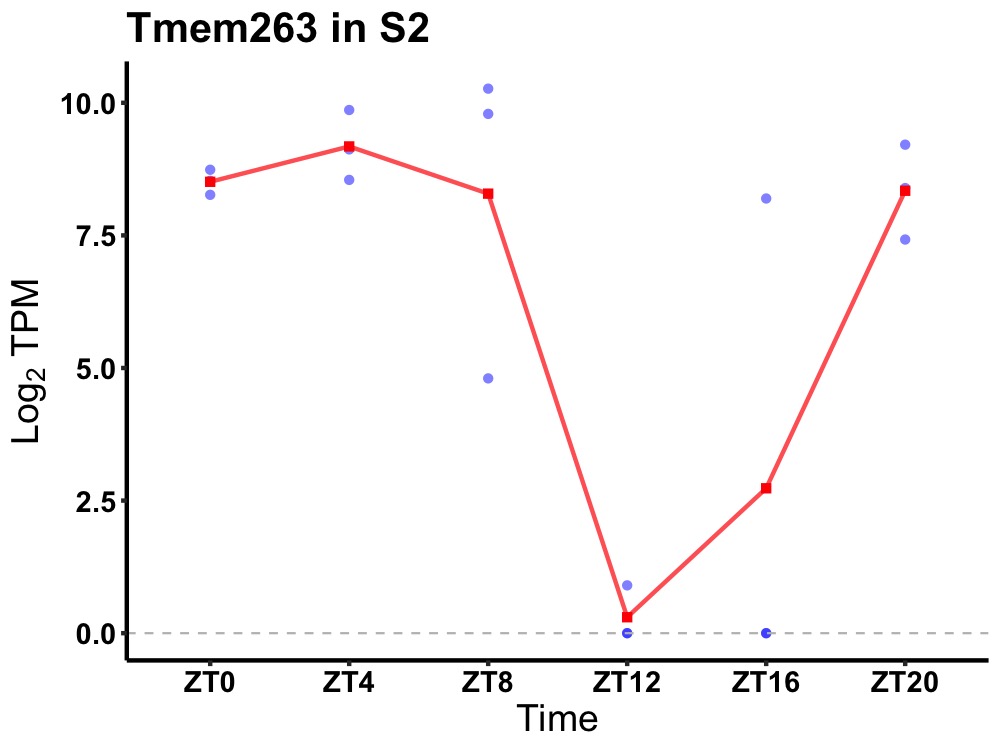 |
transmembrane protein 263 | 0.019 | 20 | 6 | 3.25 |
| ENSMUSG00000026269 | Golga1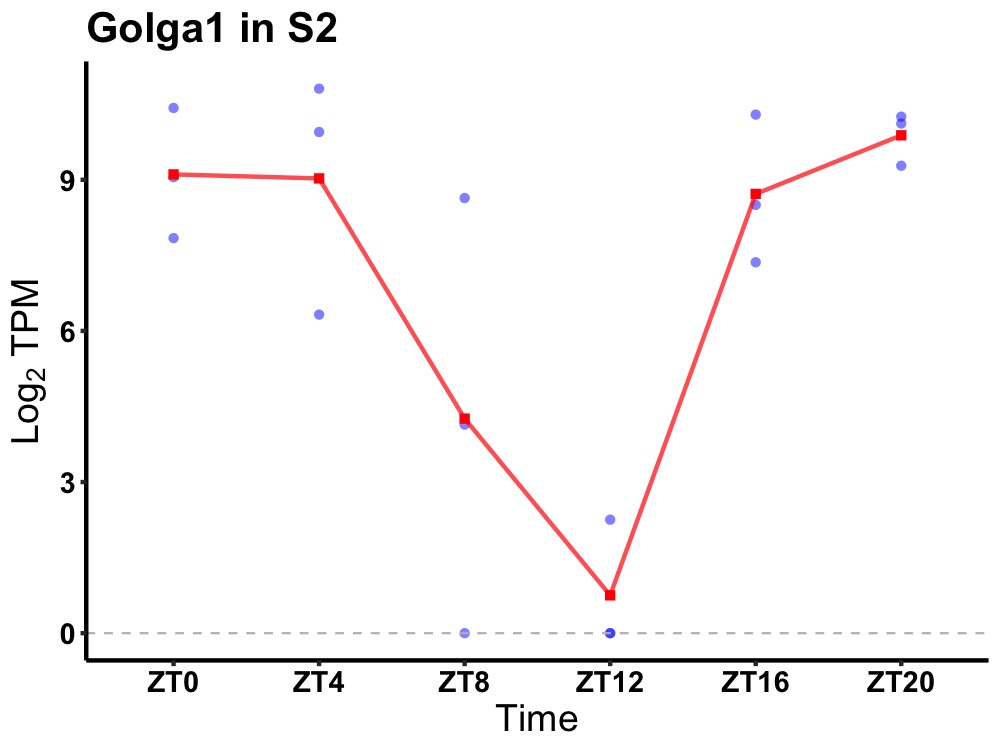 |
golgin subfamily A member 1 | 0.036 | 20 | 2 | 3.12 |
| ENSMUSG00000037780 | Mbl1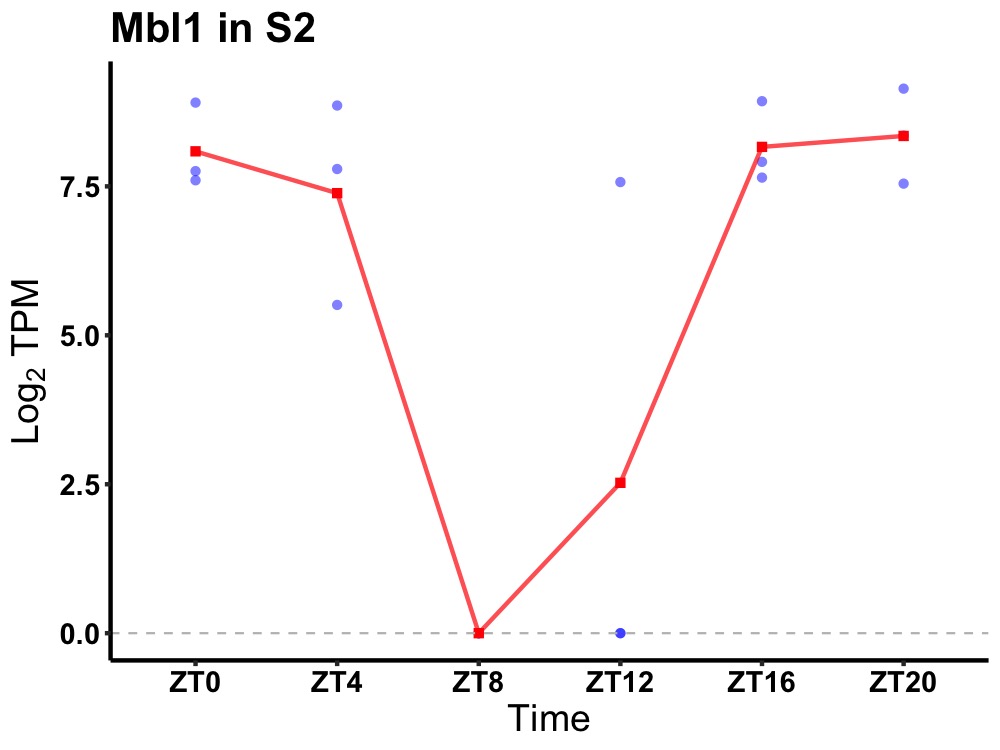 |
mannose-binding protein A | 0.049 | 24 | 22 | 3.12 |
| ENSMUSG00000020484 | Ing1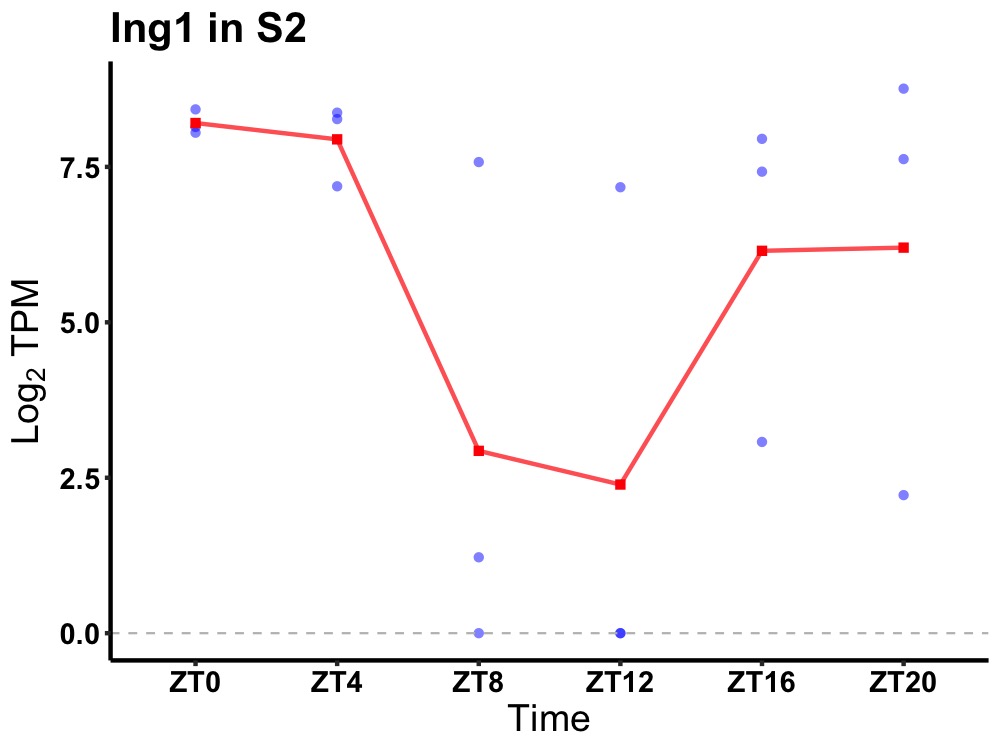 |
inhibitor of growth protein 1 | 0.027 | 24 | 0 | 3.08 |
| ENSMUSG00000030505 | Prmt3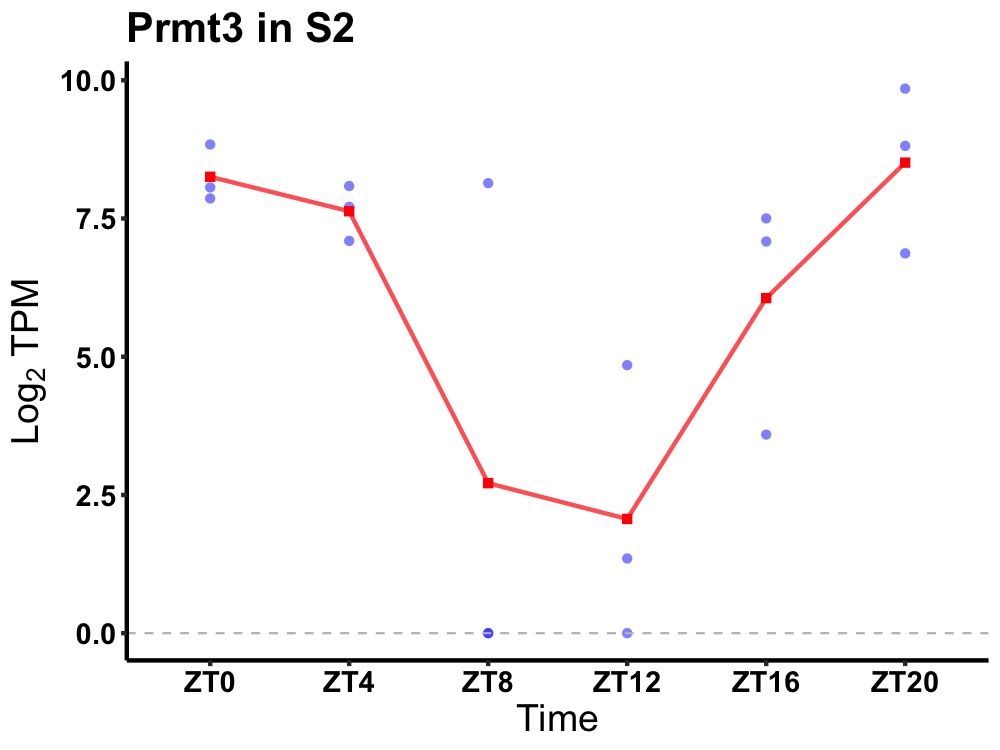 |
protein arginine N-methyltransferase 3 | 0.027 | 20 | 2 | 3.06 |
| ENSMUSG00000023460 | Rab12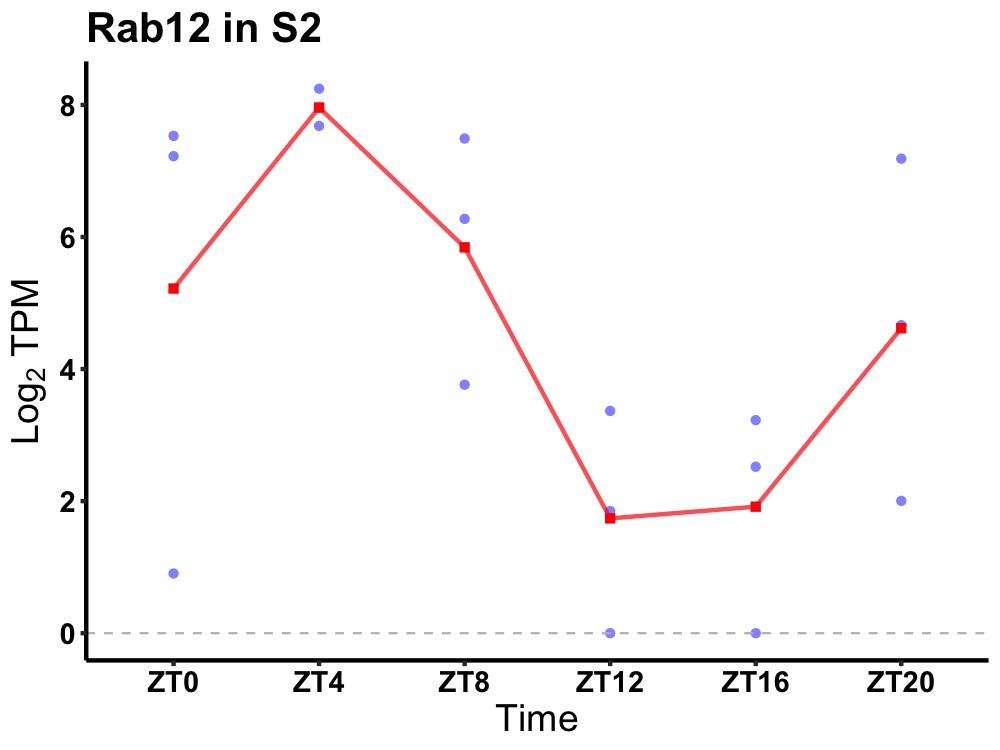 |
ras-related protein Rab-12 | 0.004 | 24 | 4 | 3.04 |
| ENSMUSG00000099083 | Atf7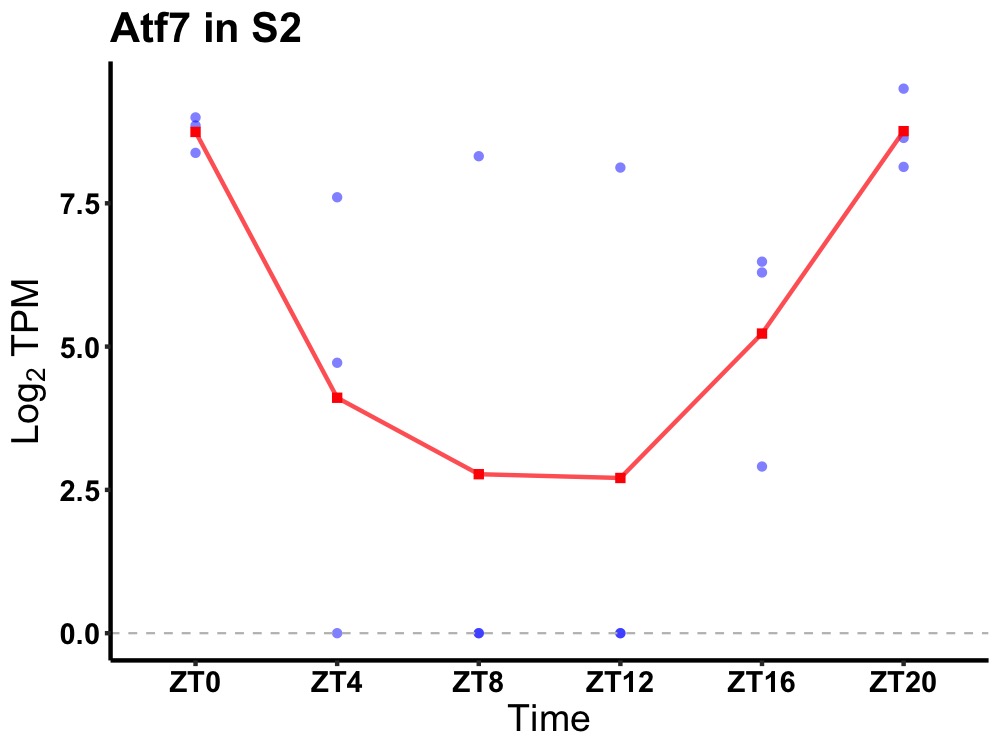 |
cyclic AMP-dependent transcription factor ATF-7 | 0.036 | 24 | 0 | 2.98 |
| ENSMUSG00000083327 | Osbpl11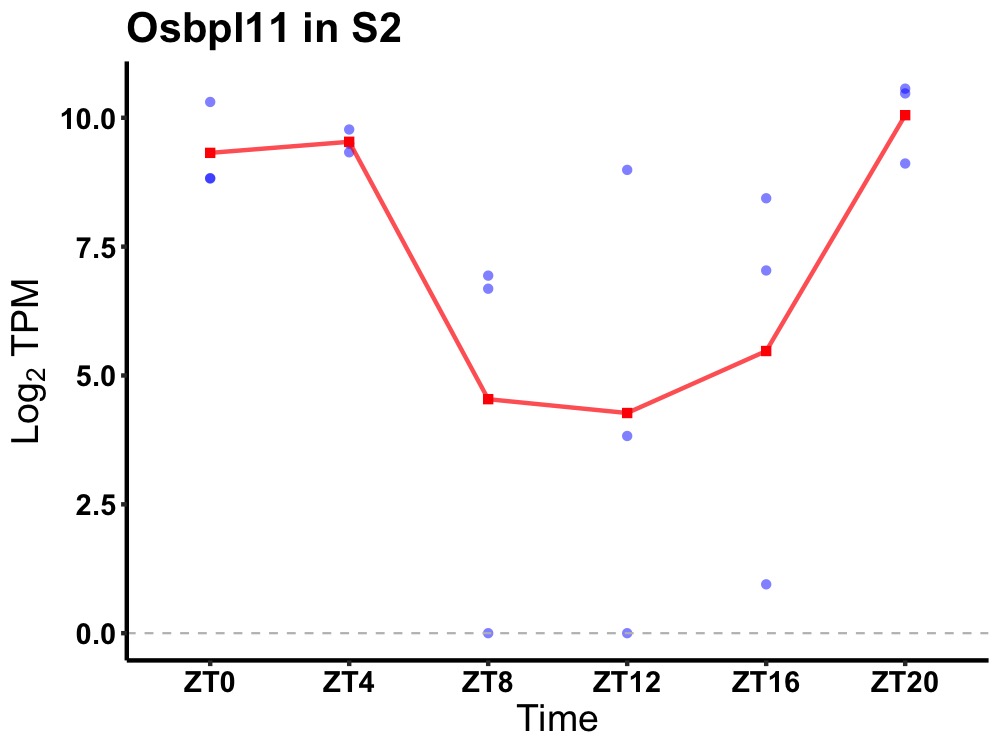 |
oxysterol-binding protein-related protein 11 | 0.006 | 20 | 2 | 2.94 |
| ENSMUSG00000070806 | Zmynd12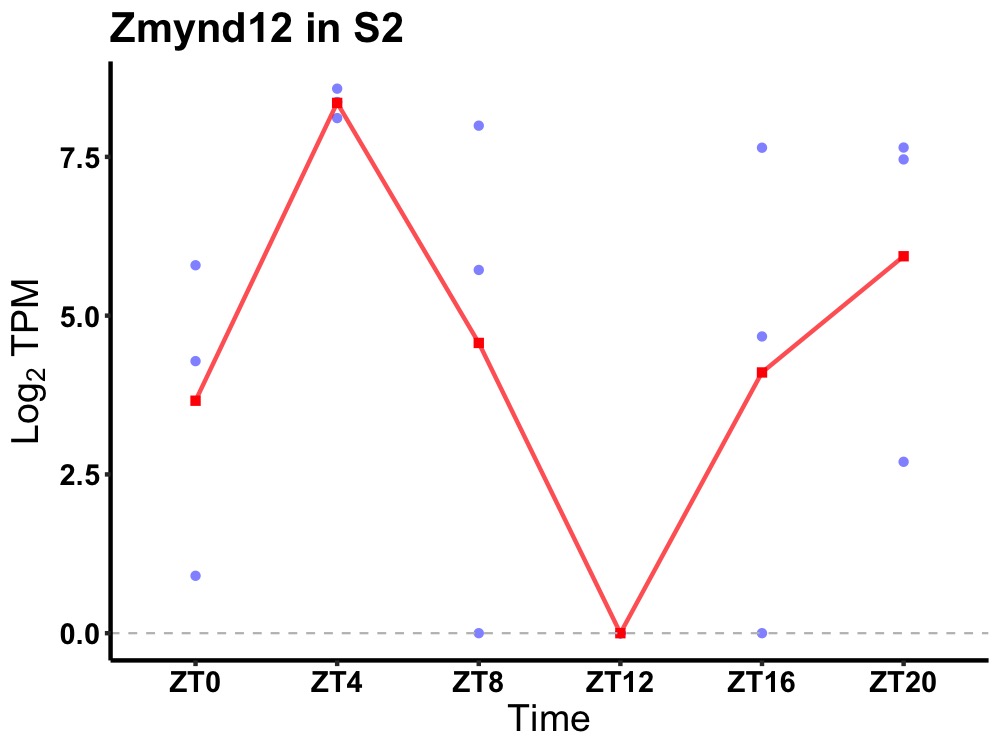 |
zinc finger MYND domain-containing protein 12 | 0.016 | 20 | 4 | 2.81 |
| ENSMUSG00000071176 | Arhgef10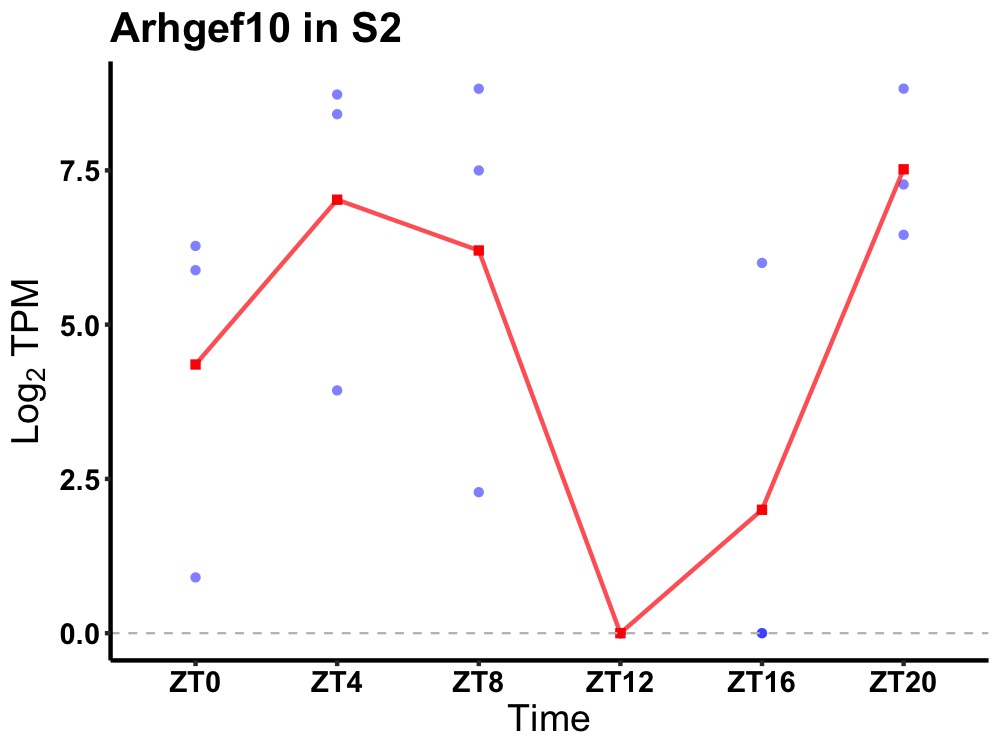 |
rho guanine nucleotide exchange factor 10 | 0.036 | 20 | 6 | 2.78 |
| ENSMUSG00000042396 | Traf6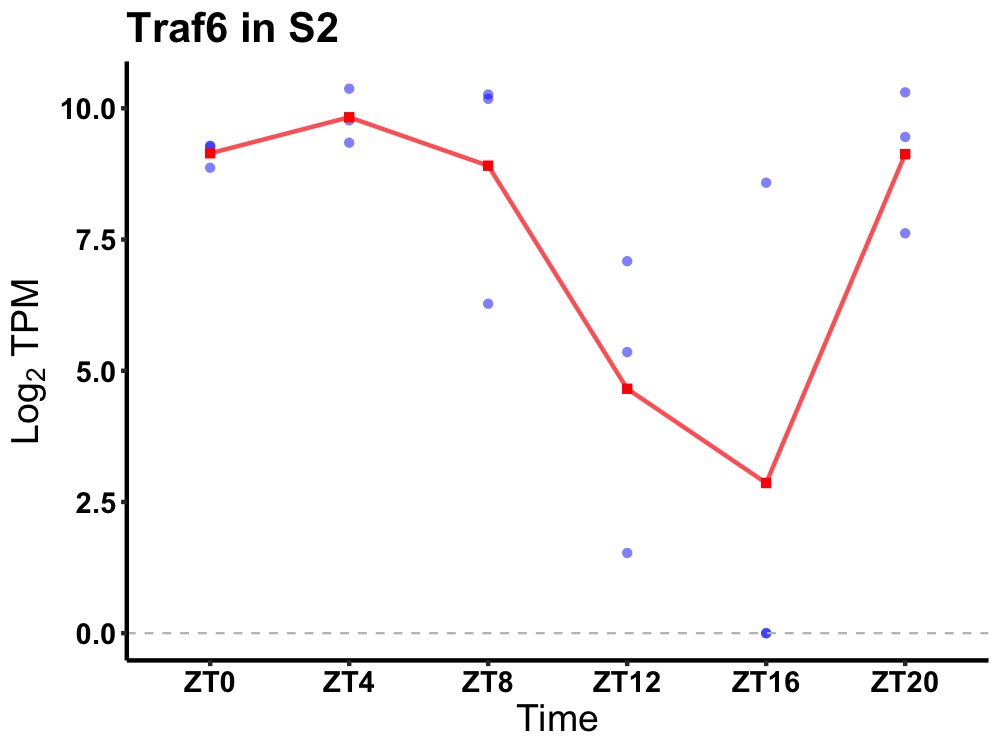 |
TNF receptor-associated factor 6 | 0.010 | 20 | 6 | 2.77 |
| ENSMUSG00000114255 | Gm10734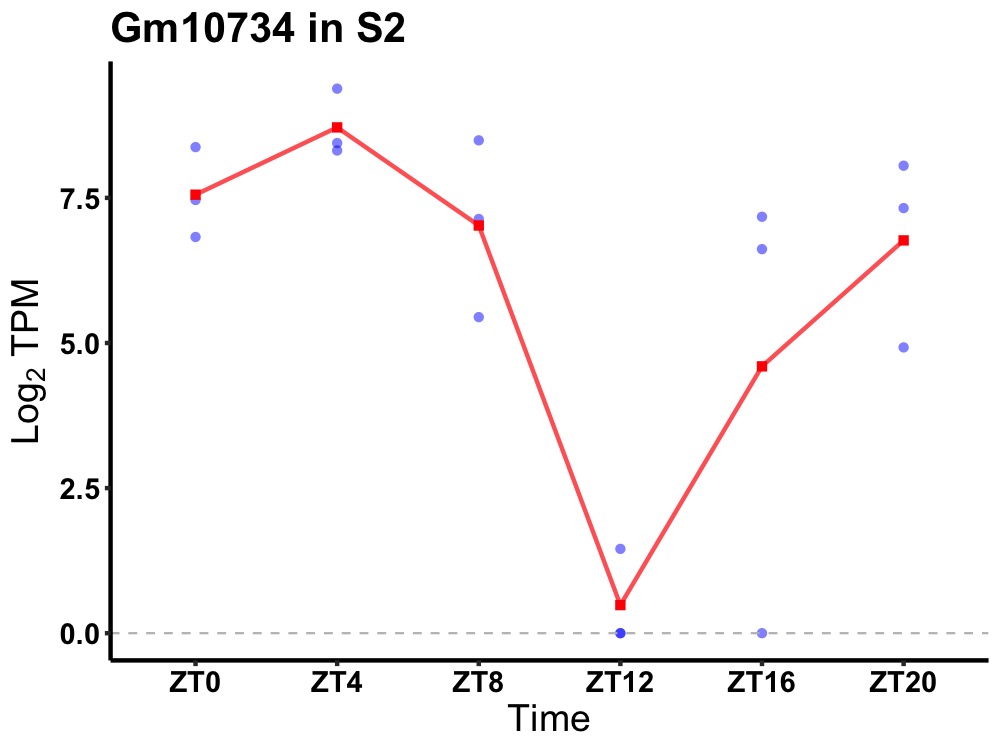 |
none | 0.019 | 20 | 4 | 2.75 |
| ENSMUSG00000053964 | Lgals4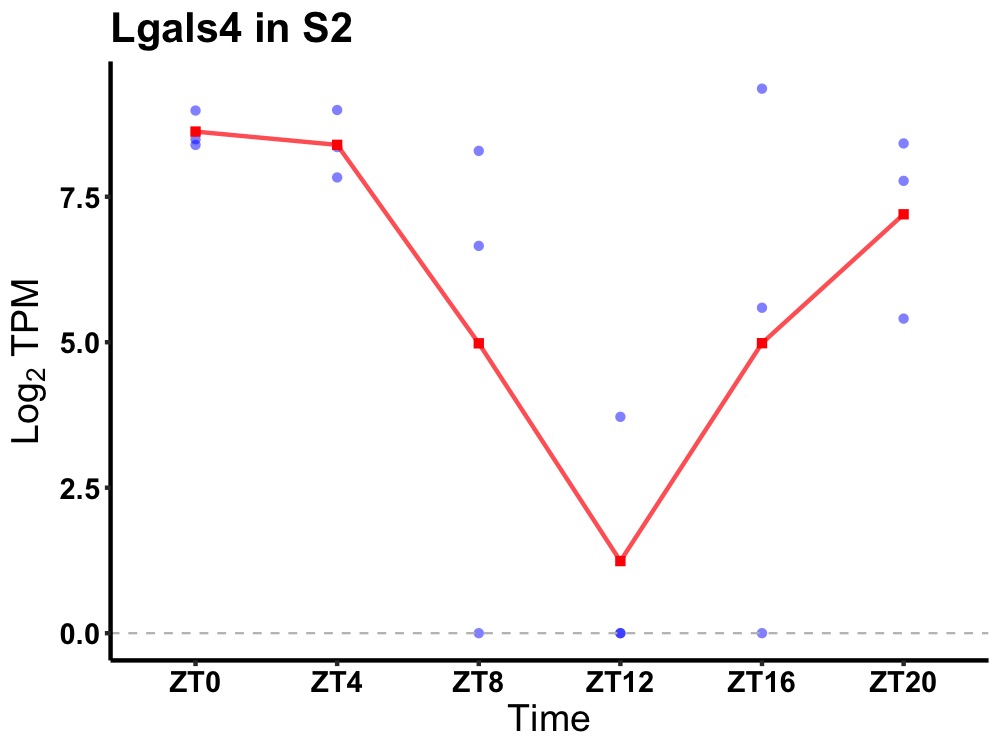 |
galectin-4 | 0.043 | 24 | 2 | 2.70 |
| ENSMUSG00000020427 | Igfbp3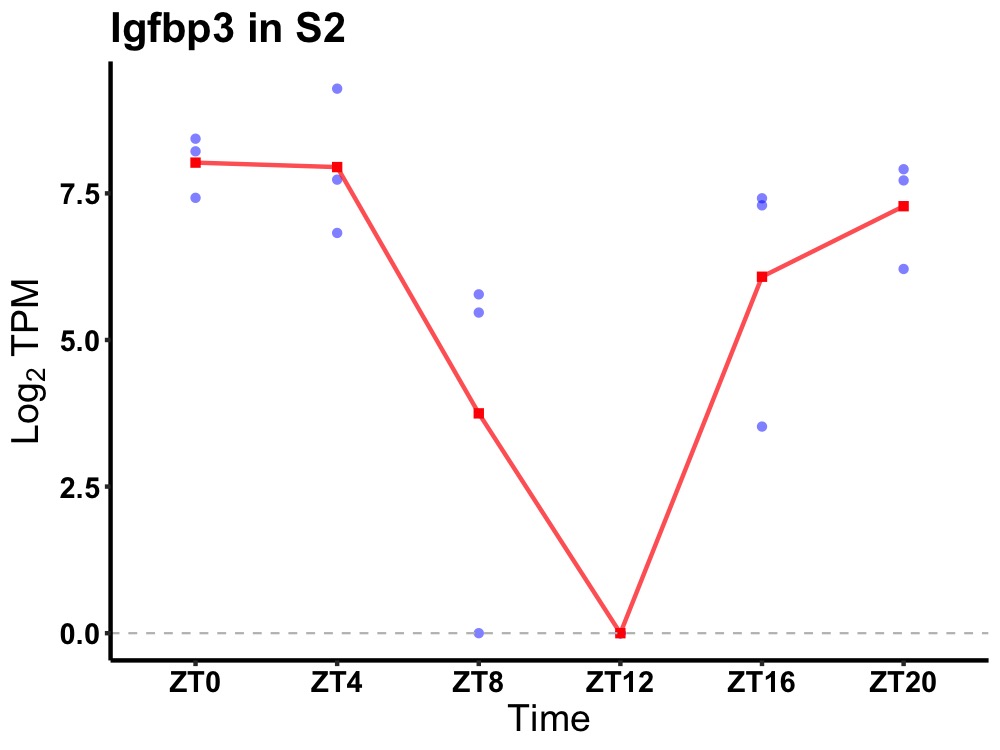 |
insulin-like growth factor-binding protein 3 | 0.002 | 24 | 0 | 2.70 |
| ENSMUSG00000047710 | Champ1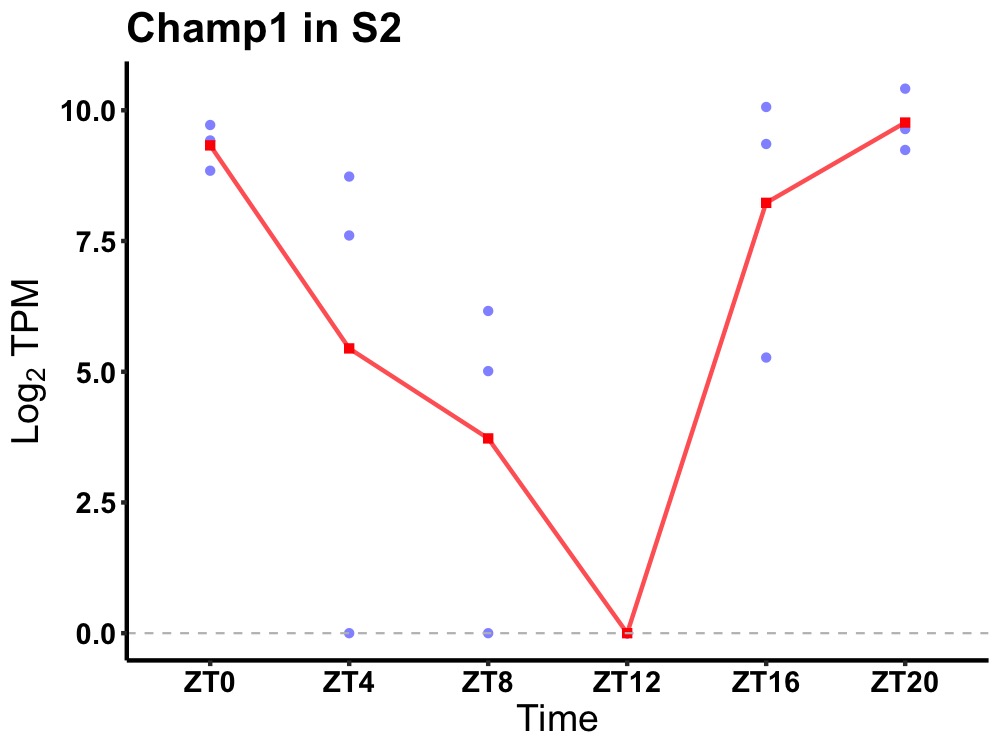 |
chromosome alignment-maintaining phosphoprotein 1 | 0.008 | 20 | 2 | 2.62 |
| ENSMUSG00000036617 | Etl4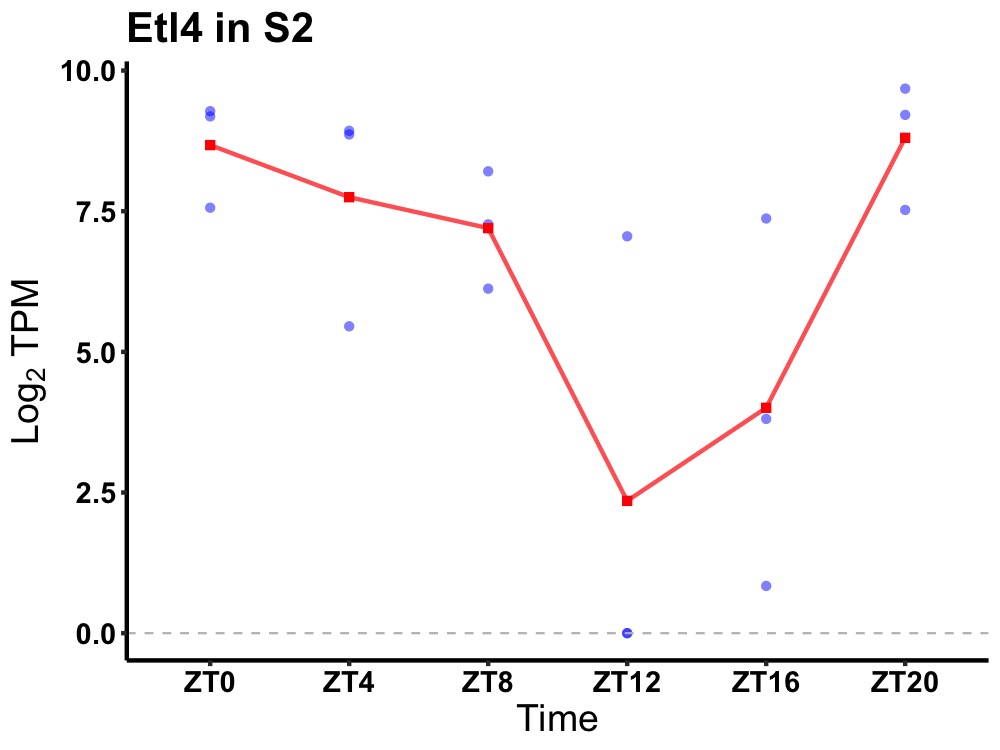 |
sickle tail protein | 0.014 | 20 | 2 | 2.52 |
| ENSMUSG00000024785 | Arhgap35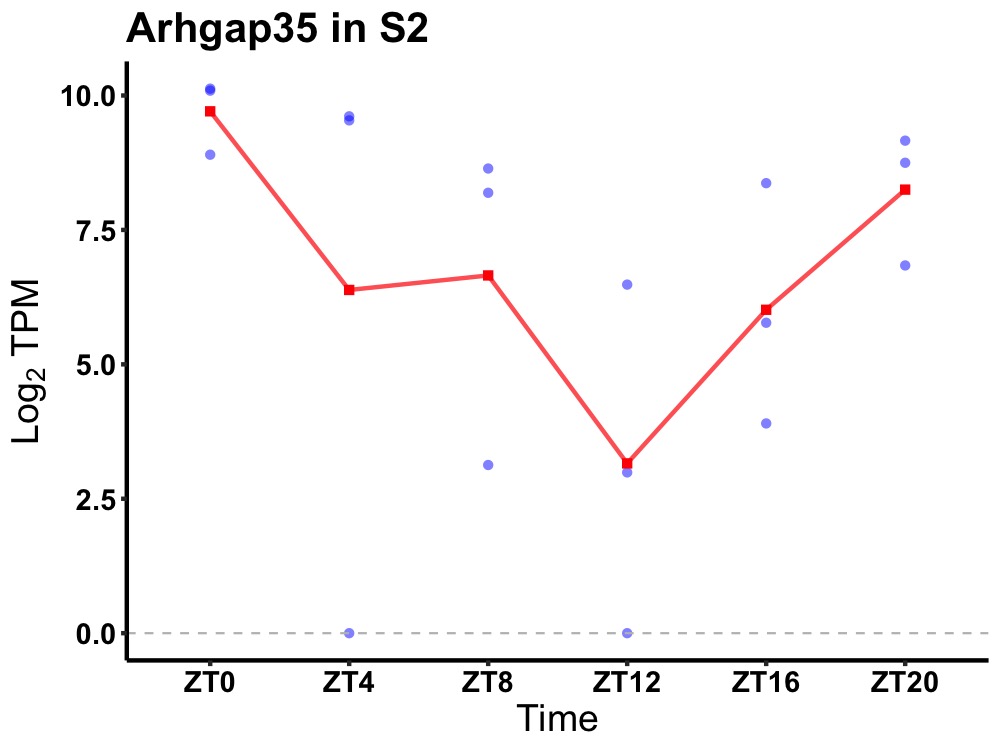 |
rho GTPase-activating protein 35 | 0.012 | 24 | 2 | 2.48 |
| ENSMUSG00000062480 | Acat3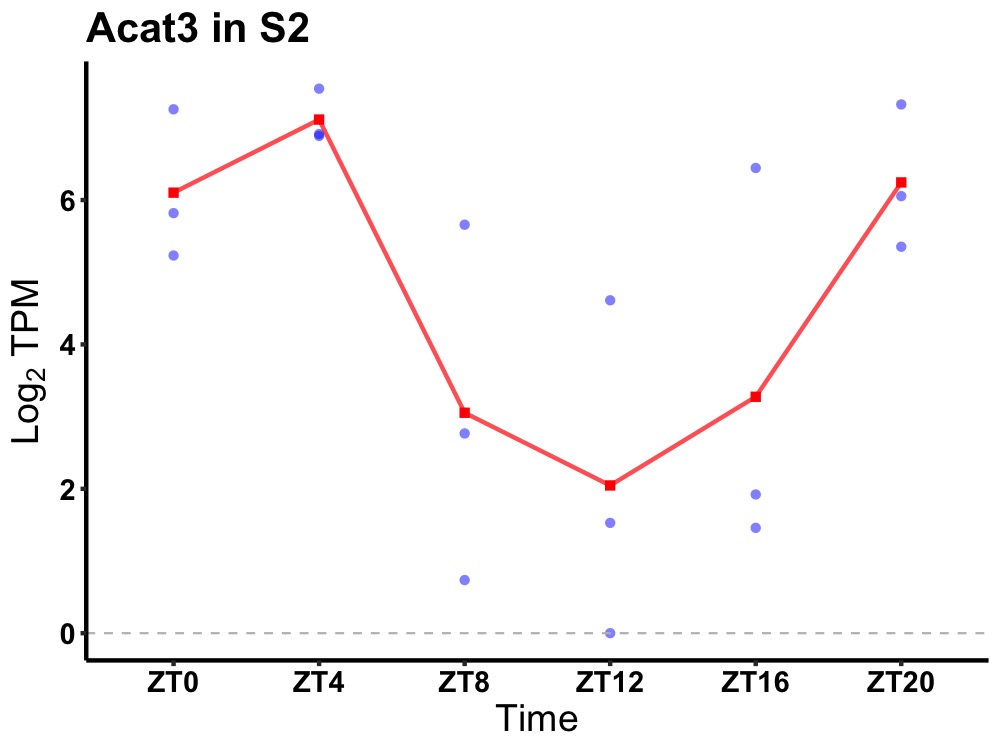 |
acetyl-Coenzyme A acetyltransferase 3 | 0.010 | 20 | 4 | 2.47 |
| ENSMUSG00000106224 | Gm43823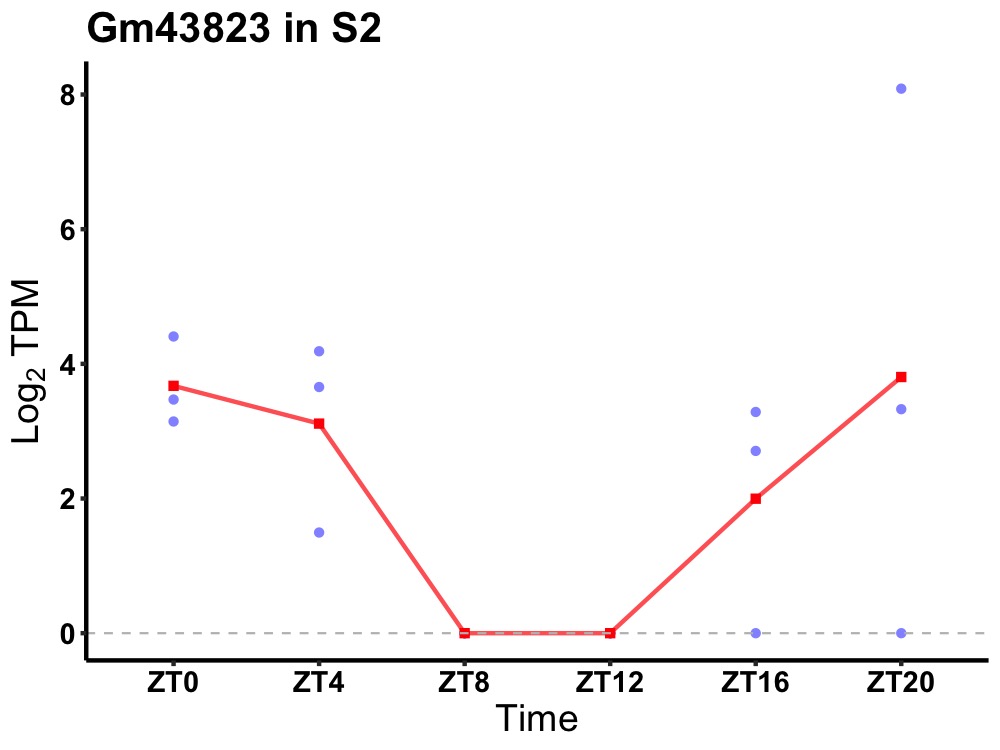 |
none | 0.049 | 24 | 0 | 2.45 |
| ENSMUSG00000031577 | Tti2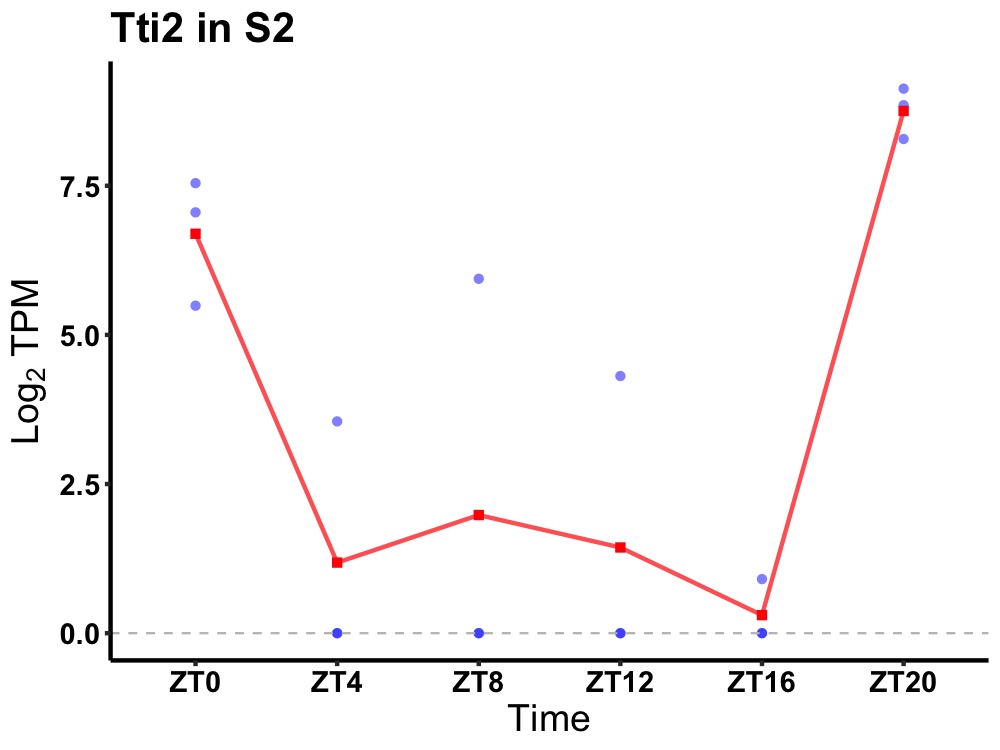 |
TELO2-interacting protein 2 | 0.049 | 20 | 2 | 2.41 |
| ENSMUSG00000052137 | Rbm12b2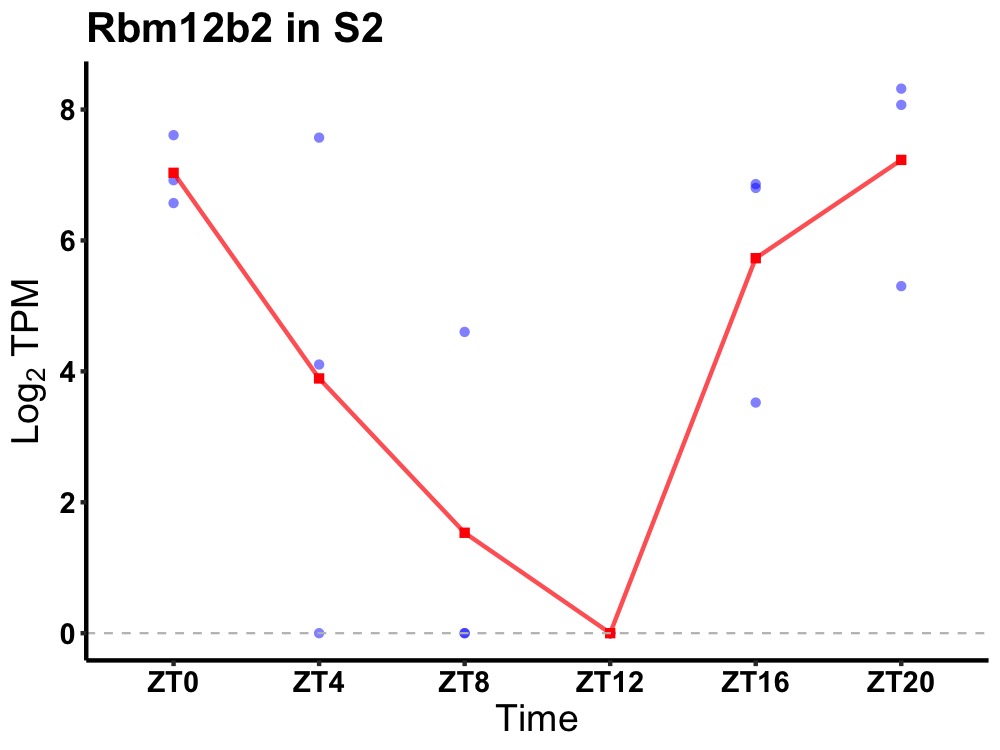 |
RNA-binding protein 12B-B | 0.008 | 20 | 2 | 2.40 |
| ENSMUSG00000037652 | Phc3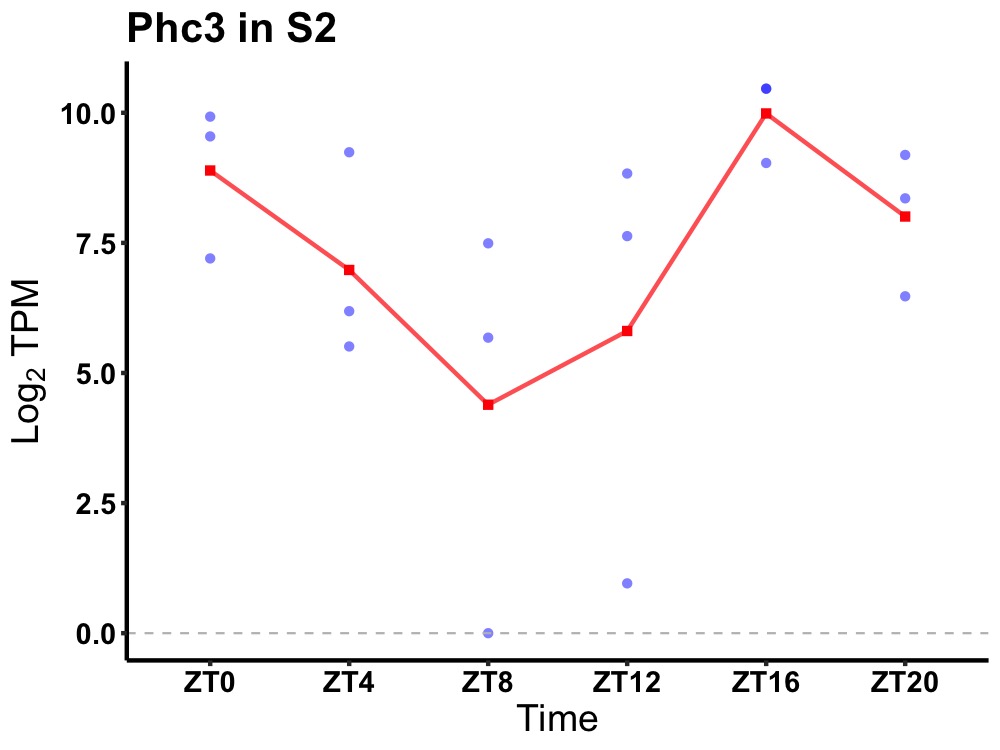 |
polyhomeotic 3 | 0.027 | 20 | 18 | 2.37 |
| ENSMUSG00000021731 | Mrps30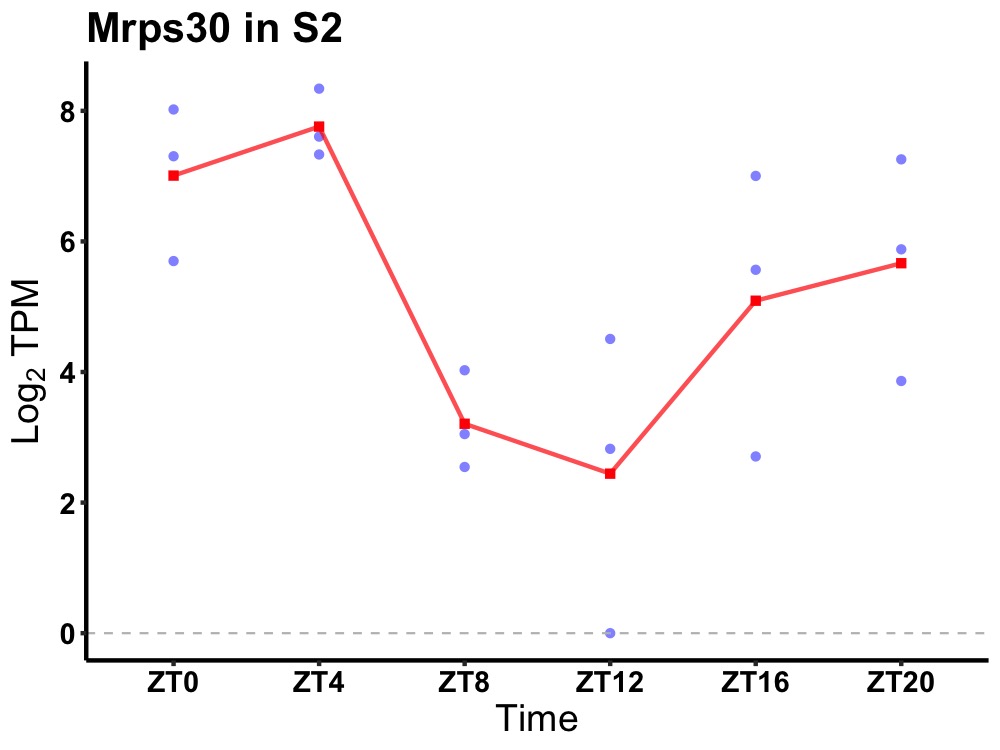 |
mitochondrial ribosomal protein S30 | 0.036 | 24 | 2 | 2.37 |
| ENSMUSG00000004896 | Rrnad1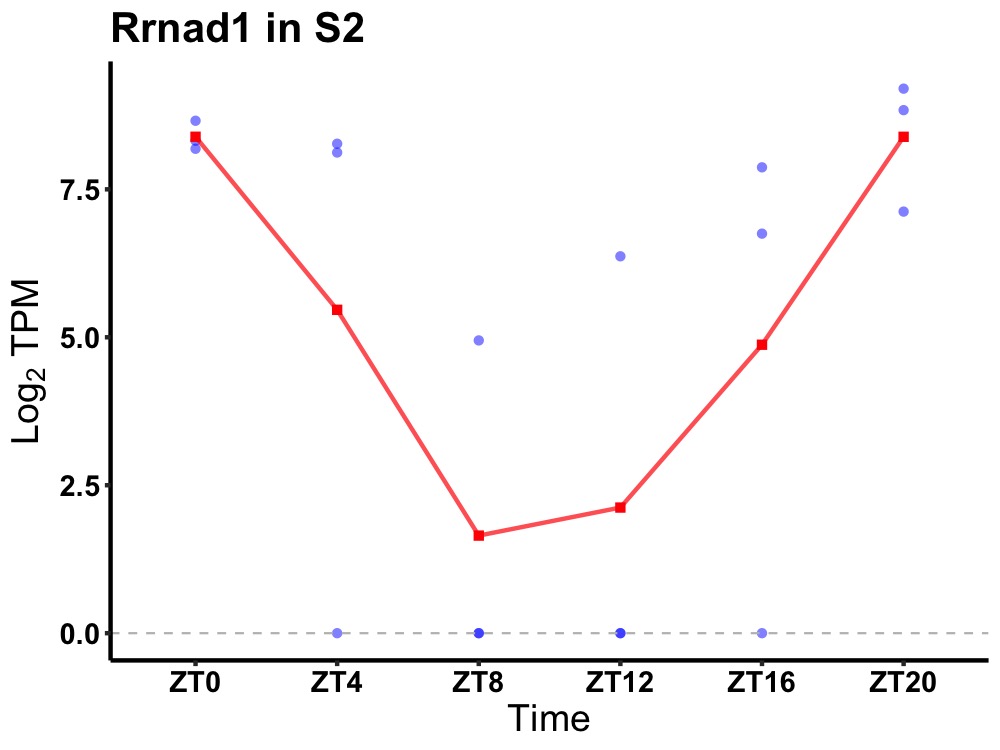 |
methyltransferase-like protein 25B | 0.006 | 20 | 2 | 2.35 |
| ENSMUSG00000039047 | Txnip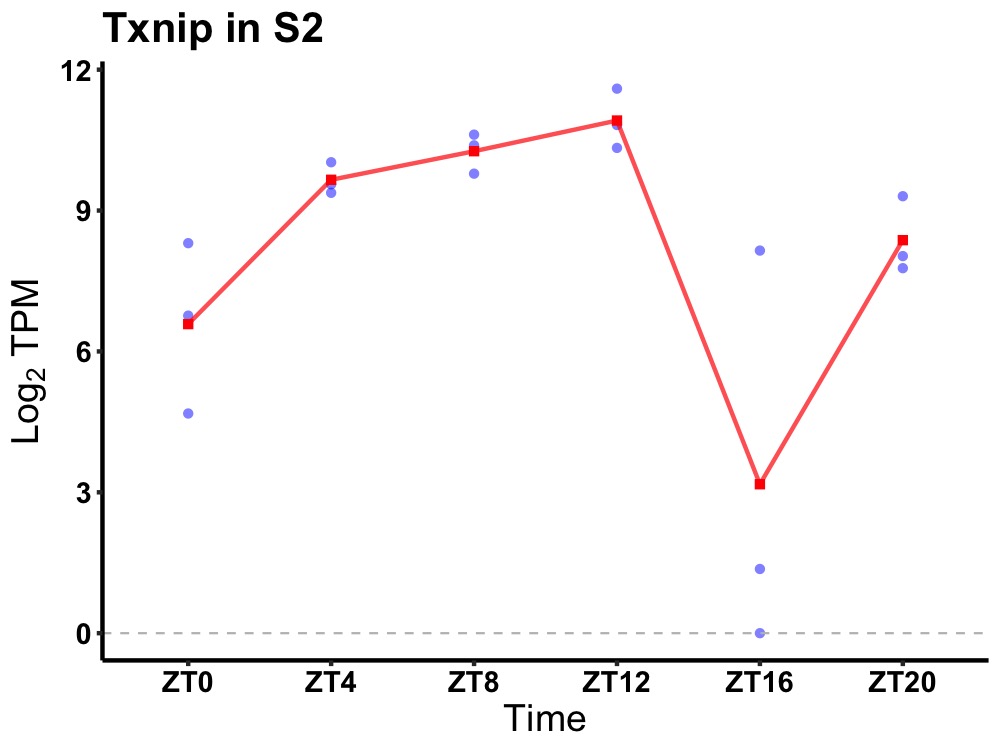 |
thioredoxin interacting protein | 0.007 | 20 | 8 | 2.33 |
| ENSMUSG00000047514 | Kbtbd4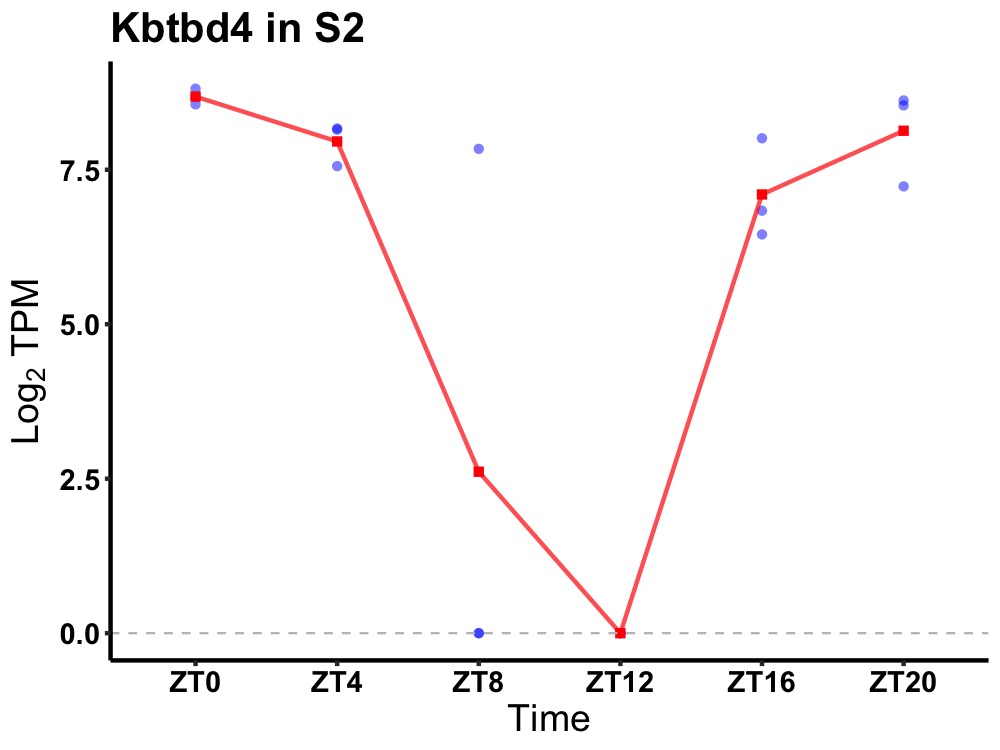 |
kelch repeat and BTB domain-containing protein 4 | 0.000 | 24 | 0 | 2.32 |
| ENSMUSG00000027589 | Arhgef10l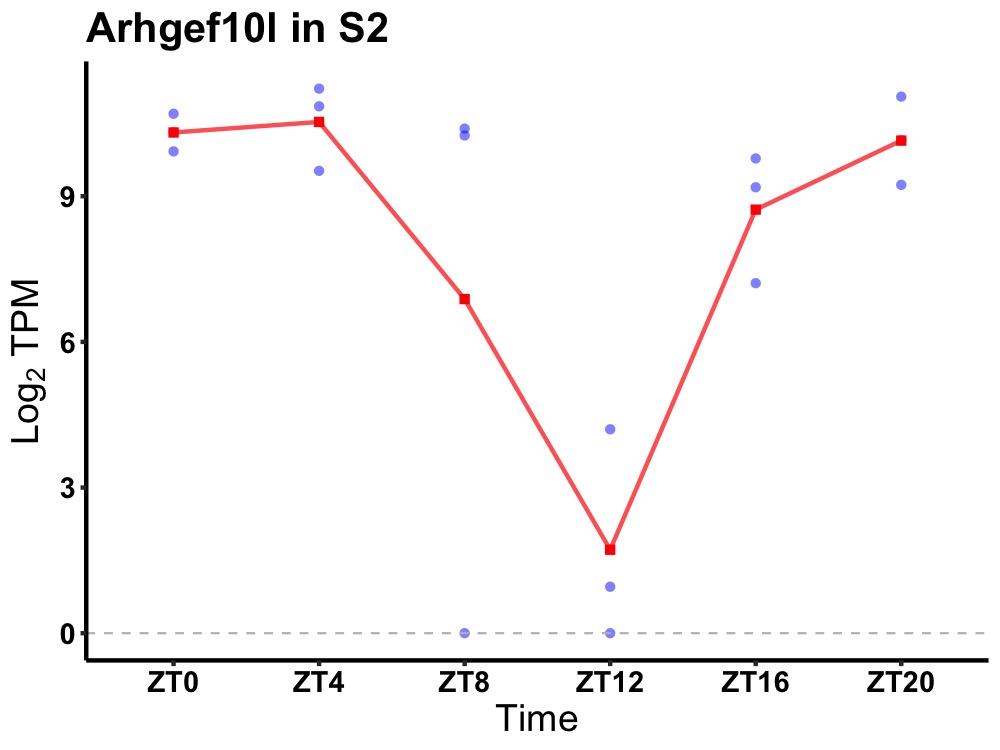 |
rho guanine nucleotide exchange factor 10-like protein | 0.031 | 20 | 4 | 2.31 |
| ENSMUSG00000026077 | Npas2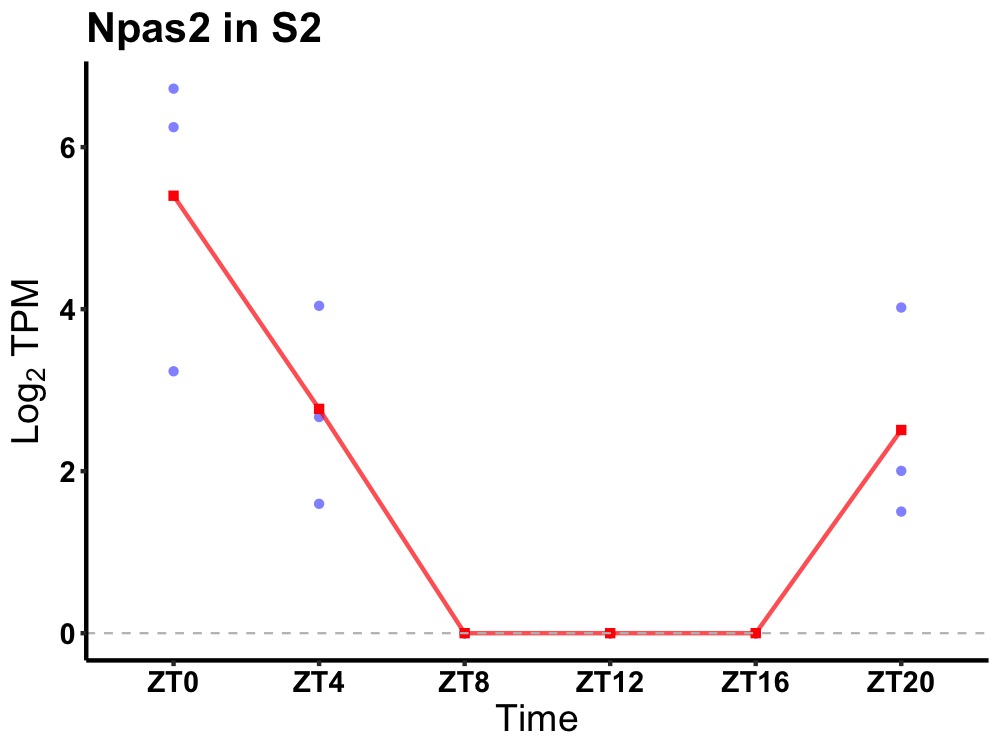 |
neuronal PAS domain-containing protein 2 | 0.001 | 24 | 2 | 2.29 |
| ENSMUSG00000073468 | Gm45871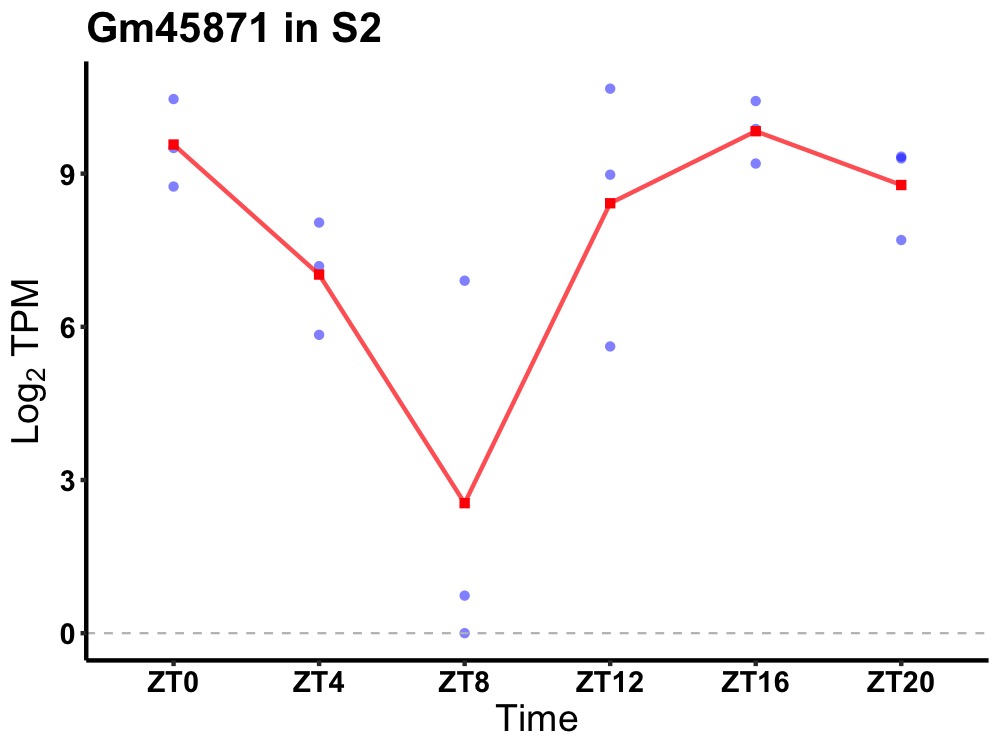 |
predicted gene 45871 | 0.014 | 20 | 18 | 2.25 |
| ENSMUSG00000014074 | Rnf168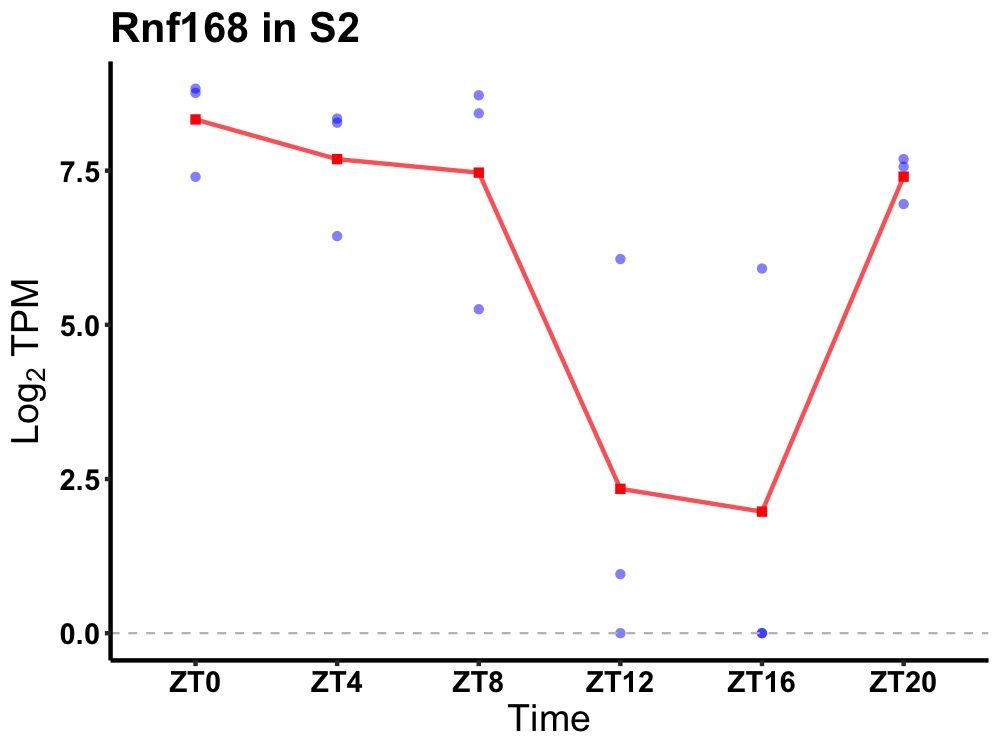 |
E3 ubiquitin-protein ligase RNF168 | 0.036 | 24 | 4 | 2.22 |
| ENSMUSG00000034274 | Thoc5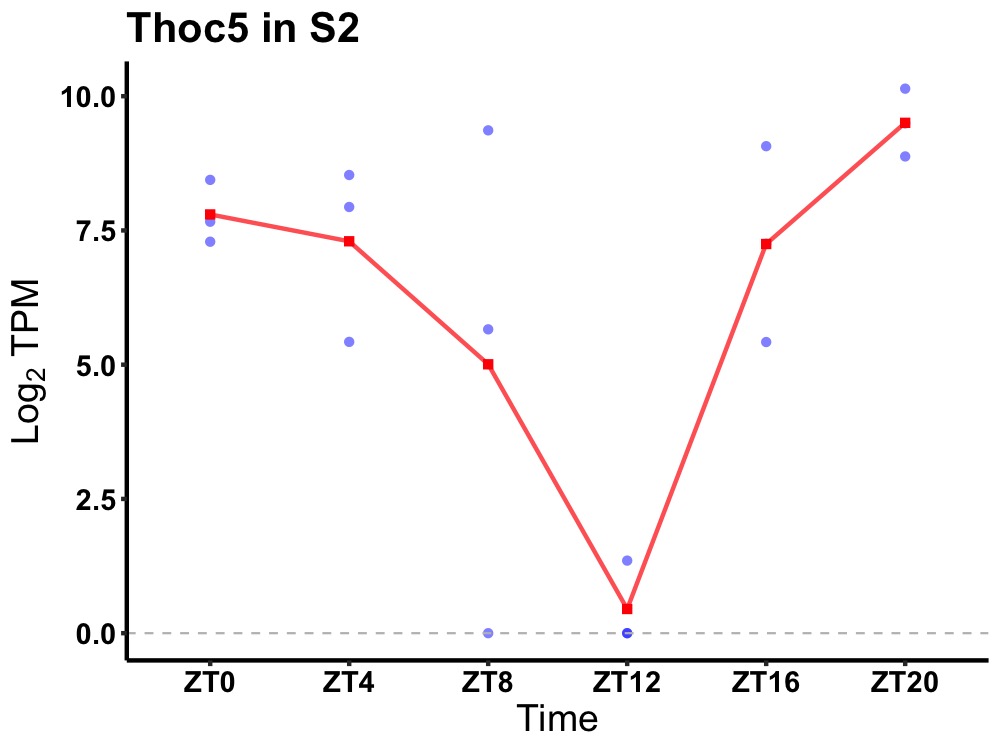 |
THO complex subunit 5 homolog | 0.027 | 20 | 2 | 2.17 |
| ENSMUSG00000036442 | Thap11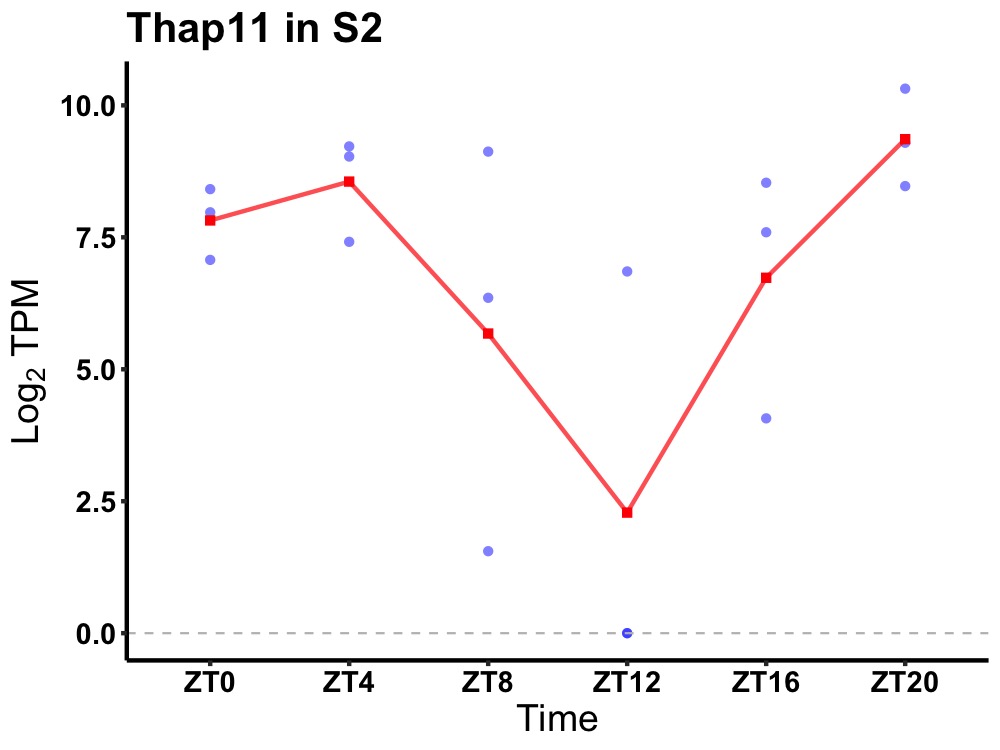 |
THAP domain-containing protein 11 | 0.049 | 20 | 2 | 1.97 |
| ENSMUSG00000020634 | Srd5a3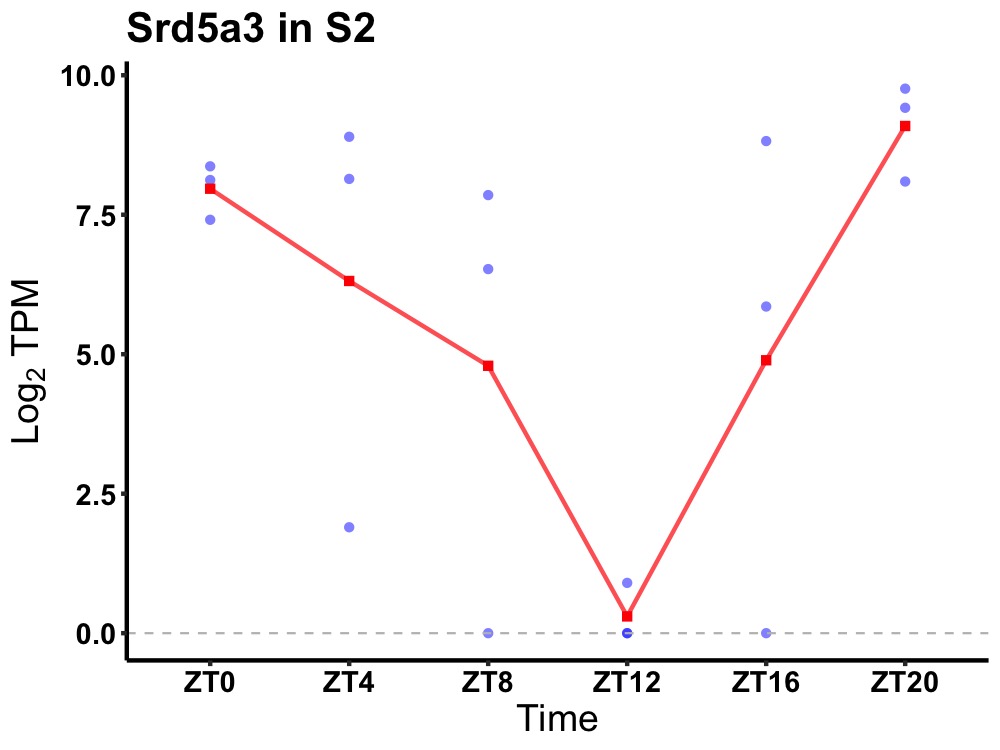 |
polyprenol reductase | 0.031 | 20 | 2 | 1.96 |
| ENSMUSG00000015127 | Unkl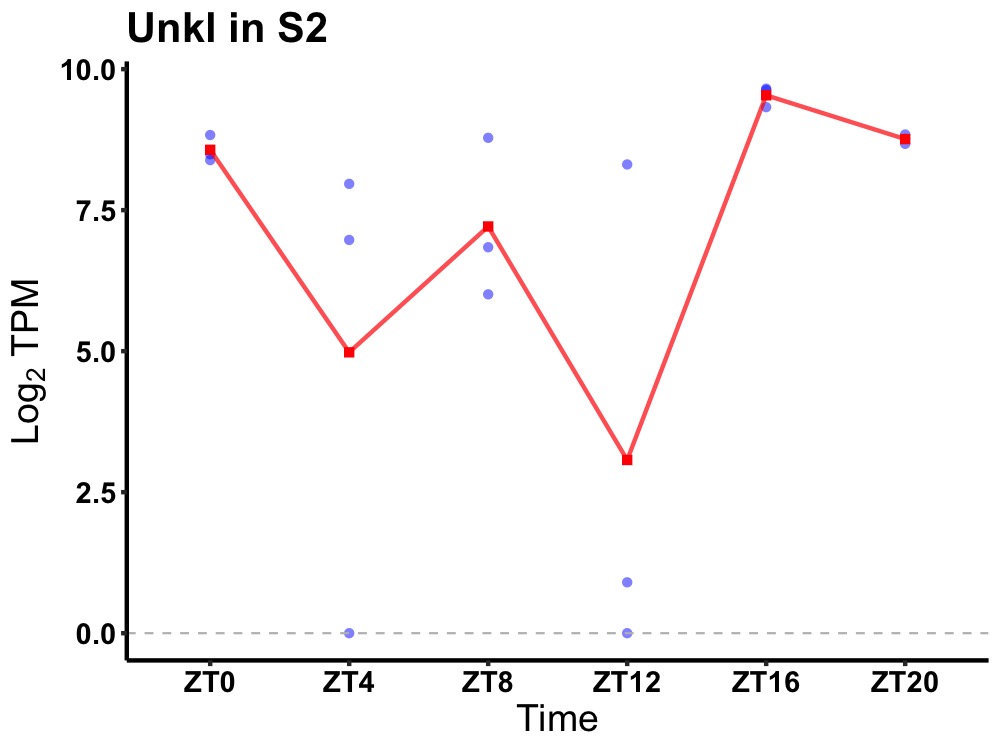 |
putative E3 ubiquitin-protein ligase UNKL | 0.031 | 20 | 18 | 1.94 |
| ENSMUSG00000066043 | Zbtb41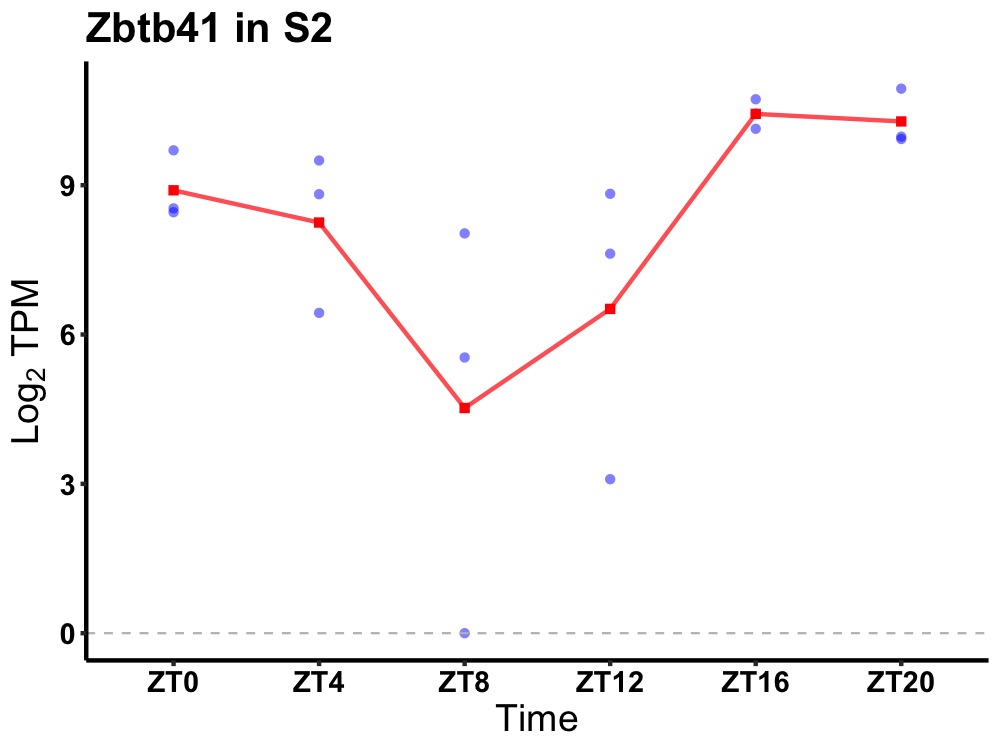 |
zinc finger and BTB domain containing 41 | 0.005 | 24 | 20 | 1.90 |
| ENSMUSG00000029198 | Rhobtb1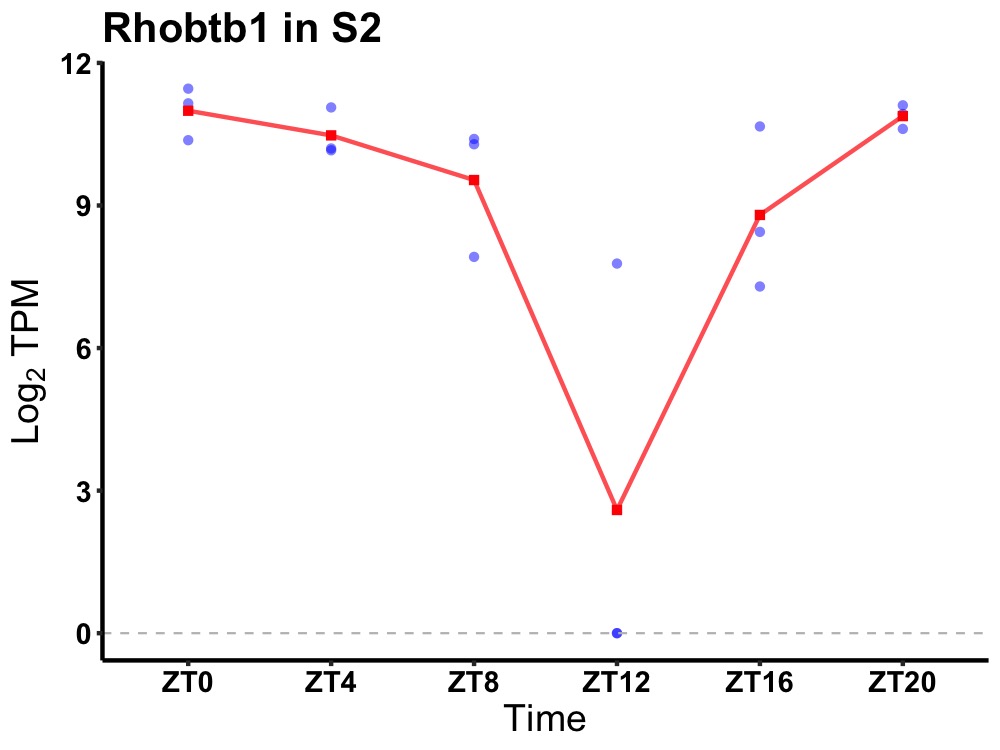 |
rho-related BTB domain-containing protein 1 | 0.003 | 24 | 0 | 1.88 |
| ENSMUSG00000020521 | Lrrc14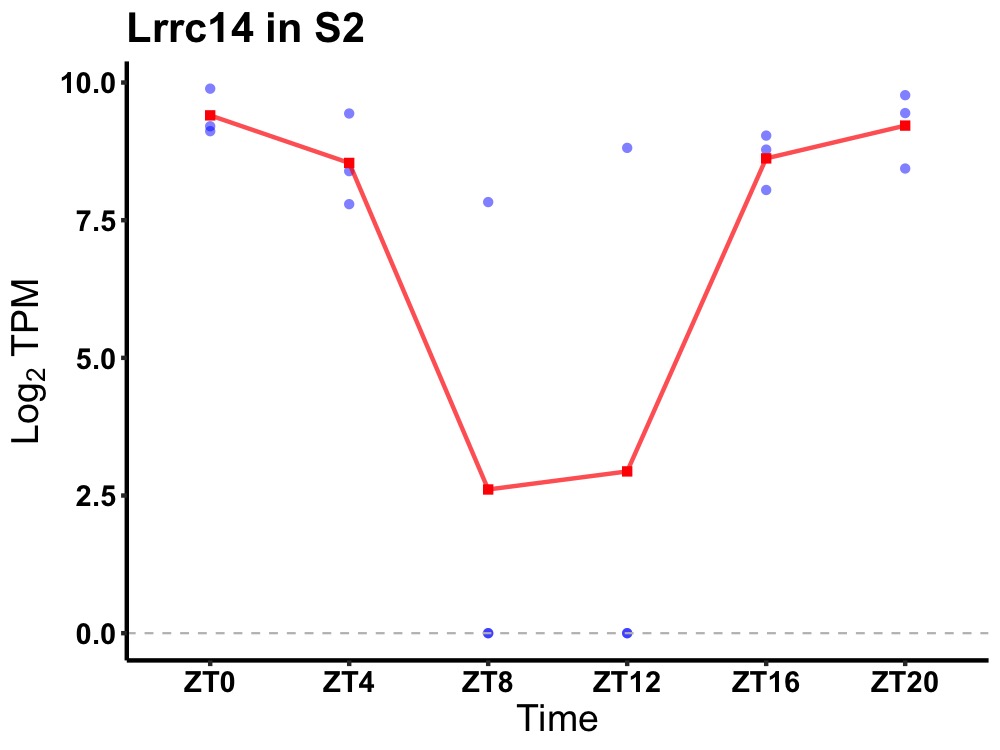 |
leucine-rich repeat-containing protein 14 | 0.007 | 20 | 0 | 1.86 |
| ENSMUSG00000045128 | Odf2l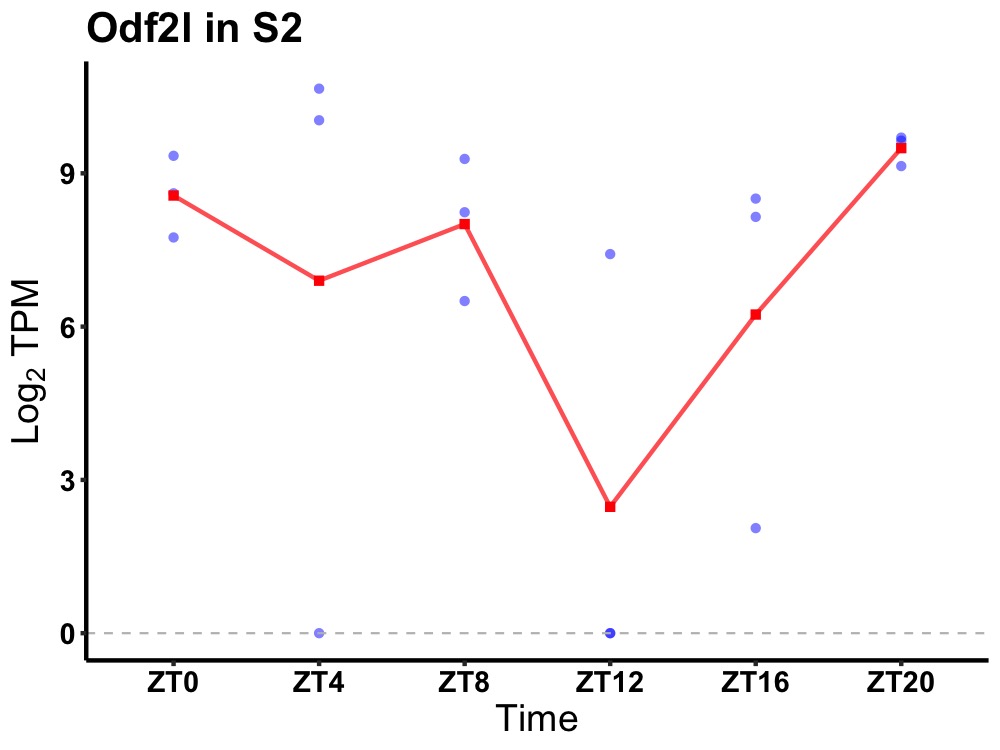 |
protein BCAP | 0.019 | 20 | 4 | 1.84 |
| ENSMUSG00000018648 | Dusp14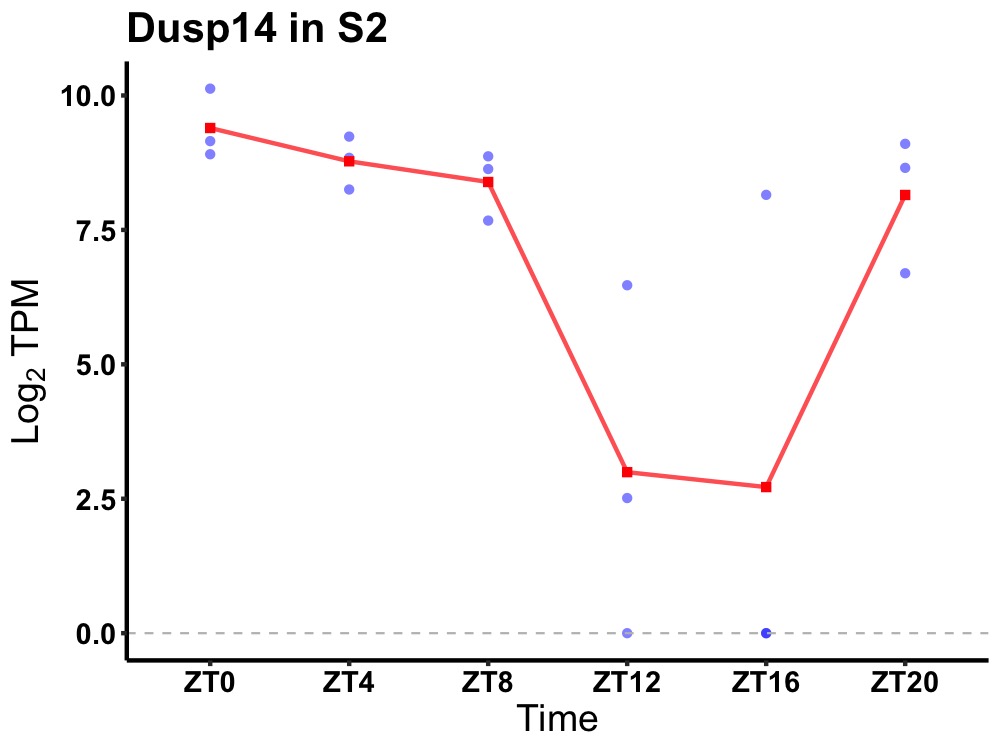 |
dual specificity protein phosphatase 14 | 0.002 | 24 | 2 | 1.82 |
| ENSMUSG00000021917 | Glis3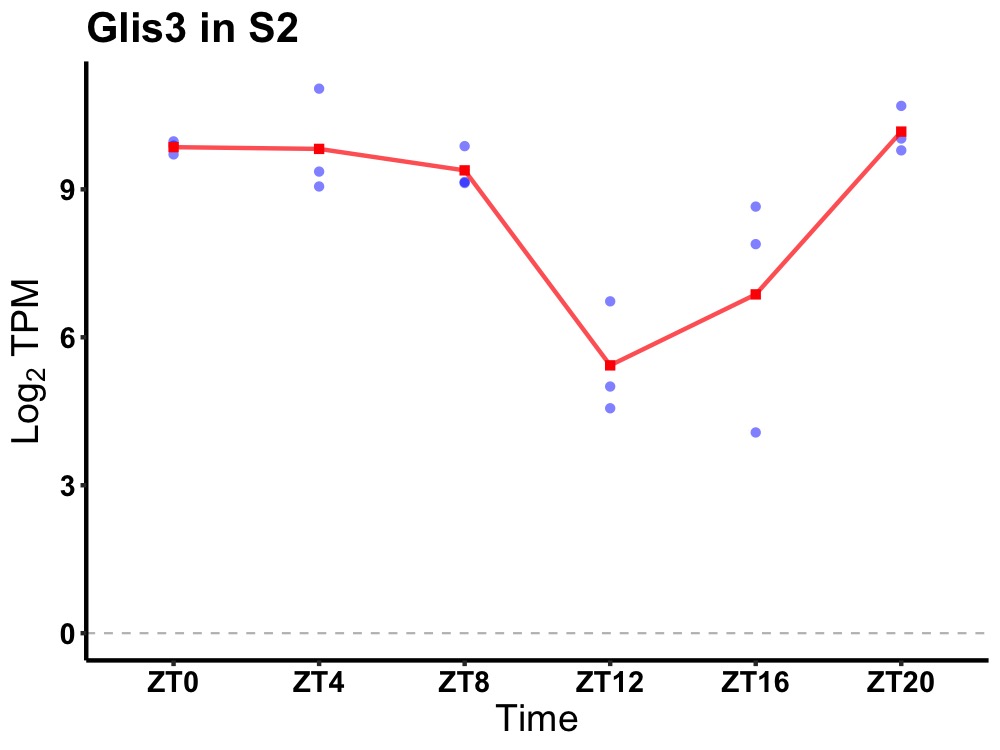 |
GLIS family zinc finger 3 | 0.007 | 20 | 4 | 1.81 |
| ENSMUSG00000025016 | Nck1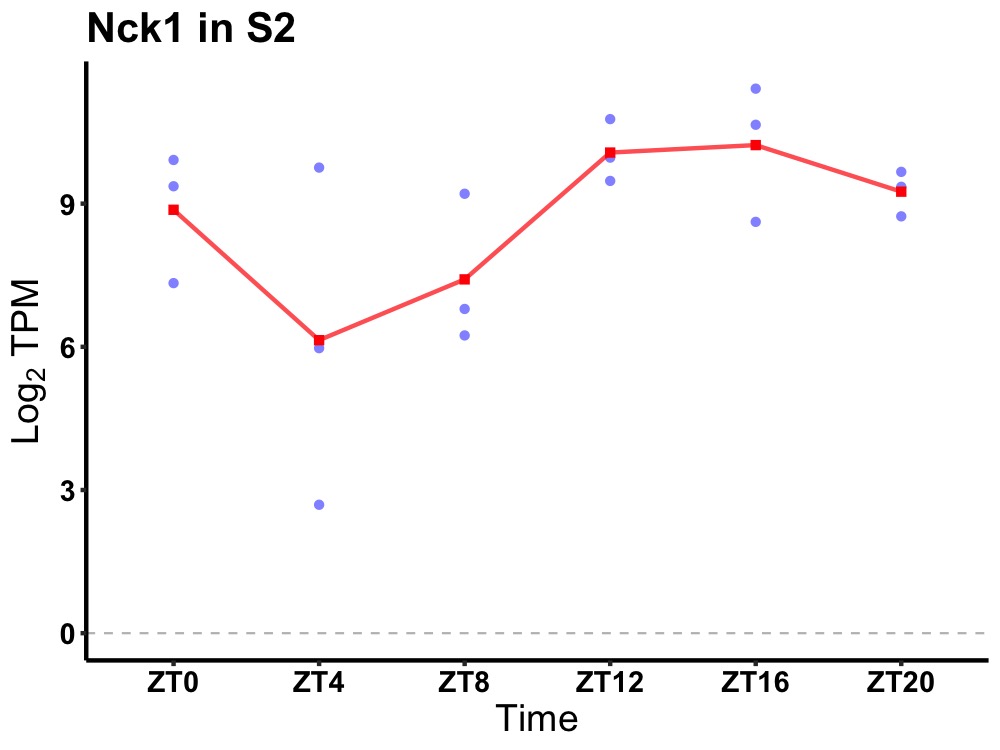 |
non-catalytic region of tyrosine kinase adaptor protein 1 | 0.036 | 20 | 16 | 1.80 |
| ENSMUSG00000028759 | Gaa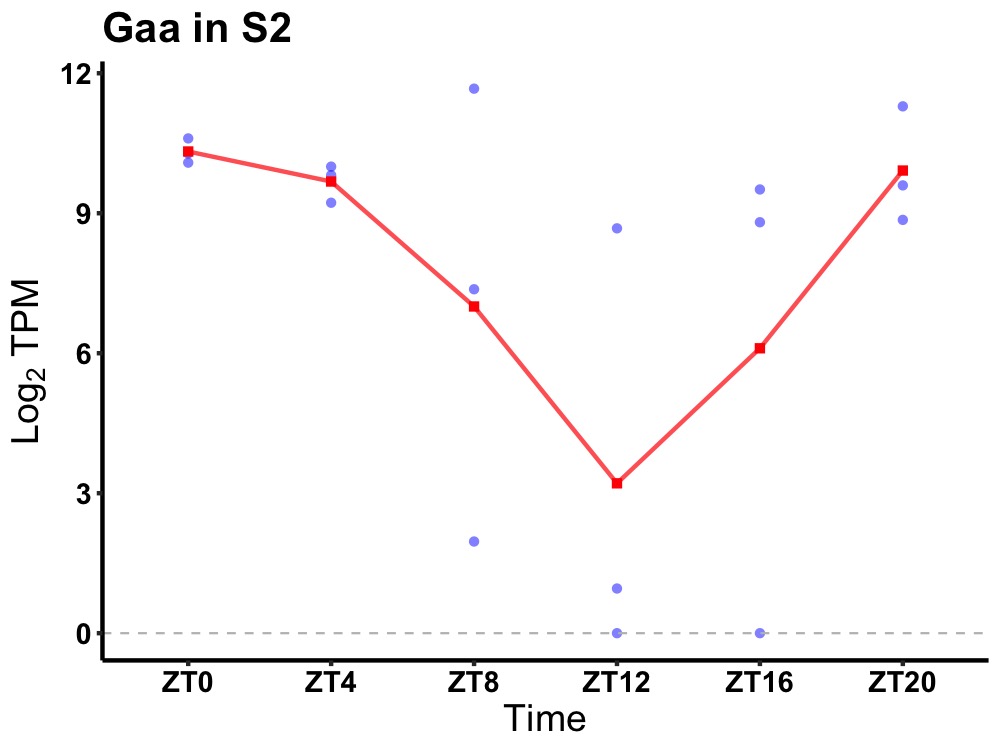 |
lysosomal alpha-glucosidase preproprotein | 0.023 | 24 | 2 | 1.76 |
| ENSMUSG00000025364 | Arhgap26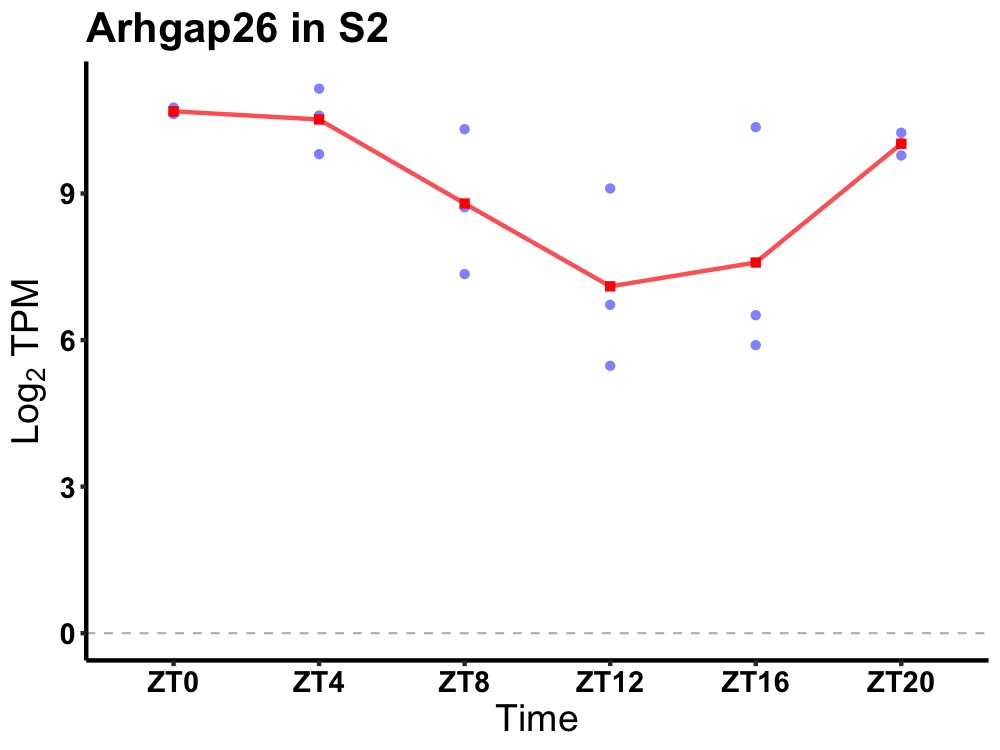 |
Rho GTPase activating protein 26 | 0.005 | 24 | 2 | 1.75 |
| ENSMUSG00000029201 | Ccdc68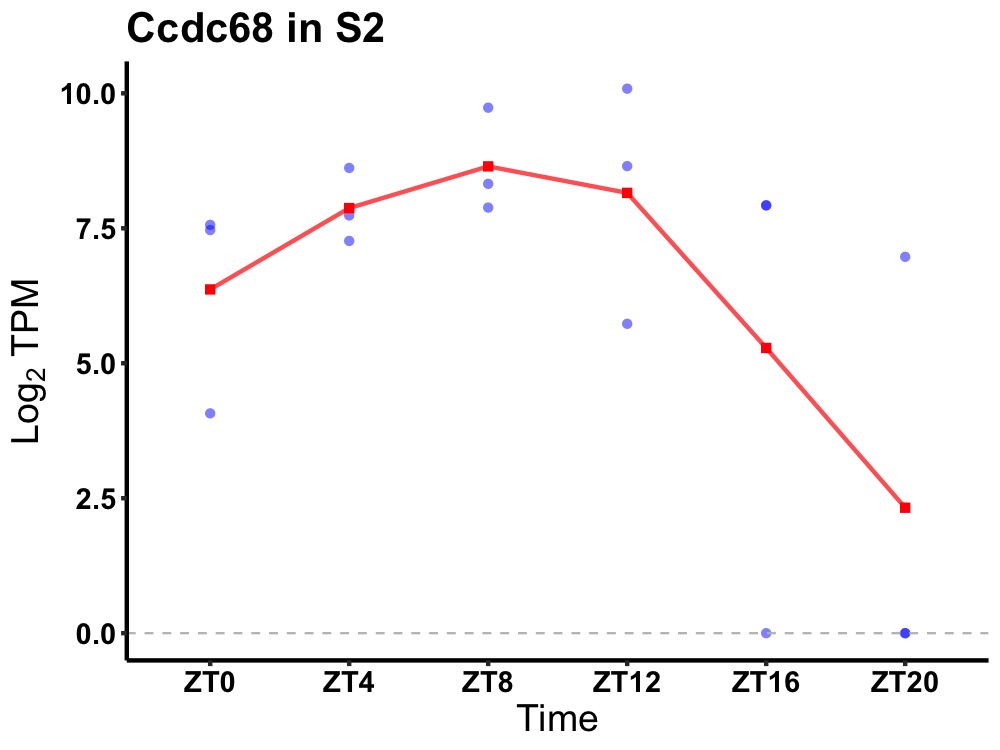 |
coiled-coil domain-containing protein 68 | 0.049 | 24 | 10 | 1.70 |
| ENSMUSG00000091020 | Gm5828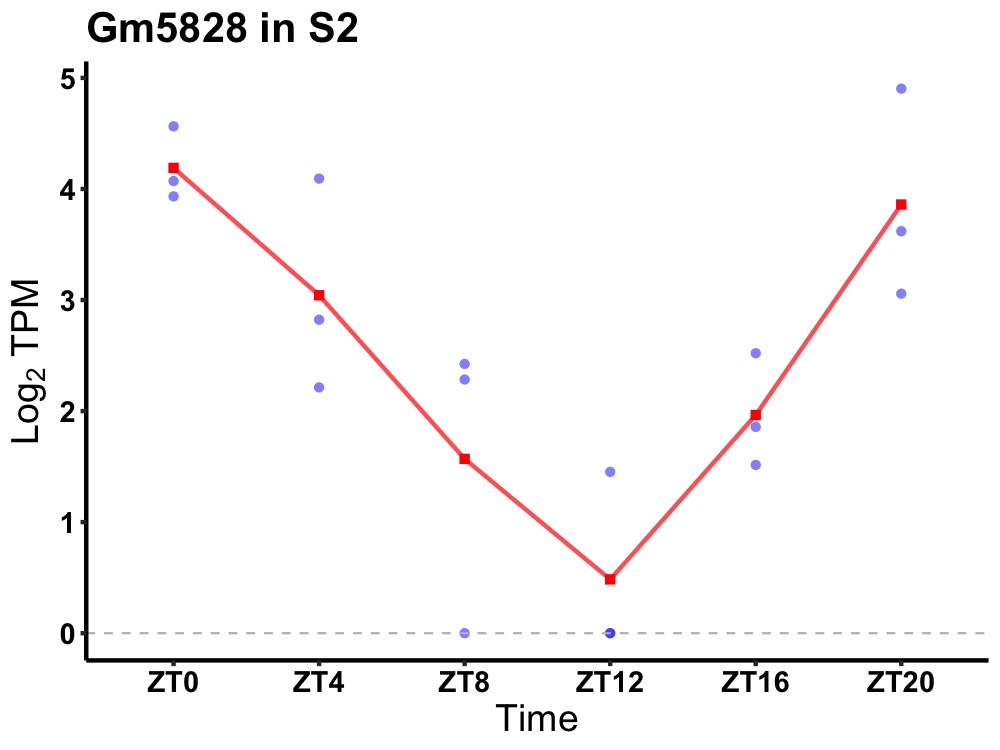 |
none | 0.000 | 24 | 0 | 1.68 |
| ENSMUSG00000118346 | Pdik1l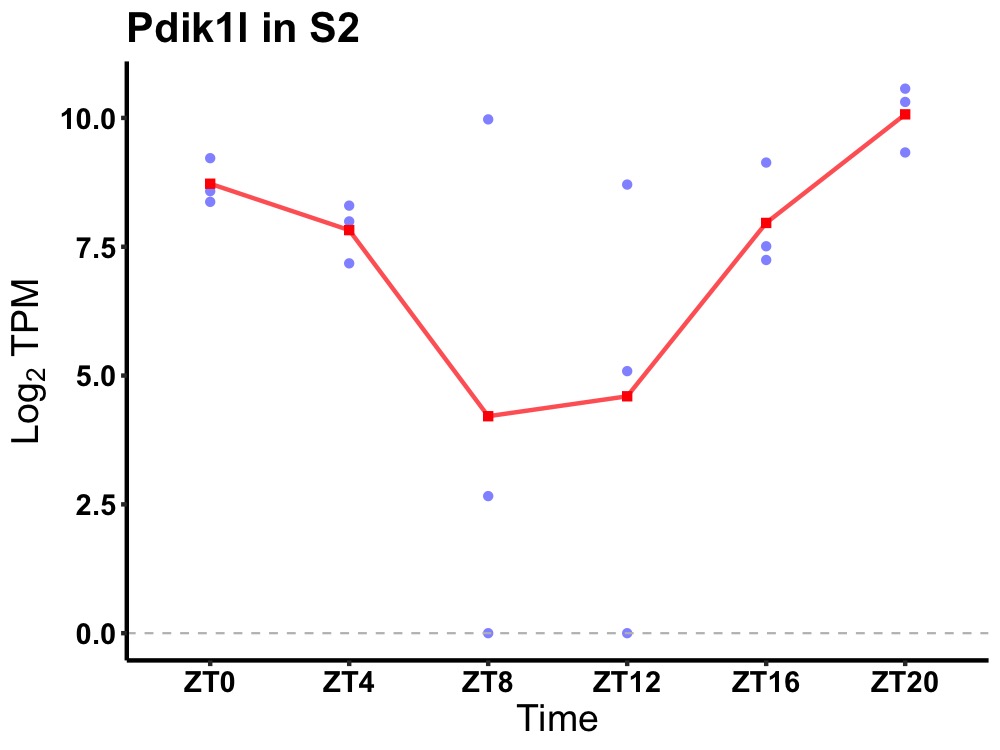 |
serine/threonine-protein kinase PDIK1L | 0.031 | 24 | 22 | 1.62 |
| ENSMUSG00000048592 | Gm5946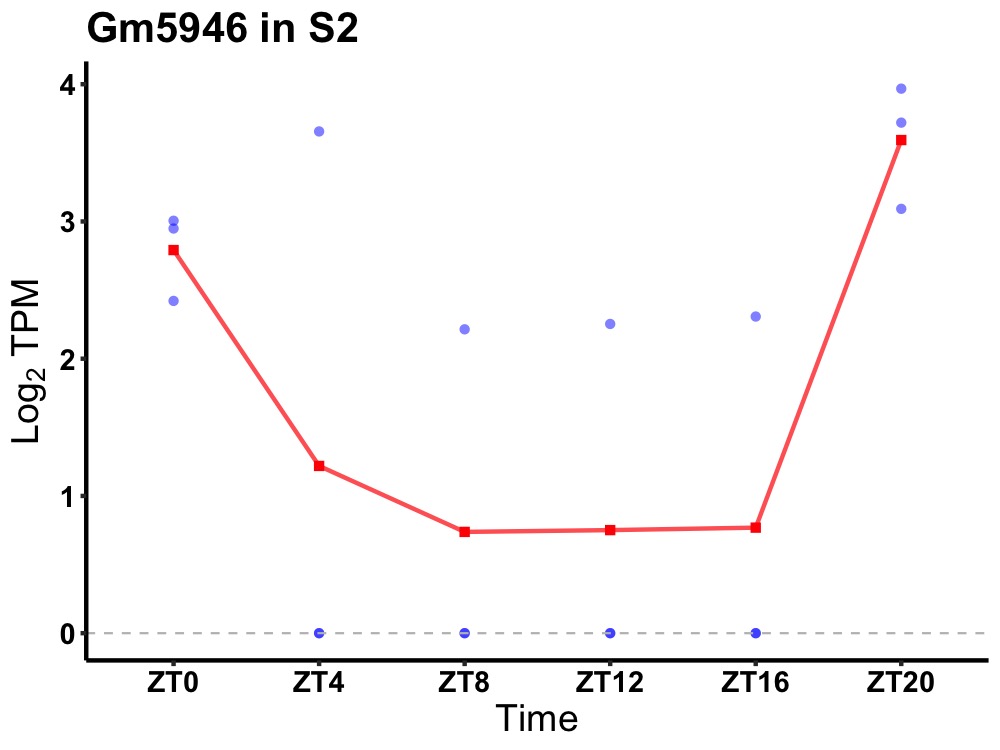 |
none | 0.049 | 24 | 22 | 1.61 |
| ENSMUSG00000030036 | Mogs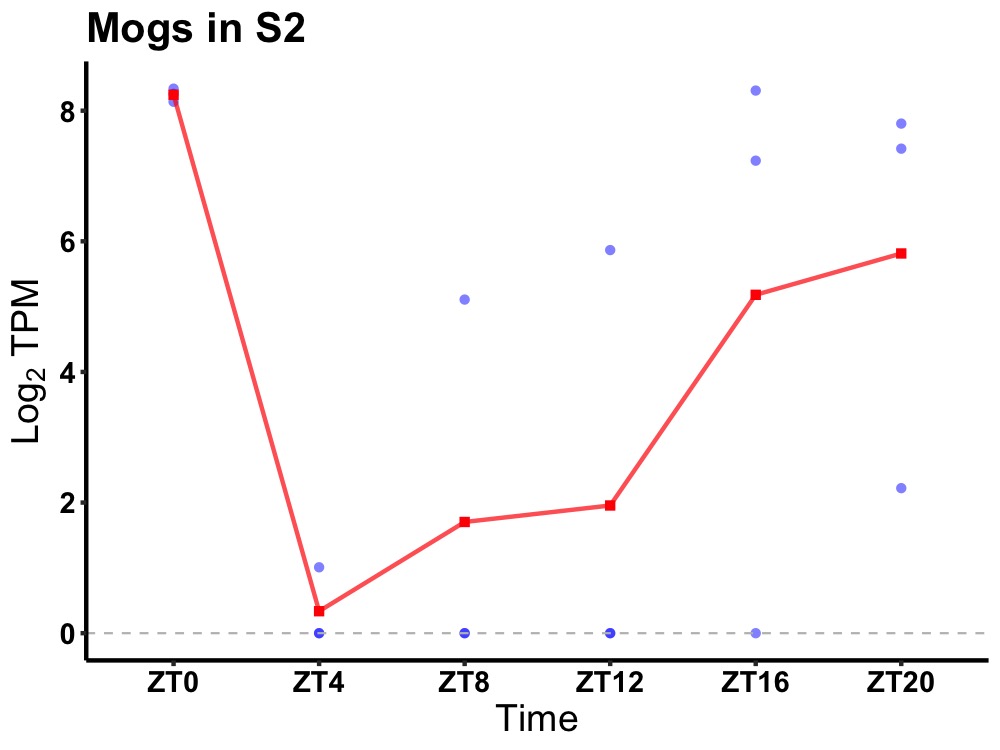 |
mannosyl-oligosaccharide glucosidase | 0.036 | 20 | 0 | 1.61 |
| ENSMUSG00000003464 | Tmem62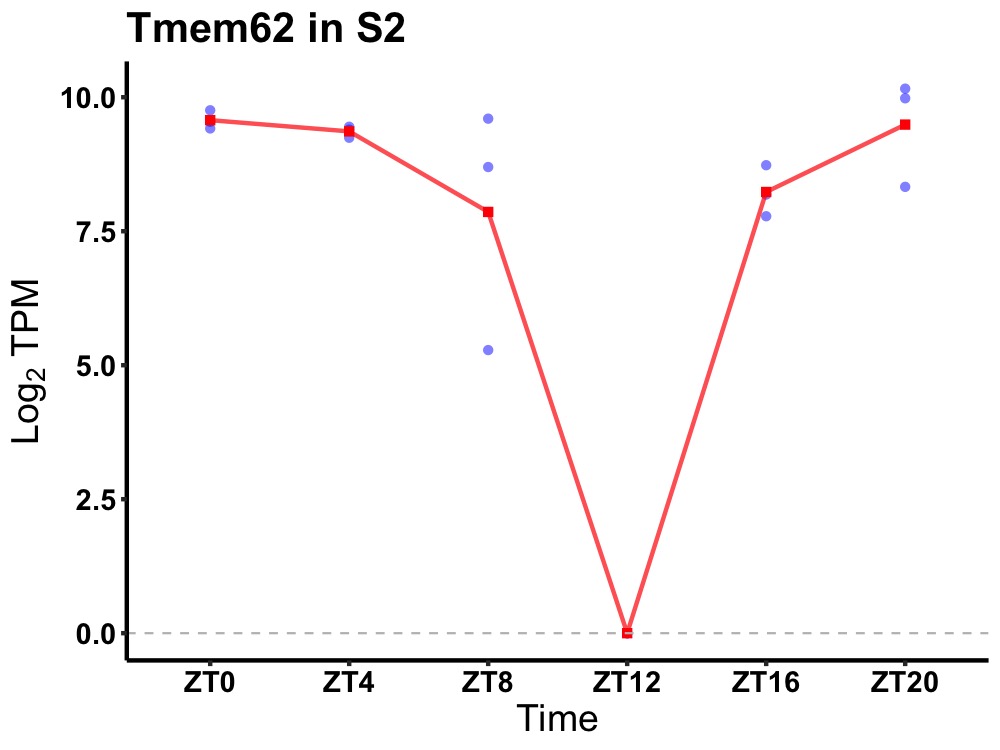 |
transmembrane protein 62 | 0.002 | 20 | 2 | 1.48 |
| ENSMUSG00000101059 | Atp13a1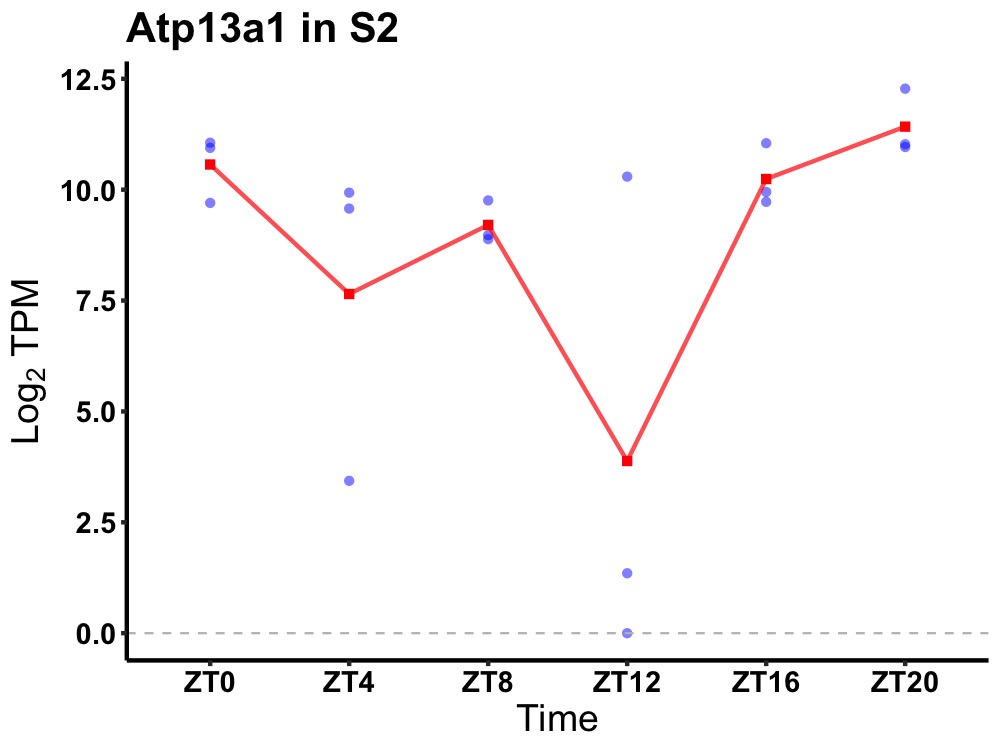 |
ATPase type 13A1 | 0.036 | 24 | 22 | 1.47 |
| ENSMUSG00000032407 | Hccs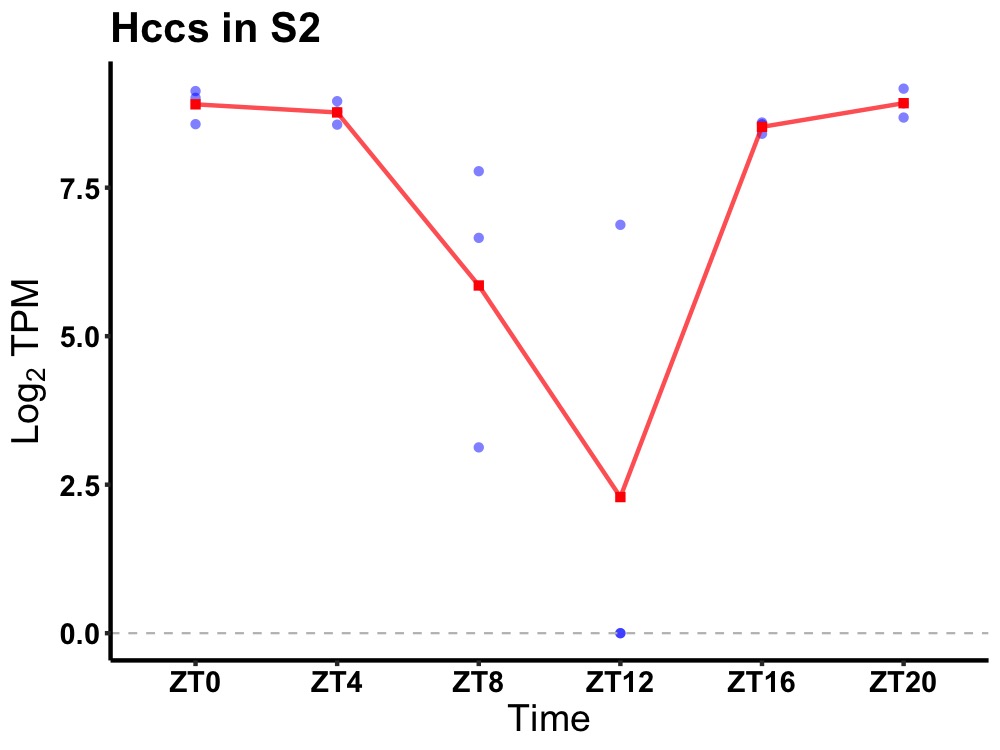 |
holocytochrome c-type synthase | 0.000 | 20 | 2 | 1.46 |
| ENSMUSG00000024975 | Gtf2h3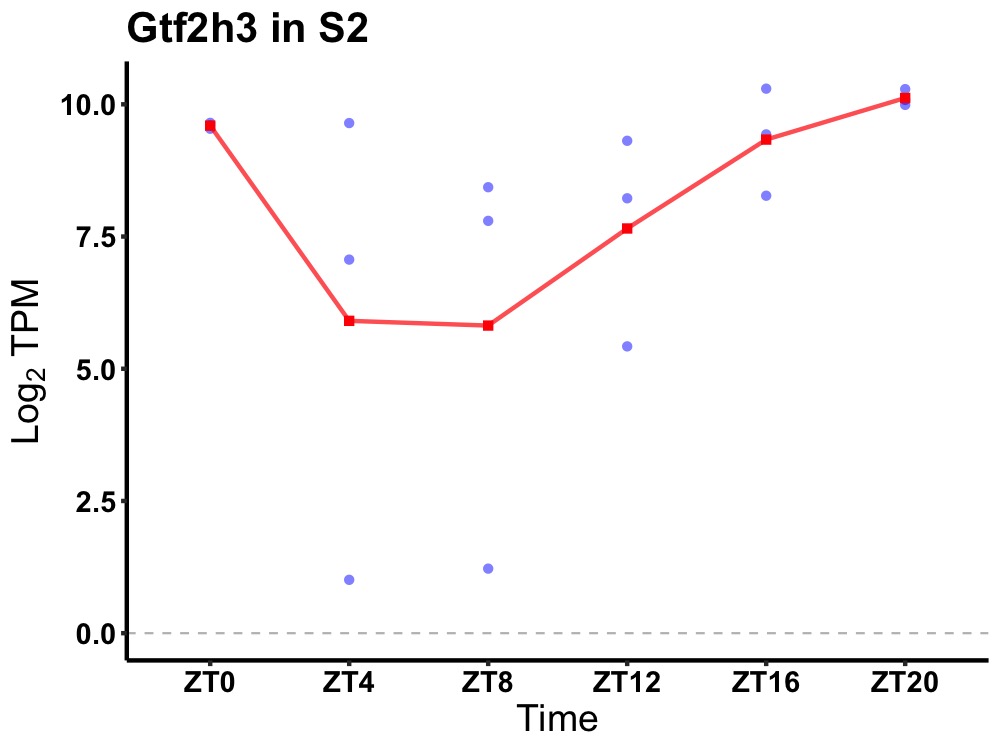 |
general transcription factor IIH, polypeptide 3 | 0.014 | 24 | 22 | 1.46 |
| ENSMUSG00000110537 | Gm4316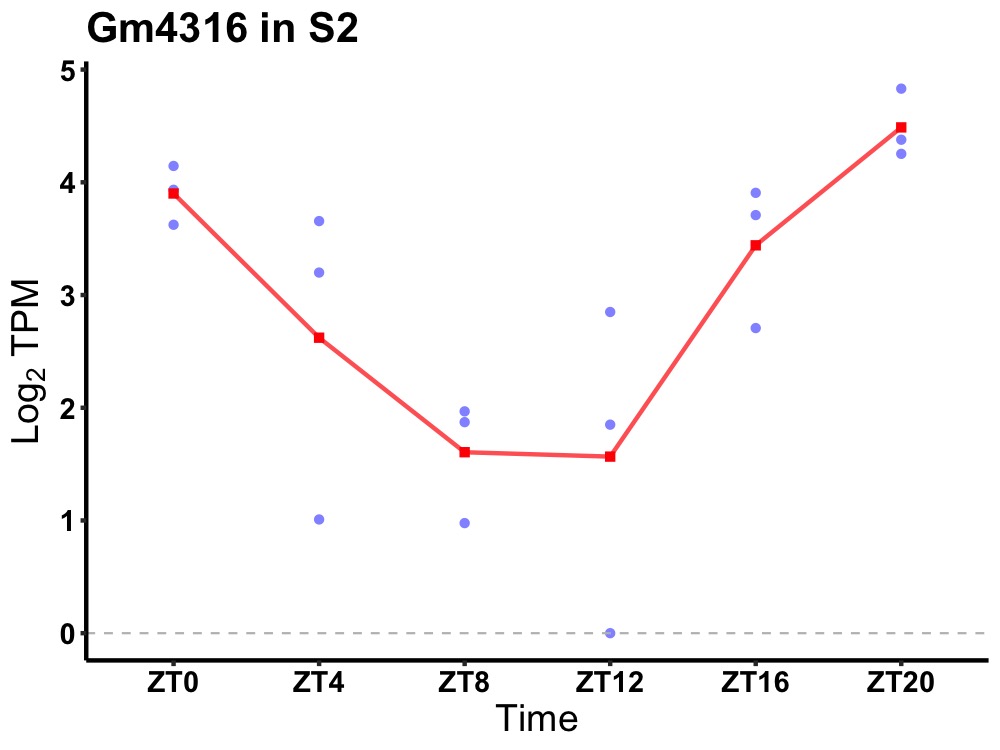 |
predicted gene 4316 | 0.000 | 24 | 22 | 1.45 |
| ENSMUSG00000085235 | Gm12576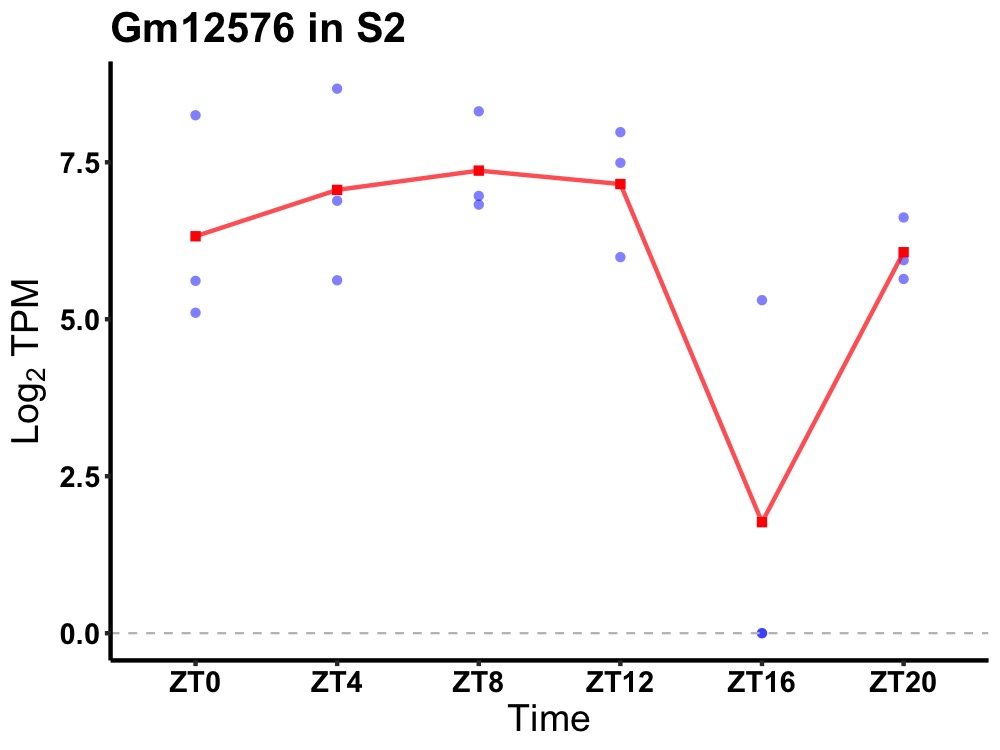 |
none | 0.036 | 20 | 8 | 1.45 |
| ENSMUSG00000000399 | Itih5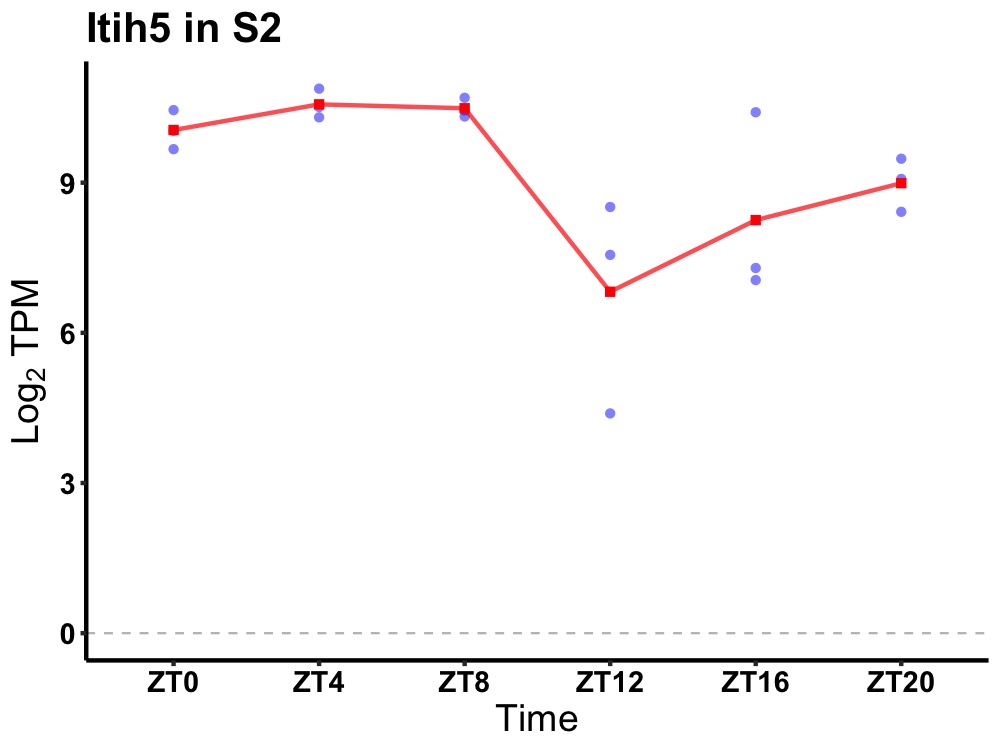 |
inter-alpha (globulin) inhibitor H5 | 0.014 | 24 | 4 | 1.44 |
| ENSMUSG00000016664 | Tox4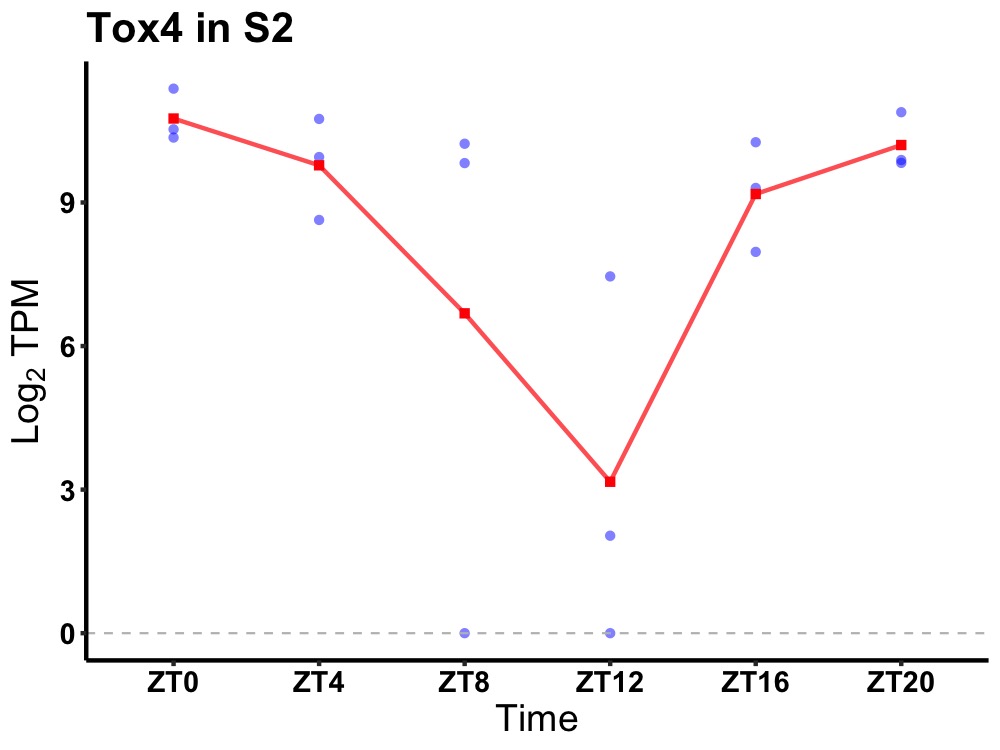 |
TOX high mobility group box family member 4 | 0.012 | 24 | 0 | 1.44 |
| ENSMUSG00000092626 | 9130230N09Rik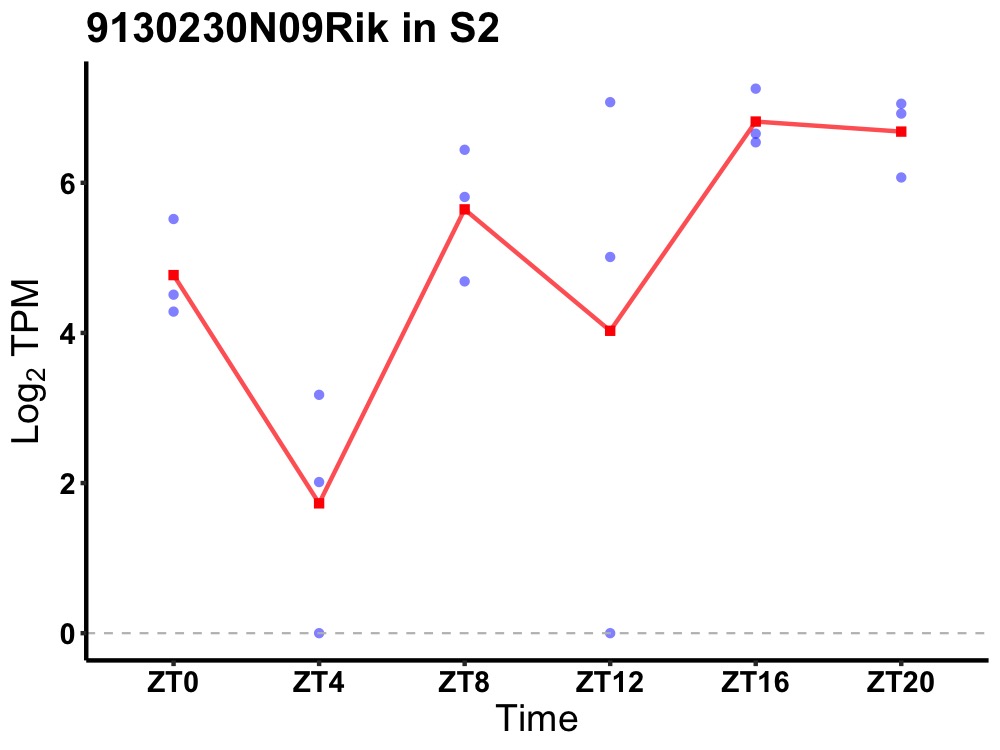 |
none | 0.016 | 24 | 16 | 1.44 |
| ENSMUSG00000084193 | Gm6823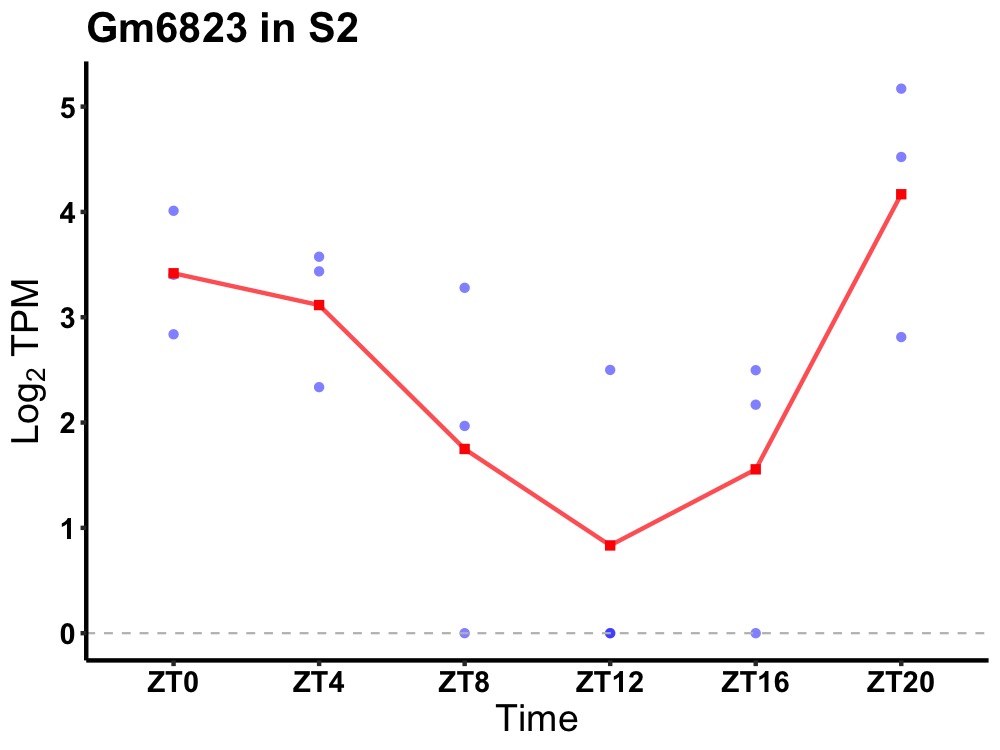 |
none | 0.016 | 20 | 2 | 1.41 |
| ENSMUSG00000012429 | Mplkip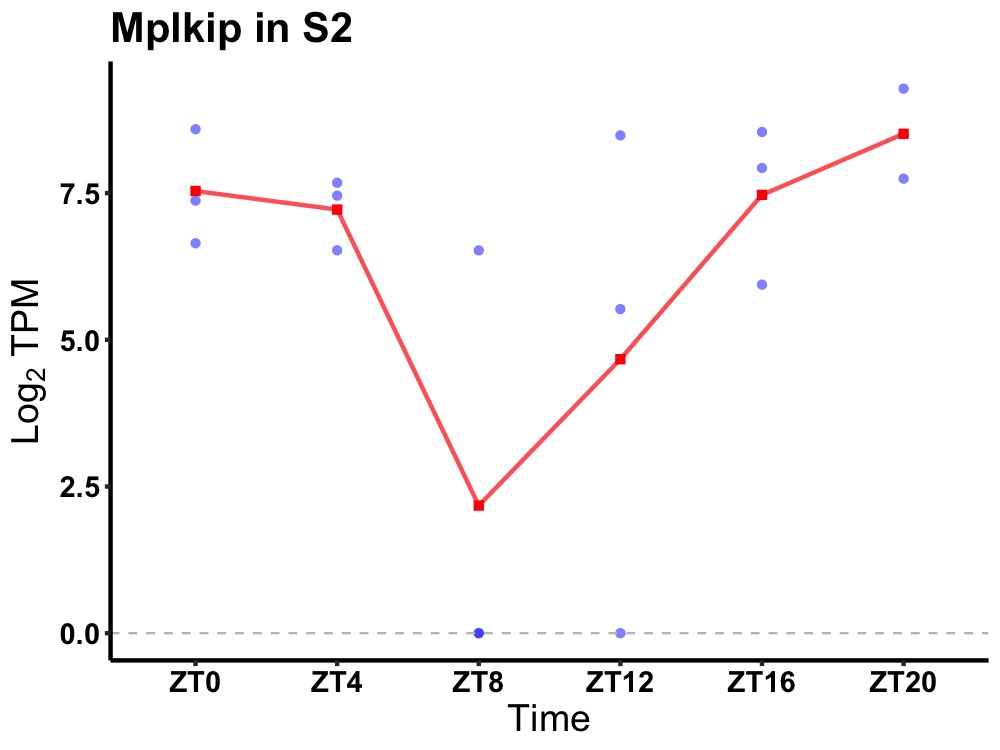 |
M-phase-specific PLK1-interacting protein | 0.036 | 24 | 22 | 1.40 |
| ENSMUSG00000032977 | Fam207a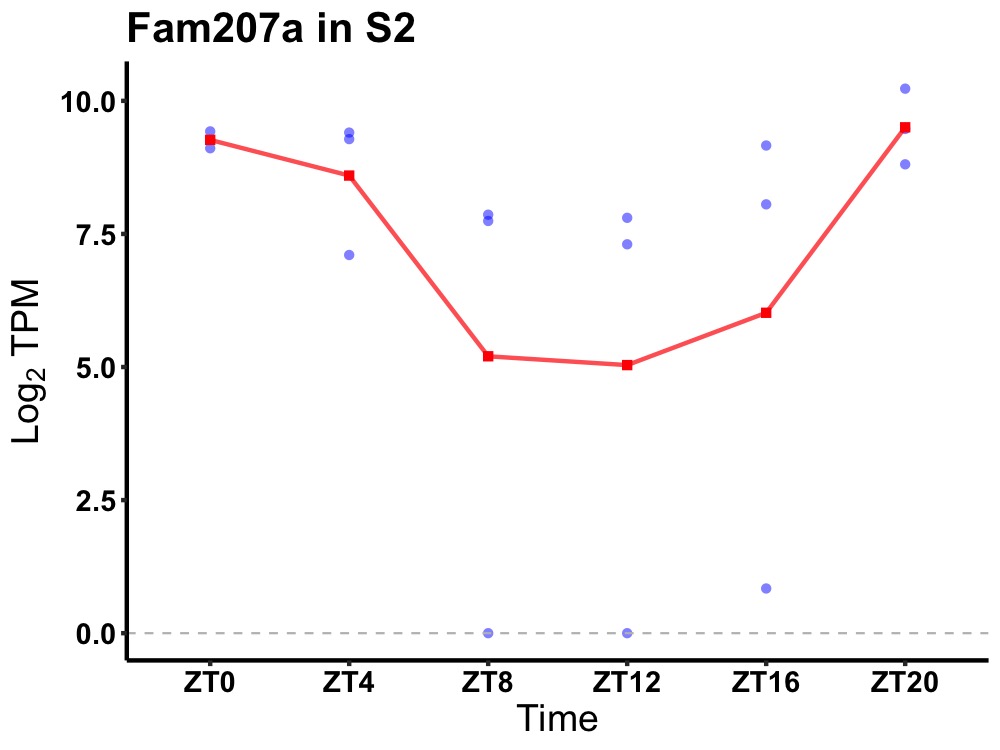 |
ribosome biogenesis protein SLX9 homolog | 0.012 | 20 | 2 | 1.39 |
| ENSMUSG00000007850 | Nr1d2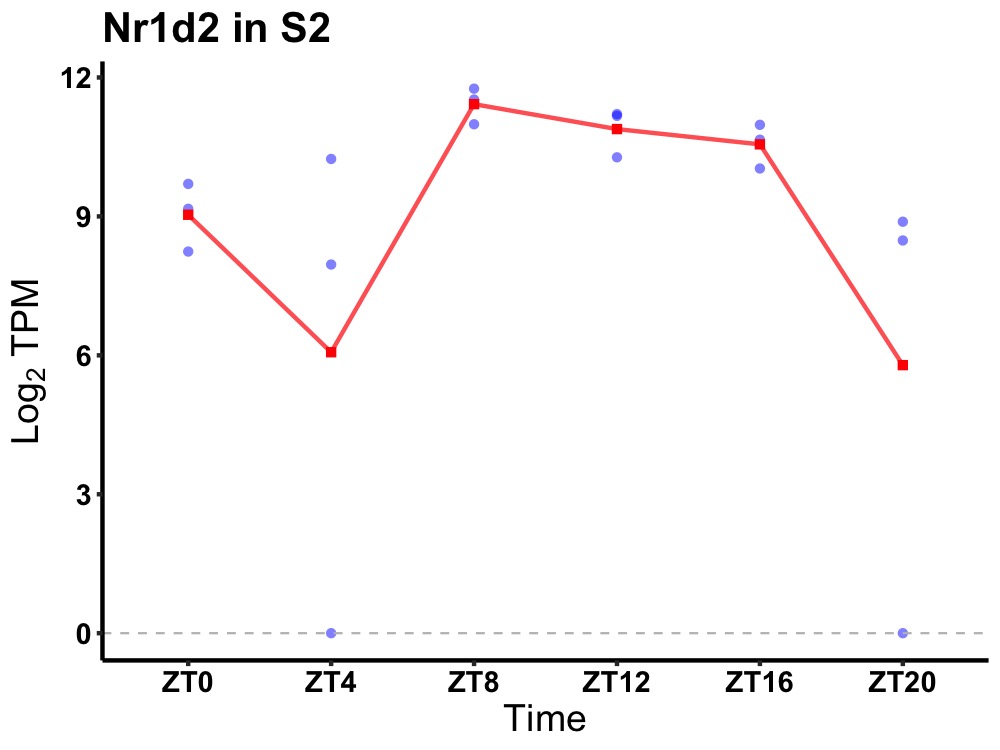 |
nuclear receptor subfamily 1 group D member 2 | 0.006 | 20 | 12 | 1.38 |
| ENSMUSG00000041740 | Upf2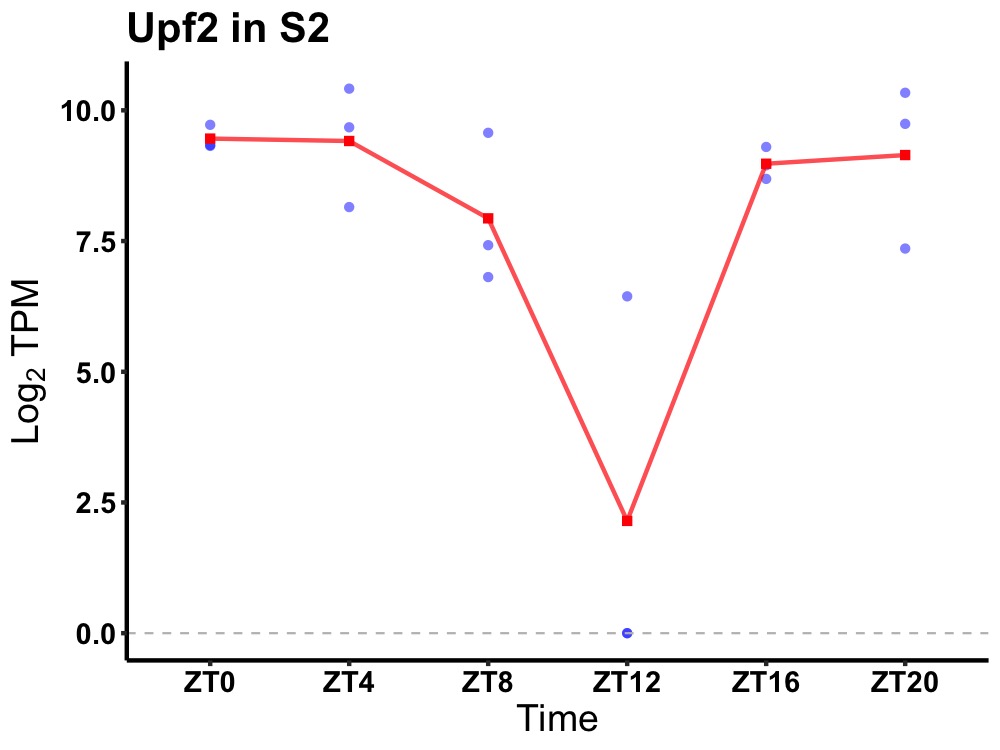 |
regulator of nonsense transcripts 2 | 0.027 | 20 | 2 | 1.37 |
| ENSMUSG00000026307 | Rubcn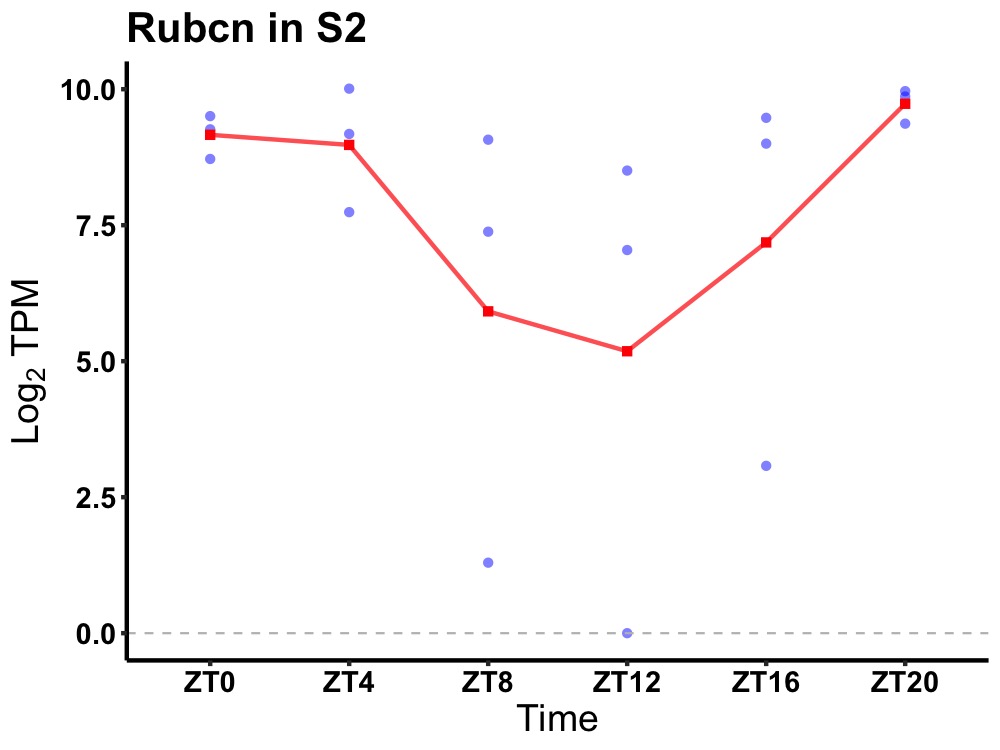 |
RUN domain and cysteine-rich domain containing, Beclin 1-interacting protein | 0.019 | 20 | 2 | 1.36 |
| ENSMUSG00000023852 | Chd1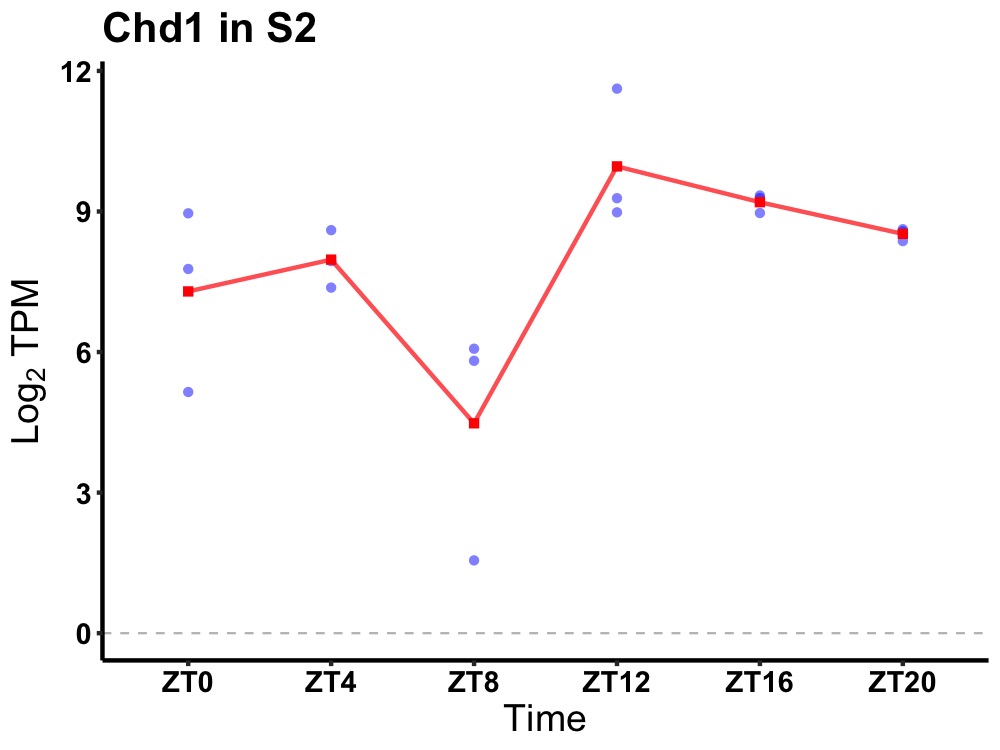 |
chromodomain helicase DNA binding protein 1 | 0.049 | 20 | 16 | 1.36 |
| ENSMUSG00000097328 | Tnfsf12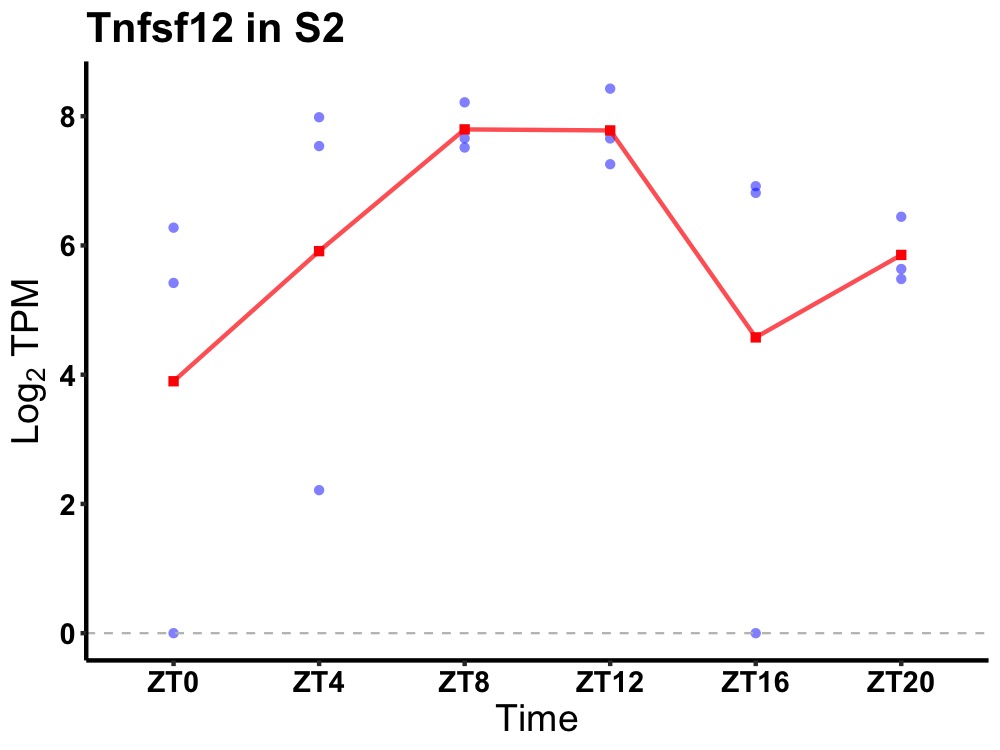 |
tumor necrosis factor ligand superfamily member 12 | 0.012 | 20 | 10 | 1.35 |
| ENSMUSG00000041426 | Tmem185a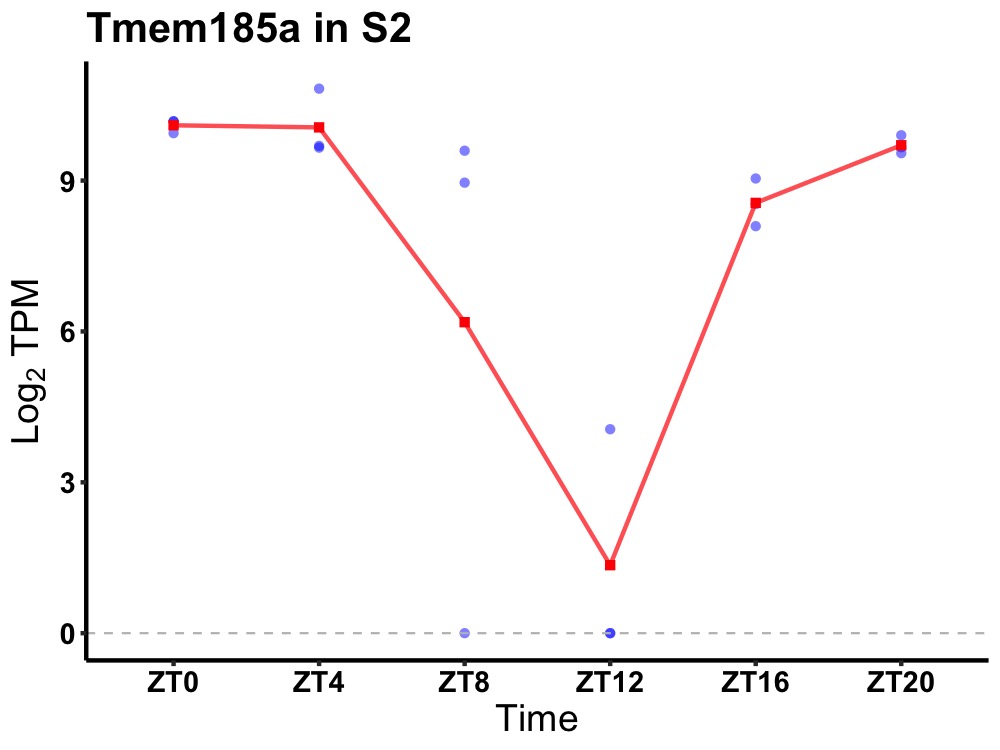 |
transmembrane protein 185A | 0.000 | 24 | 2 | 1.34 |
| ENSMUSG00000000738 | Slc16a6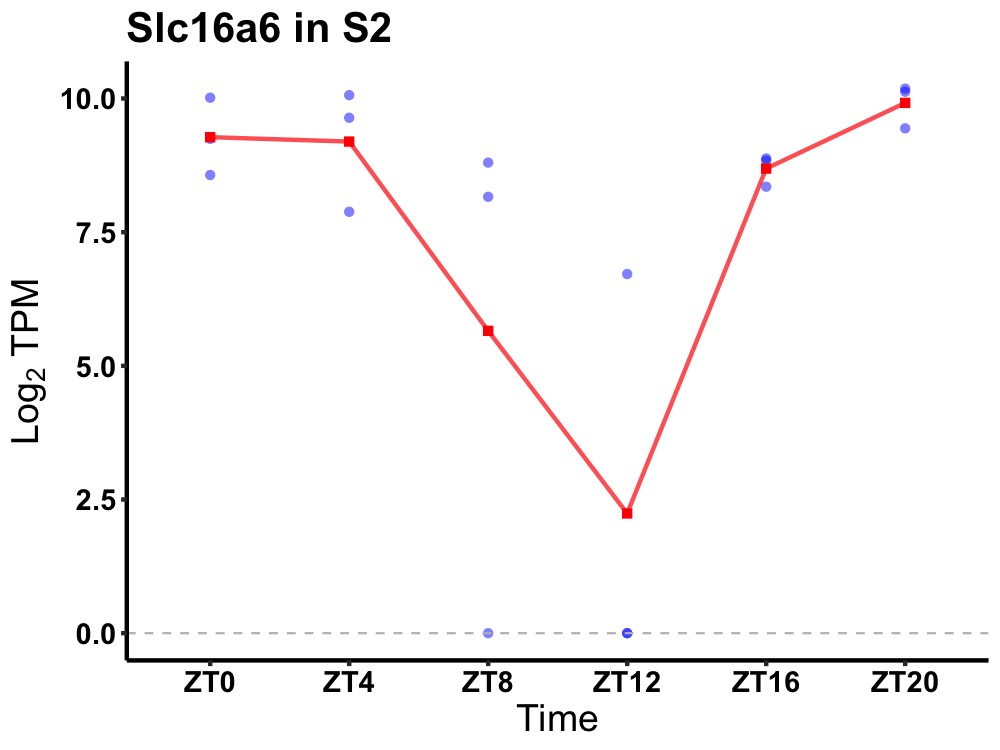 |
monocarboxylate transporter 7 | 0.001 | 20 | 2 | 1.31 |
| ENSMUSG00000036959 | Bcorl1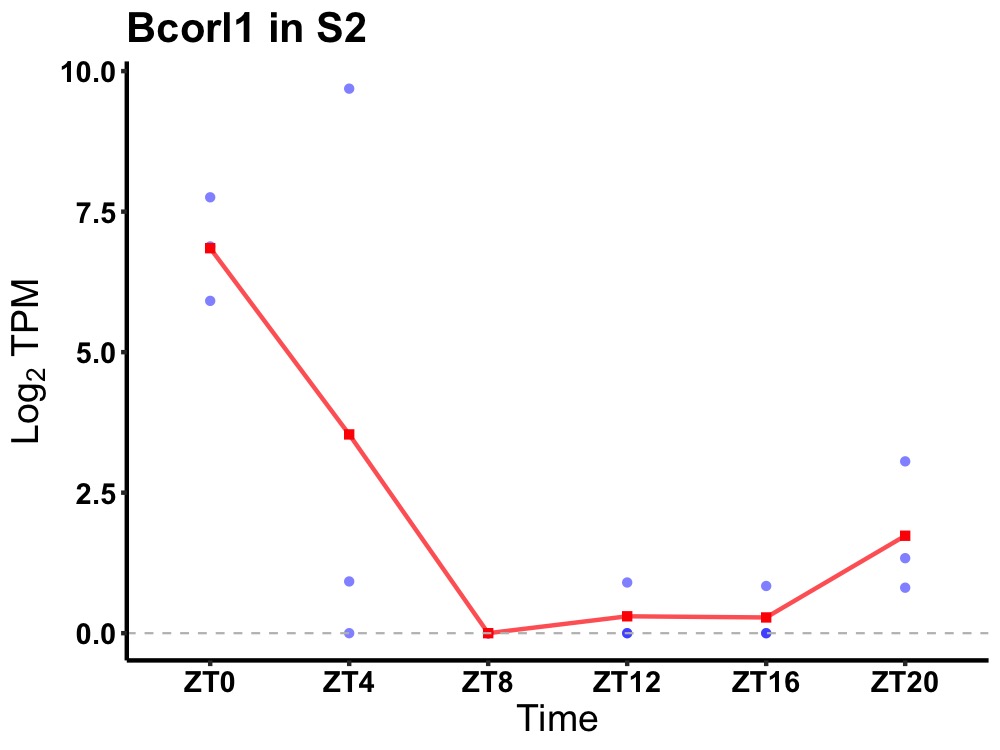 |
BCL-6 corepressor-like protein 1 | 0.043 | 24 | 0 | 1.30 |
| ENSMUSG00000083139 | Gm12418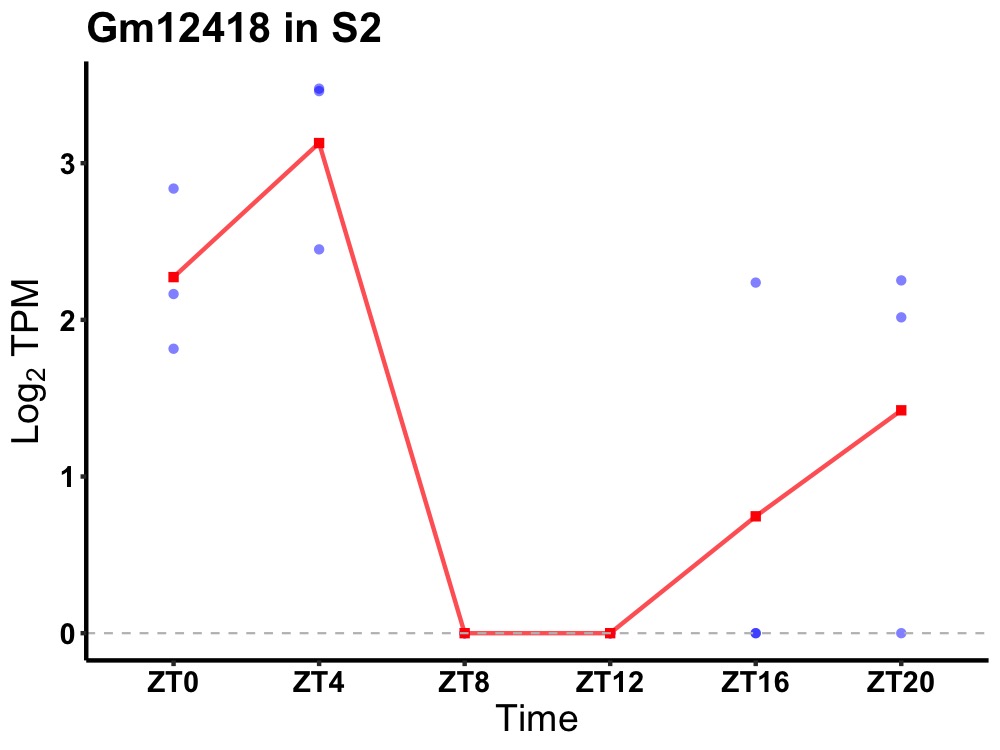 |
none | 0.043 | 20 | 4 | 1.28 |
| ENSMUSG00000051650 | B3gnt2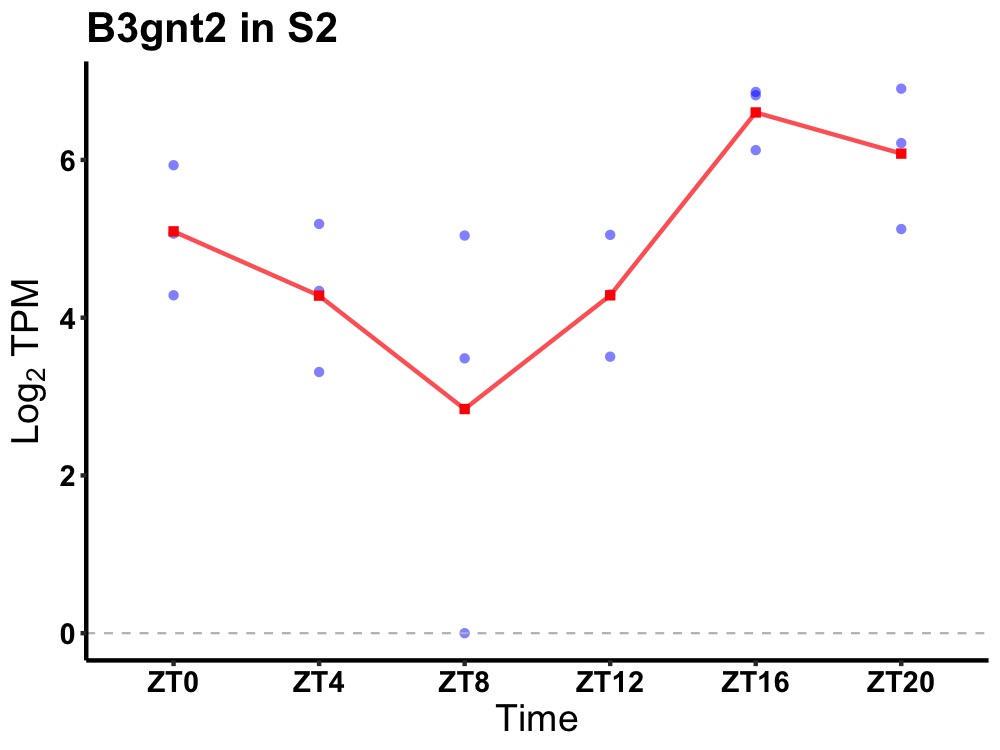 |
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 2 | 0.007 | 24 | 20 | 1.28 |
| ENSMUSG00000006315 | Sec23ip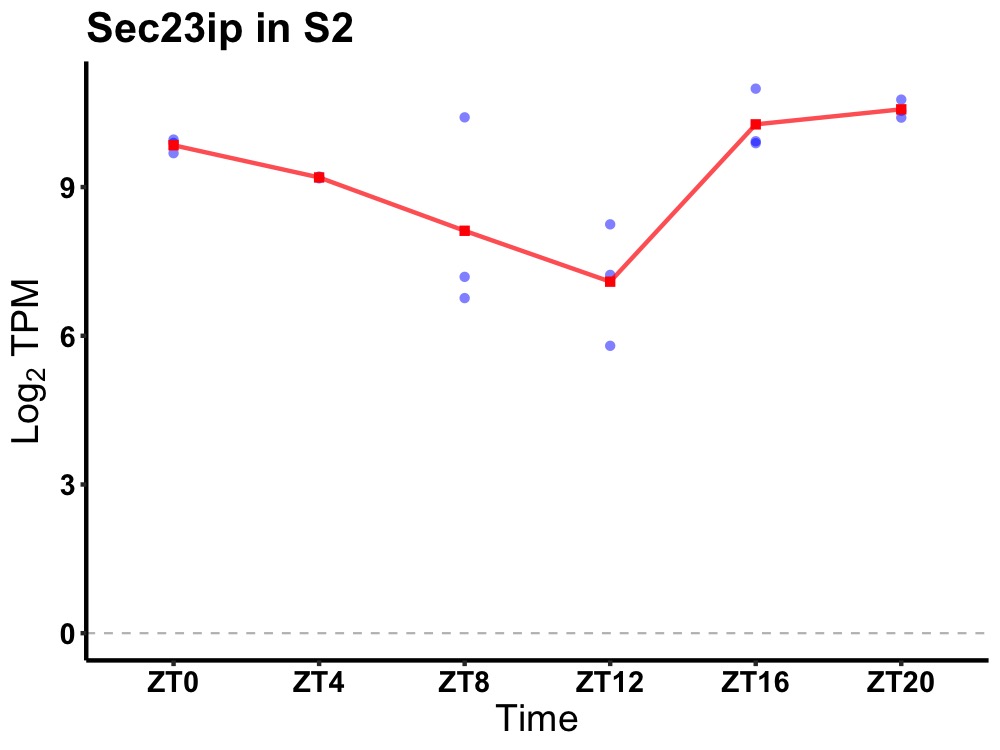 |
Sec23 interacting protein | 0.007 | 24 | 22 | 1.27 |
| ENSMUSG00000020308 | Spats2l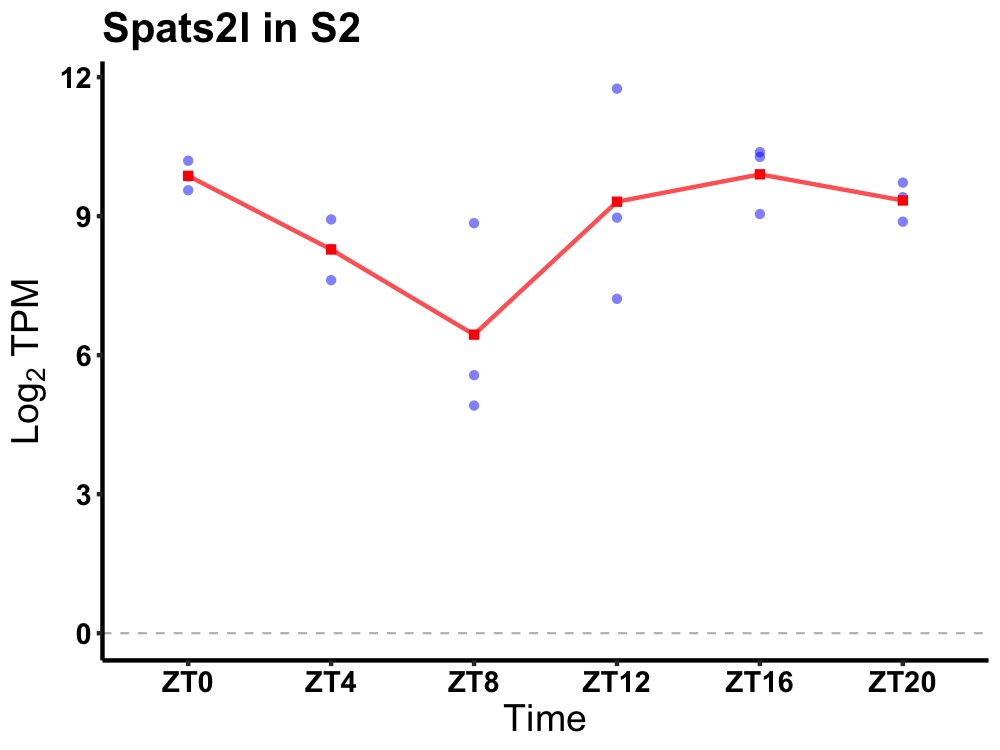 |
spermatogenesis associated, serine-rich 2-like | 0.007 | 20 | 18 | 1.25 |
| ENSMUSG00000039640 | Itga6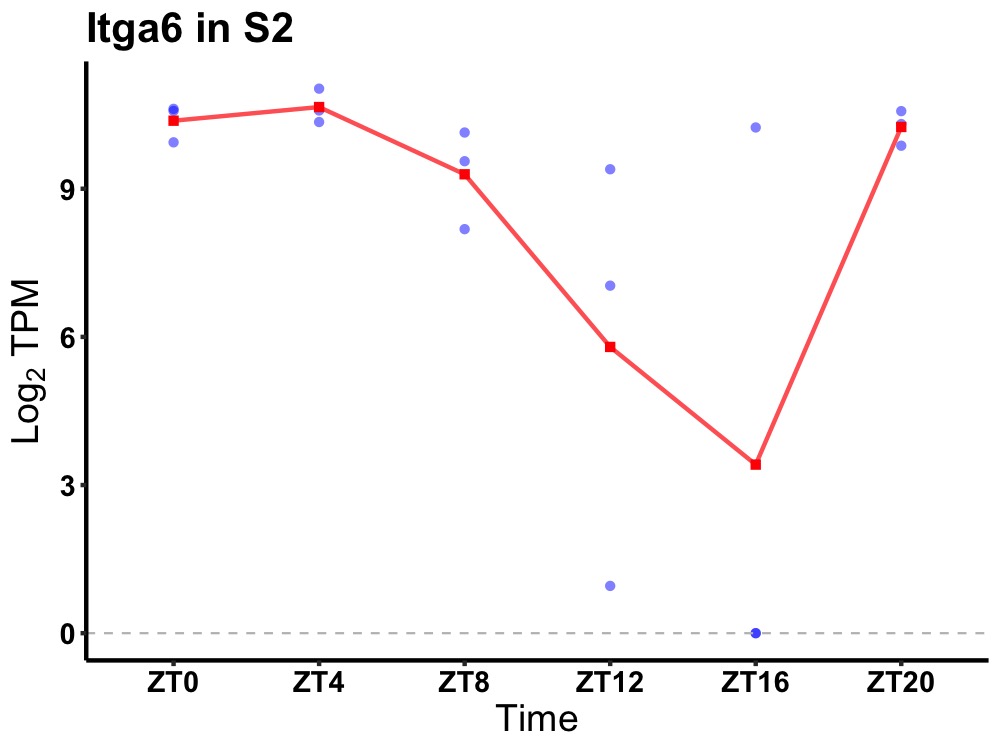 |
integrin alpha-6 | 0.007 | 24 | 2 | 1.25 |
| ENSMUSG00000107390 | Gm43323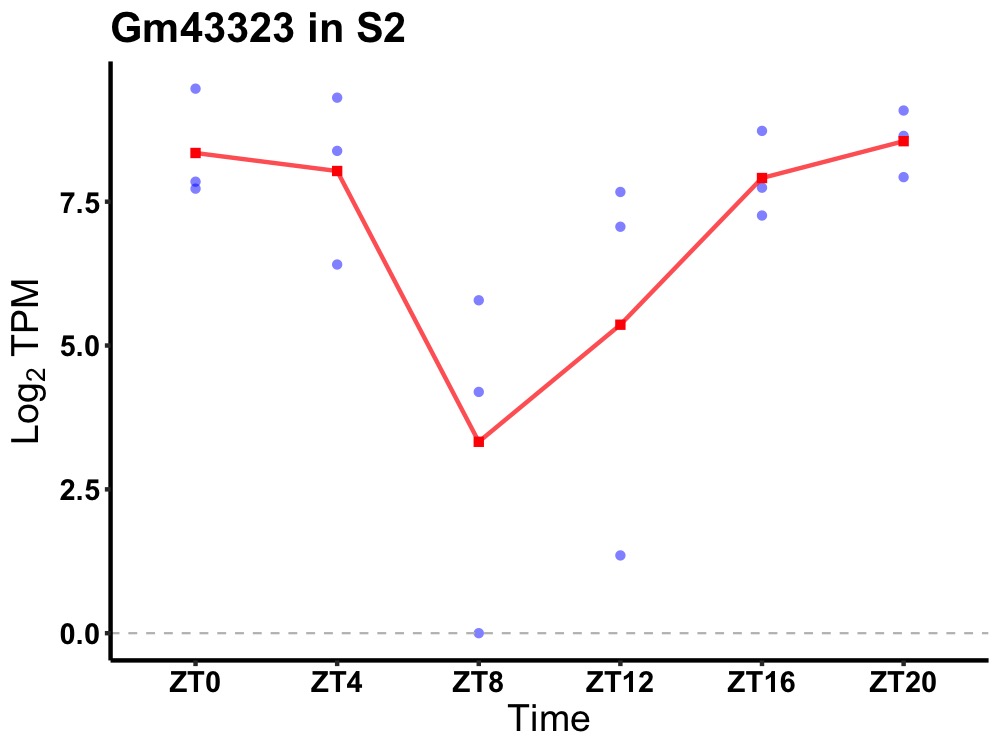 |
predicted gene 43323 | 0.019 | 24 | 22 | 1.23 |
| ENSMUSG00000050628 | Entpd7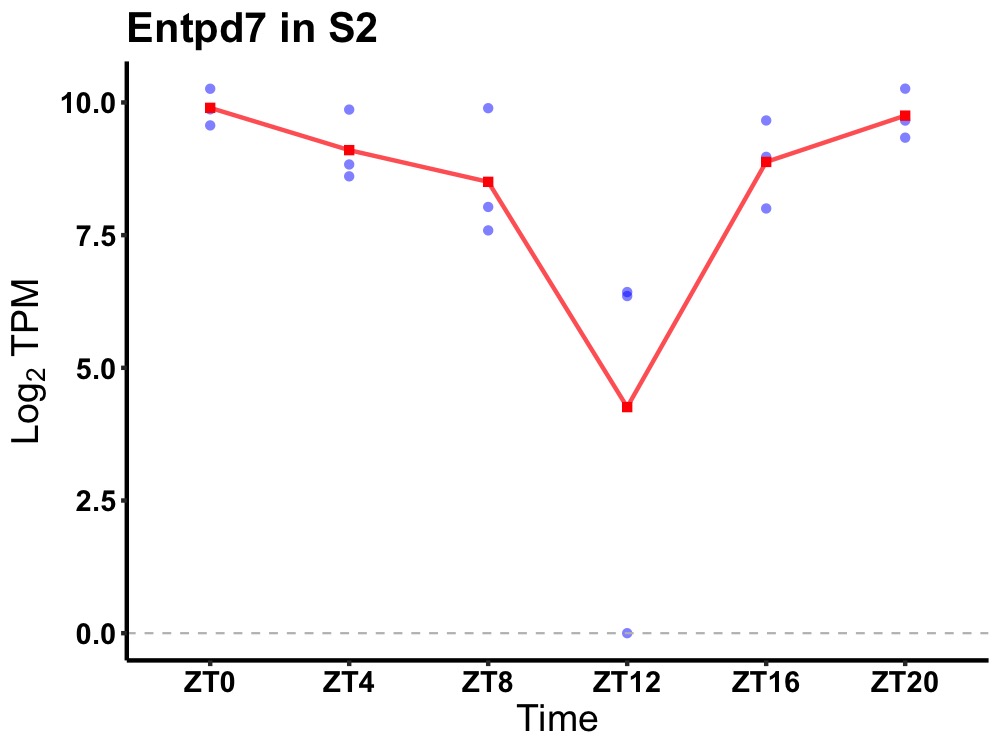 |
ectonucleoside triphosphate diphosphohydrolase 7 | 0.014 | 24 | 0 | 1.22 |
| ENSMUSG00000061684 | Zzz3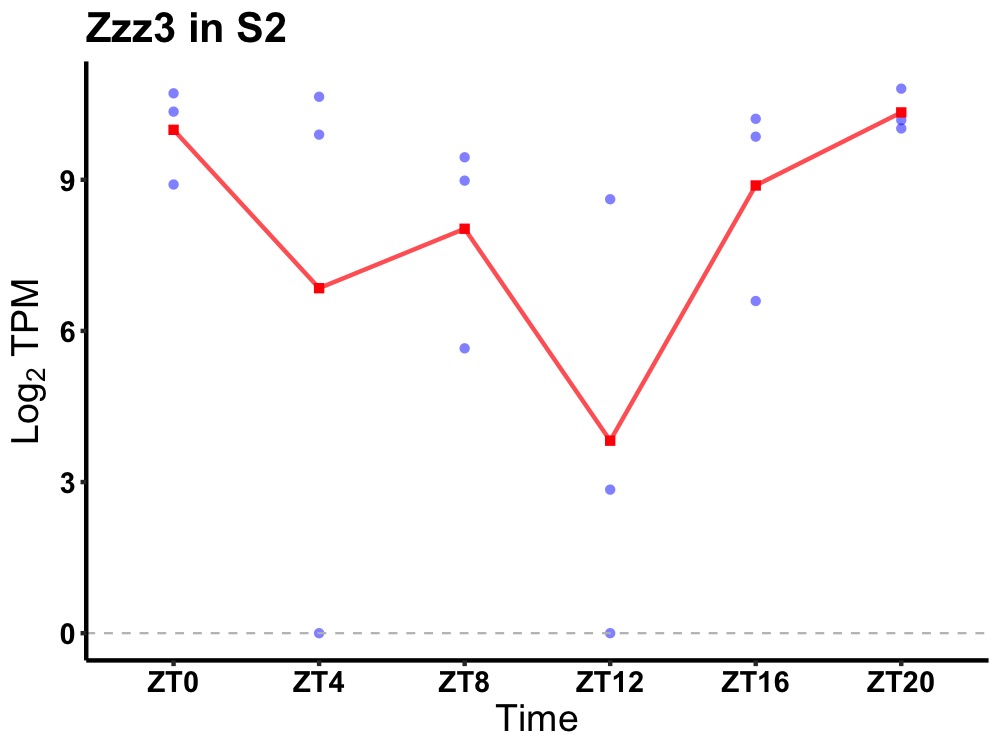 |
ZZ-type zinc finger-containing protein 3 | 0.043 | 20 | 2 | 1.20 |
| ENSMUSG00000084170 | Slc25a25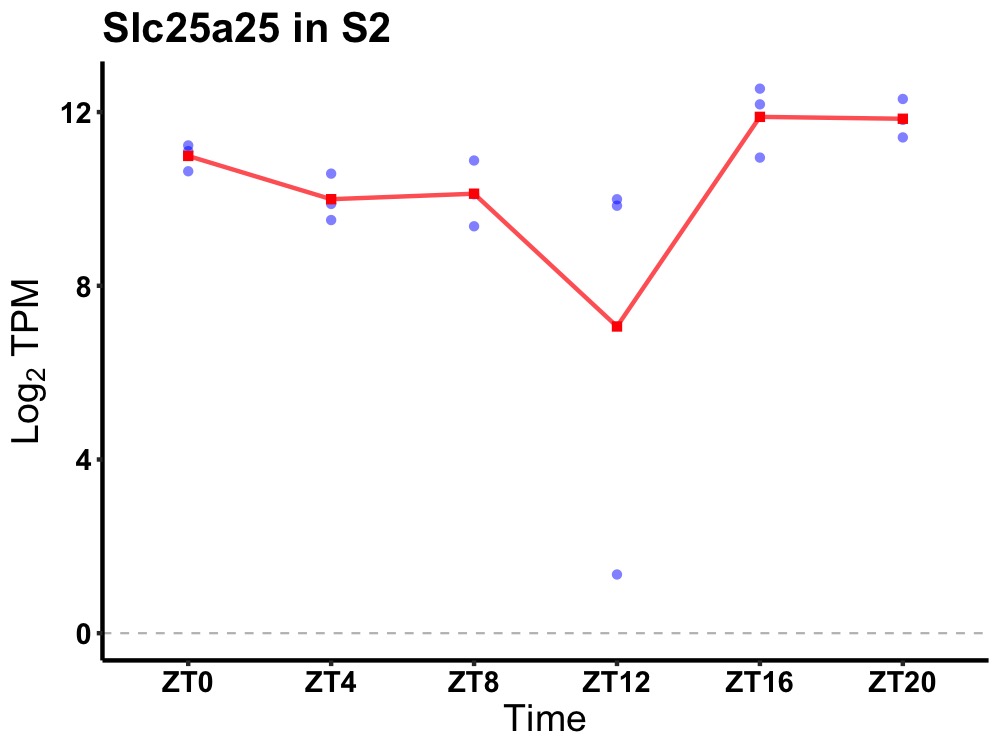 |
solute carrier family 25 (mitochondrial carrier, phosphate carrier), member 25 | 0.019 | 24 | 20 | 1.20 |
| ENSMUSG00000046096 | Mosmo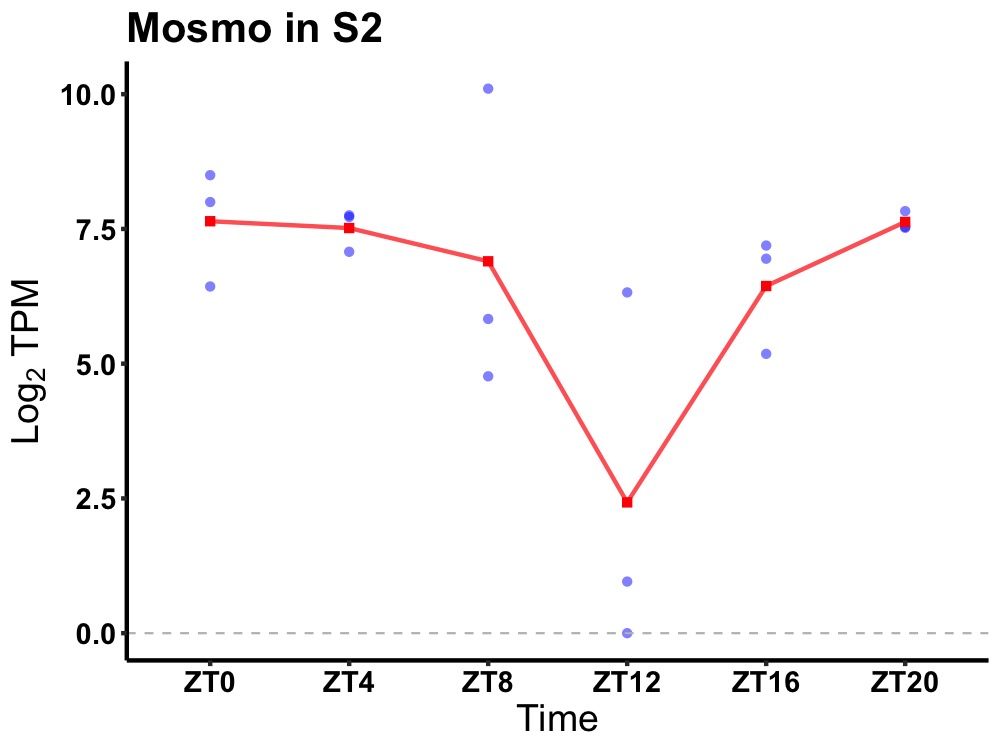 |
modulator of smoothened | 0.049 | 24 | 0 | 1.20 |
| ENSMUSG00000114252 | Gm5630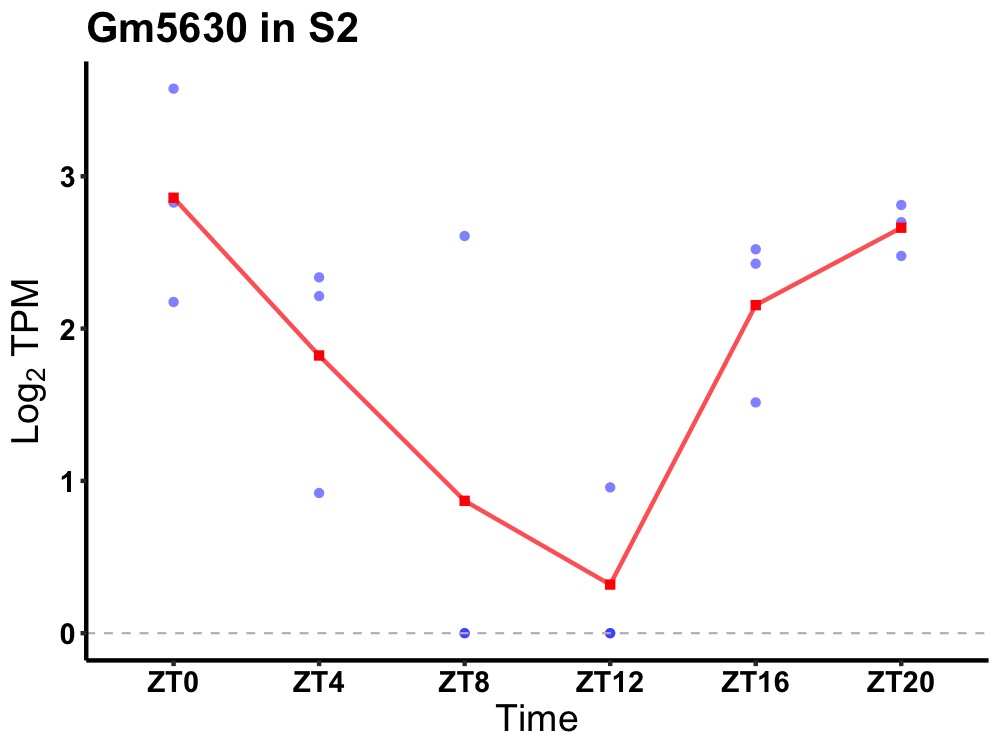 |
none | 0.010 | 20 | 0 | 1.19 |
| ENSMUSG00000026277 | Sec24d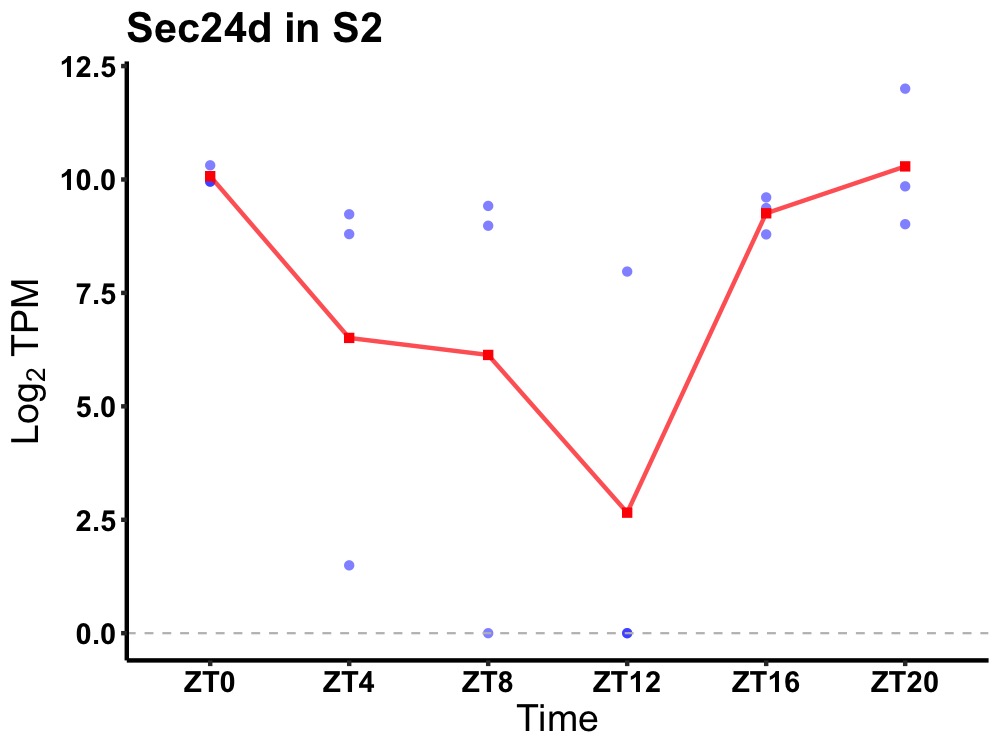 |
protein transport protein Sec24D | 0.014 | 20 | 0 | 1.19 |
| ENSMUSG00000022197 | Cdc23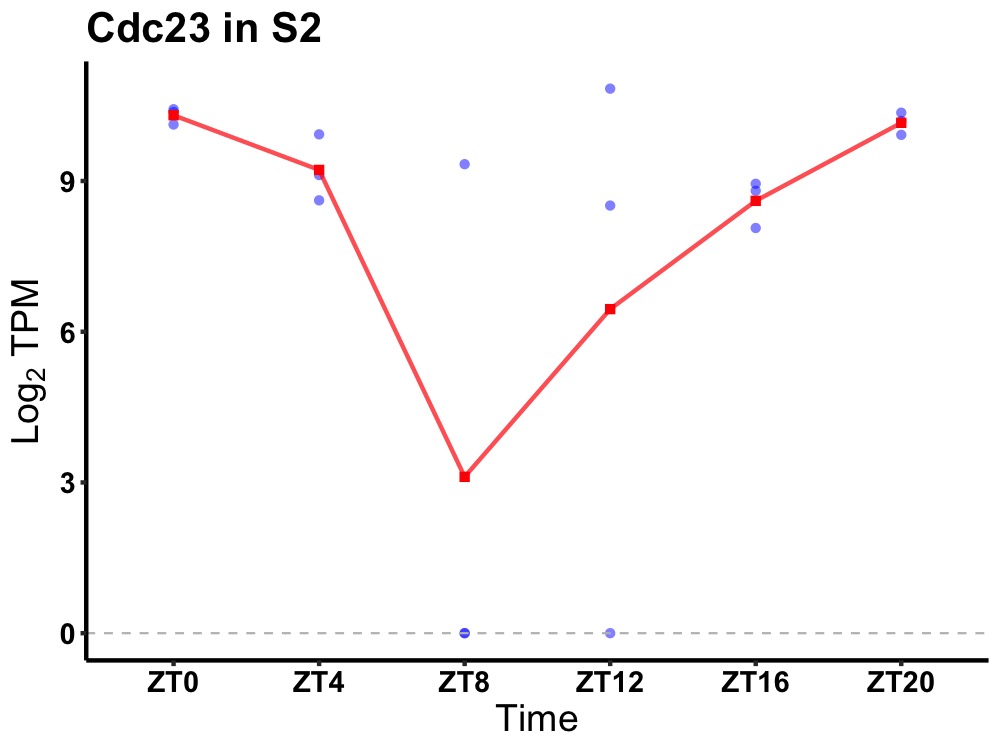 |
cell division cycle protein 23 homolog | 0.049 | 24 | 0 | 1.18 |
| ENSMUSG00000080950 | Cep85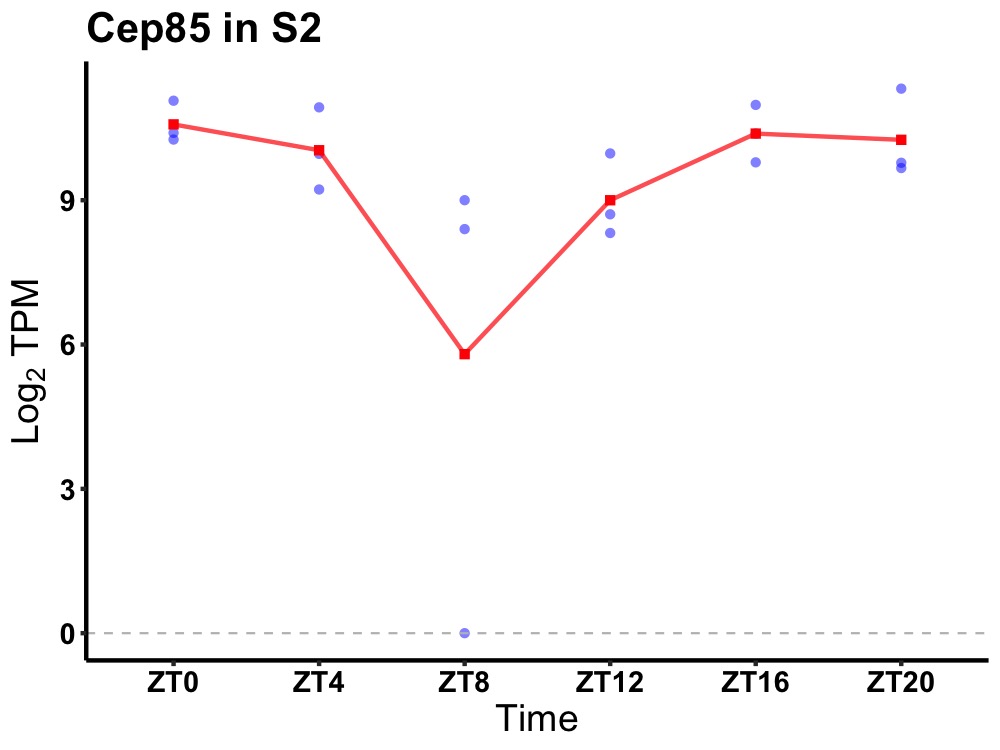 |
centrosomal protein 85 | 0.049 | 20 | 0 | 1.16 |
| ENSMUSG00000115662 | Hook1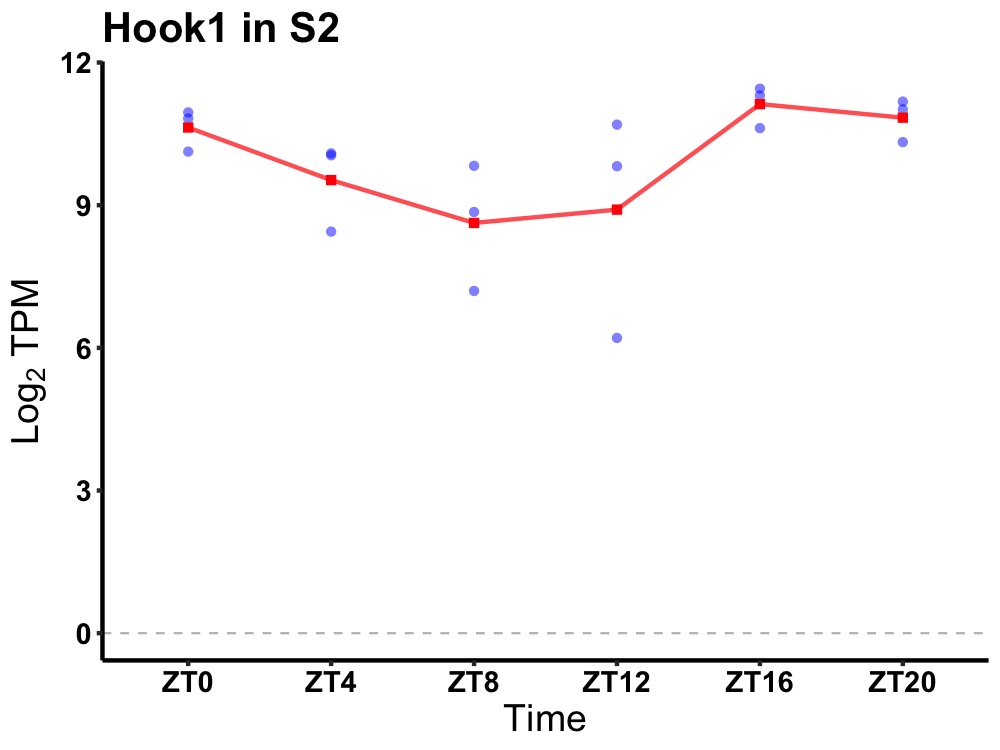 |
hook microtubule tethering protein 1 | 0.014 | 20 | 18 | 1.15 |
| ENSMUSG00000056050 | Ostm1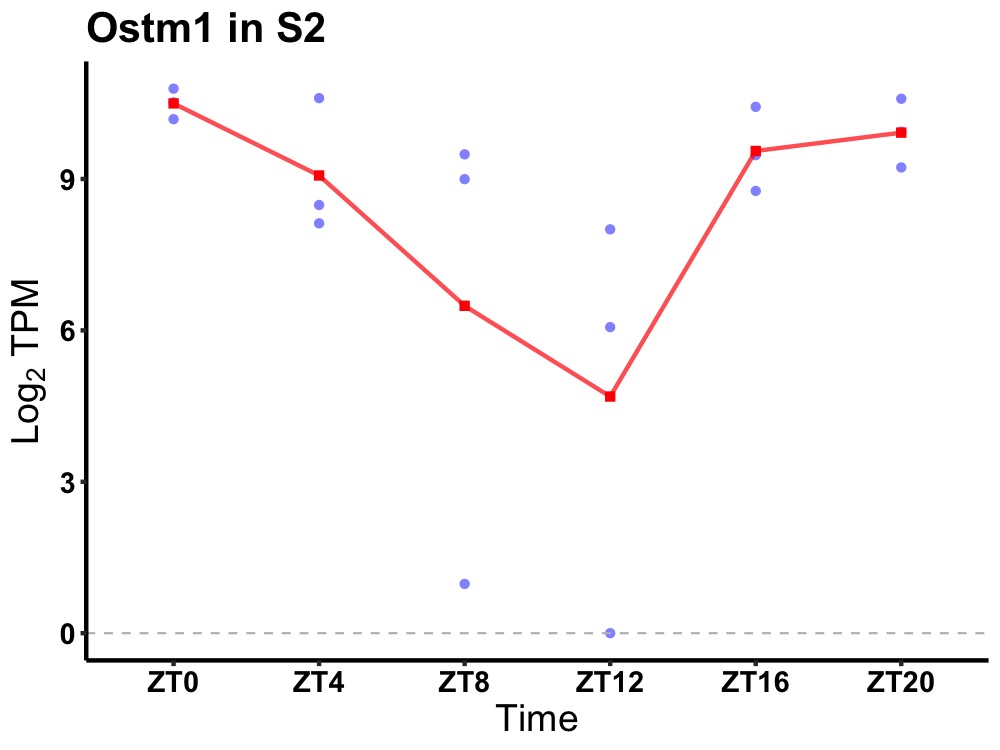 |
osteopetrosis associated transmembrane protein 1 | 0.027 | 24 | 0 | 1.13 |
| ENSMUSG00000055416 | Fam136b-ps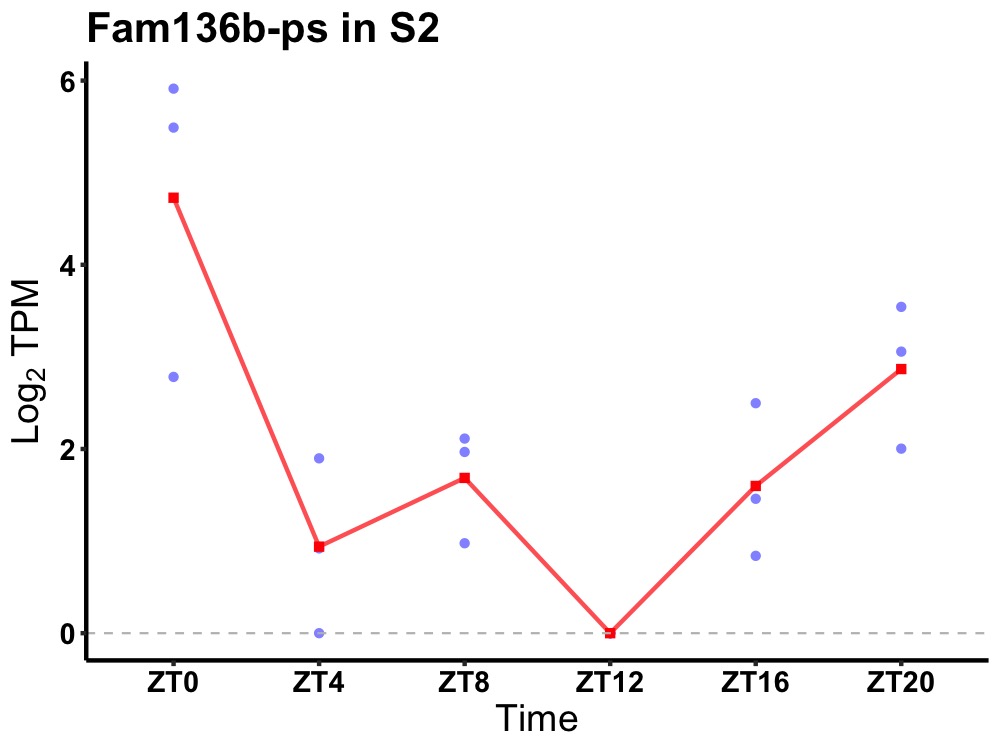 |
none | 0.012 | 24 | 0 | 1.11 |
| ENSMUSG00000082336 | Gm8401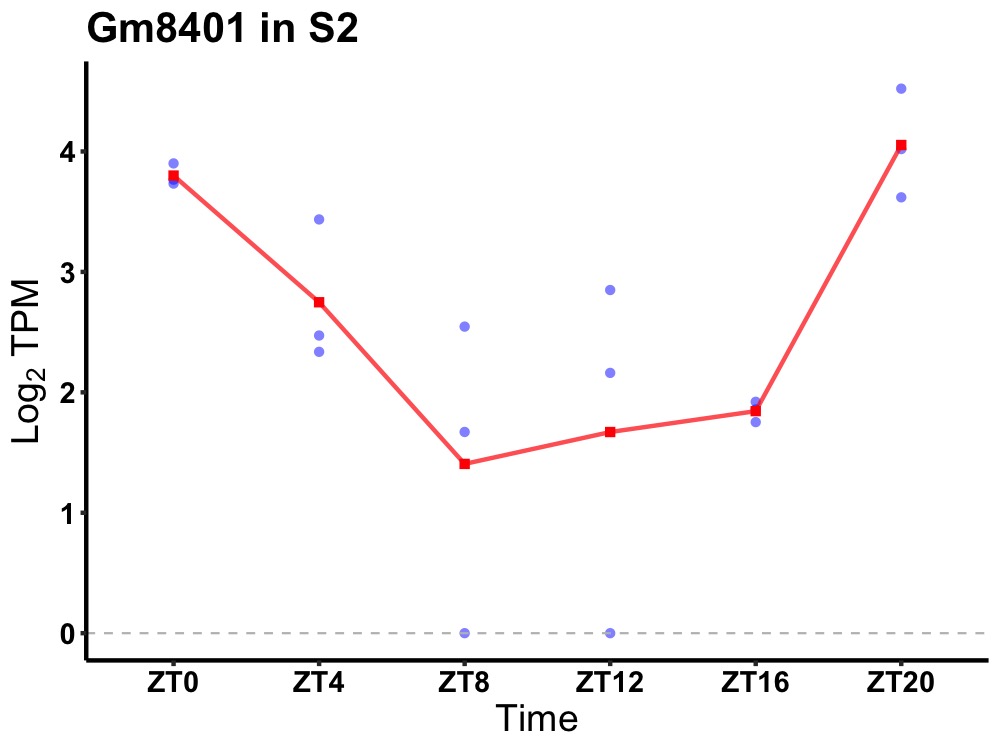 |
none | 0.004 | 20 | 2 | 1.10 |
| ENSMUSG00000103998 | Gm38025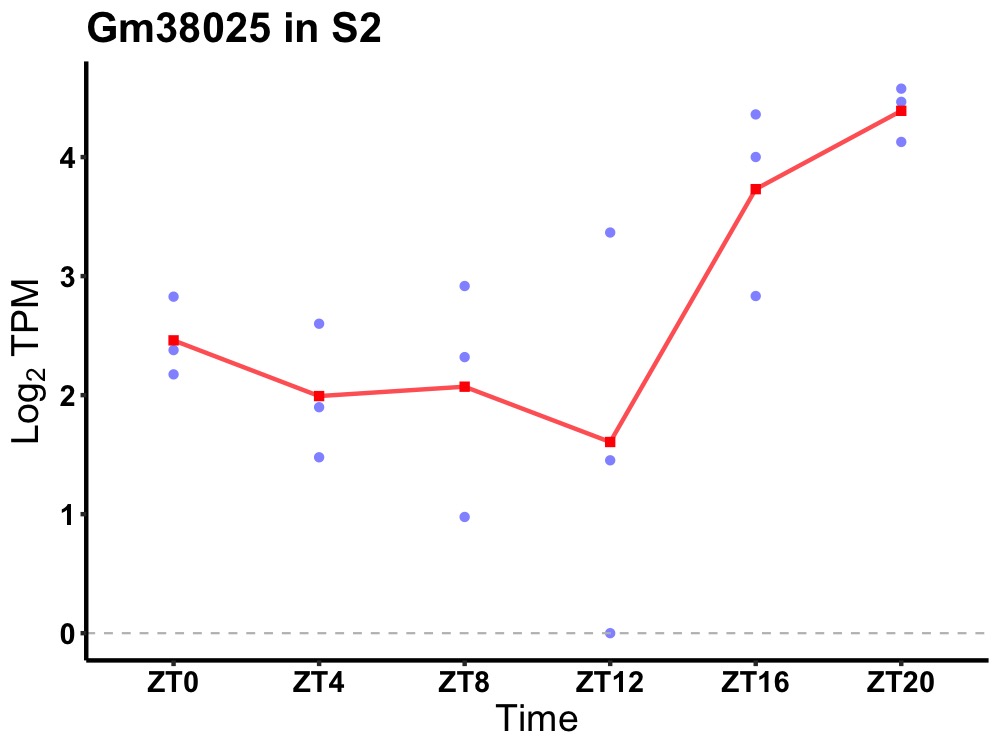 |
predicted gene, 38025 | 0.036 | 24 | 20 | 1.09 |
| ENSMUSG00000004393 | Trpv4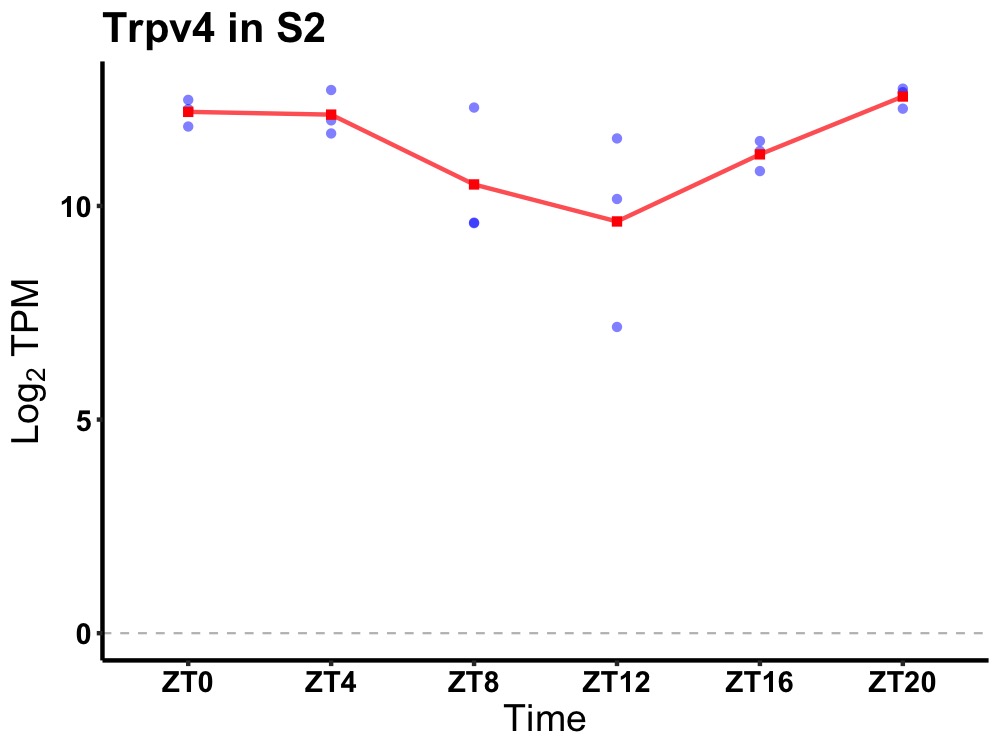 |
transient receptor potential cation channel, subfamily V, member 4 | 0.005 | 20 | 2 | 1.09 |
| ENSMUSG00000081789 | Gm11893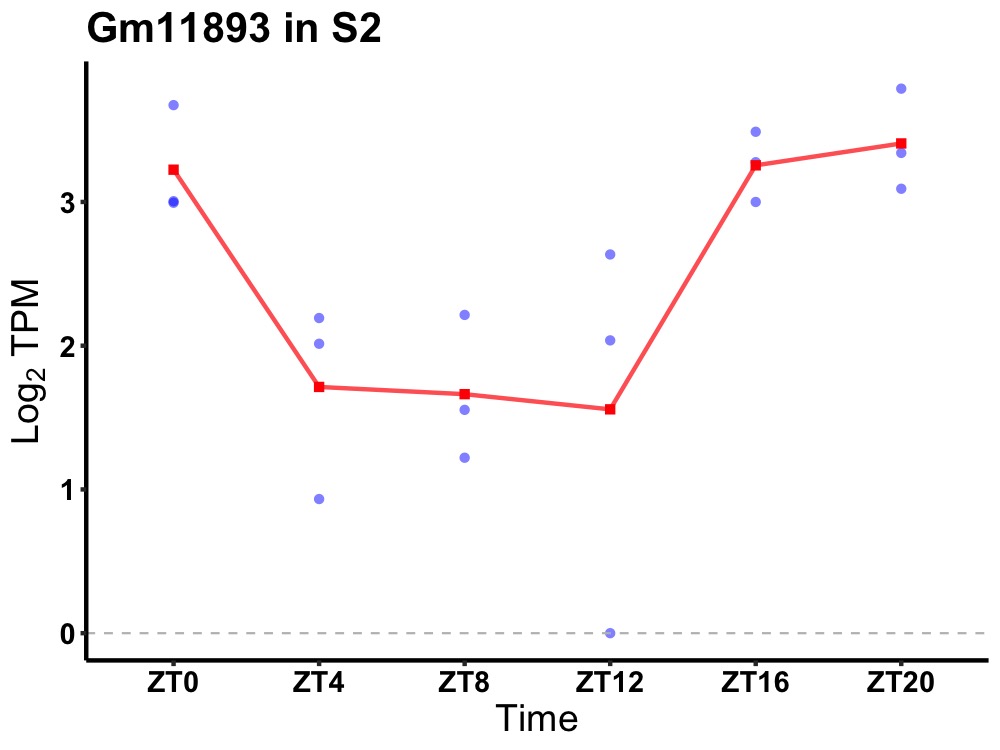 |
predicted gene 11893 | 0.005 | 24 | 20 | 1.09 |
| ENSMUSG00000036106 | Clcn7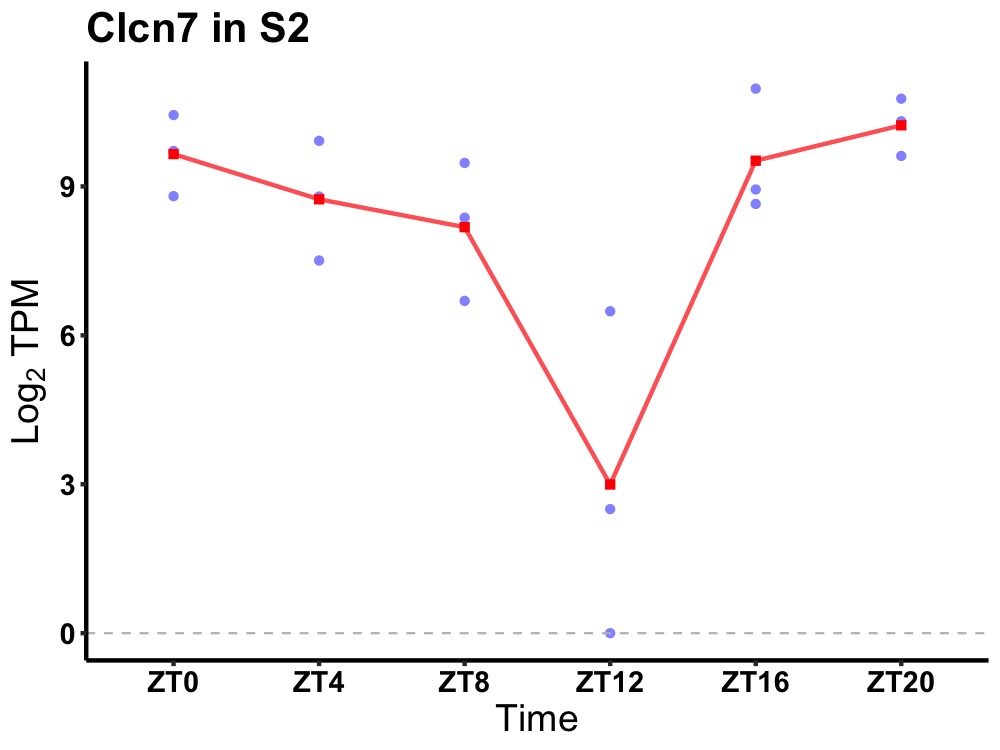 |
chloride channel, voltage-sensitive 7 | 0.014 | 20 | 2 | 1.08 |
| ENSMUSG00000019158 | Tmem160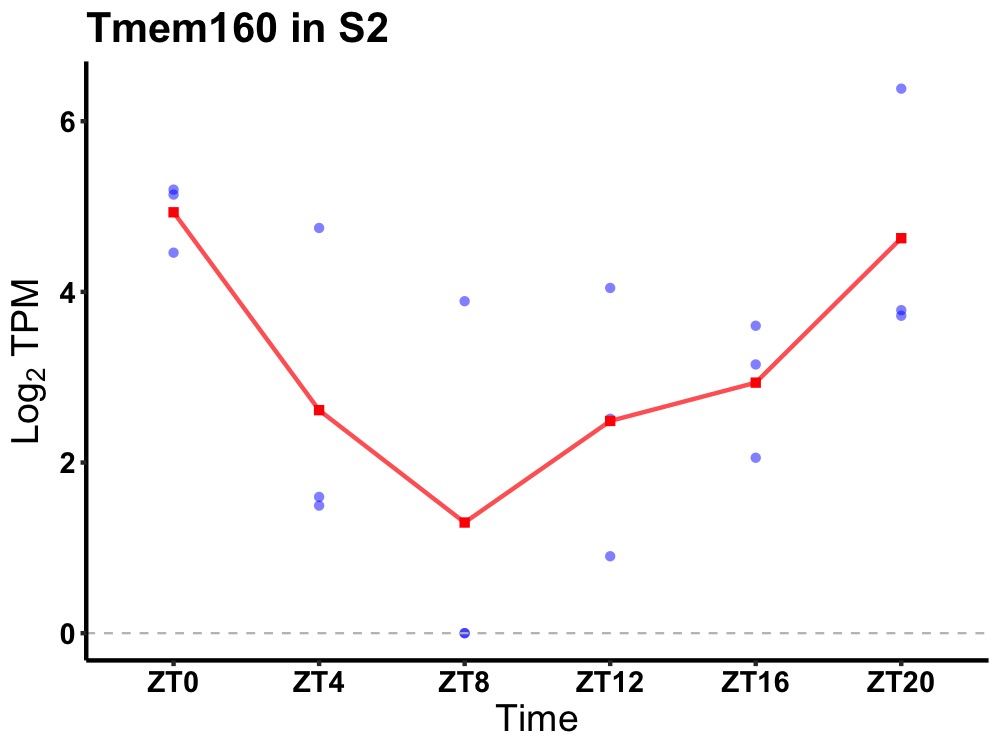 |
transmembrane protein 160 | 0.036 | 20 | 0 | 1.07 |
| ENSMUSG00000061486 | Gm5161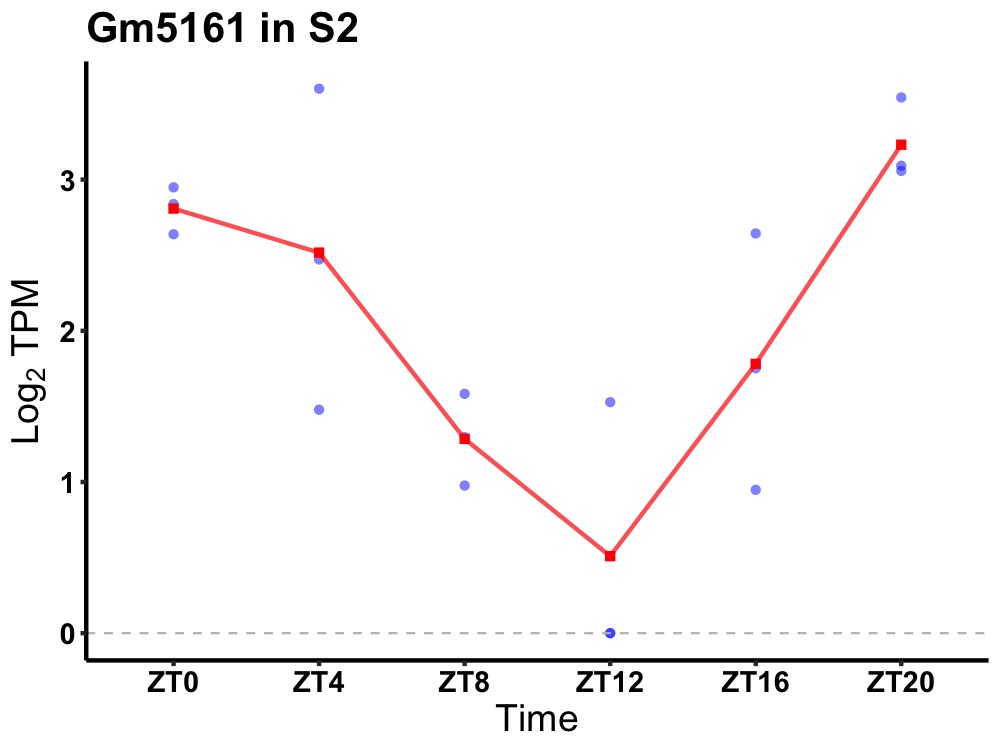 |
U1 small nuclear ribonucleoprotein A-like | 0.001 | 20 | 2 | 1.05 |
| ENSMUSG00000082245 | Gm6292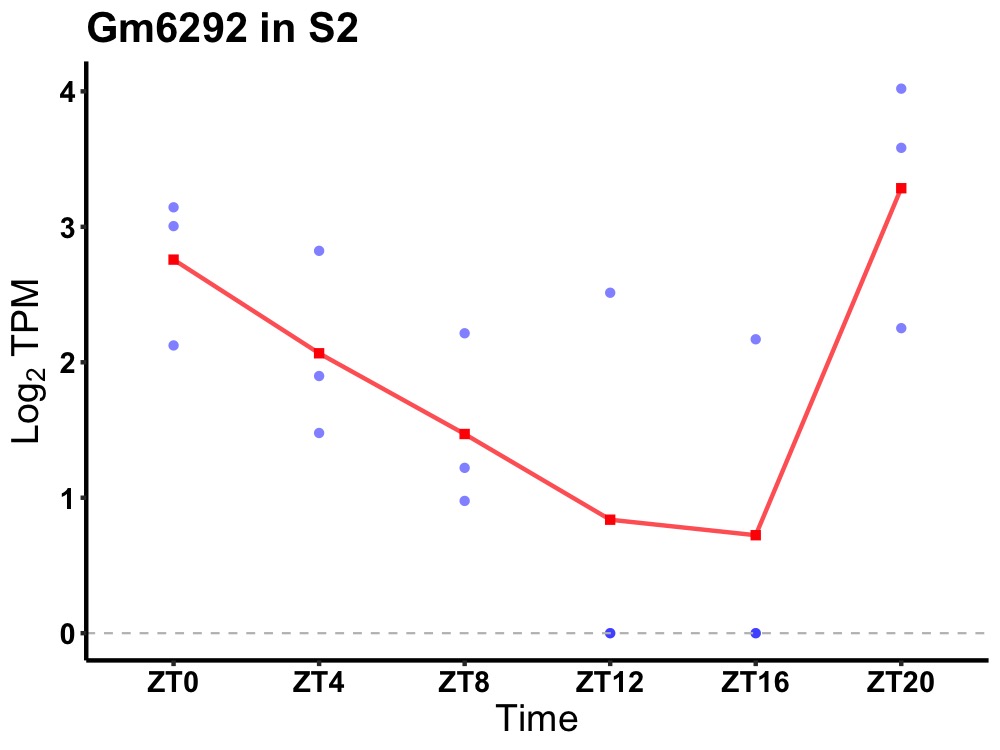 |
none | 0.049 | 20 | 2 | 1.05 |
| ENSMUSG00000048371 | Washc1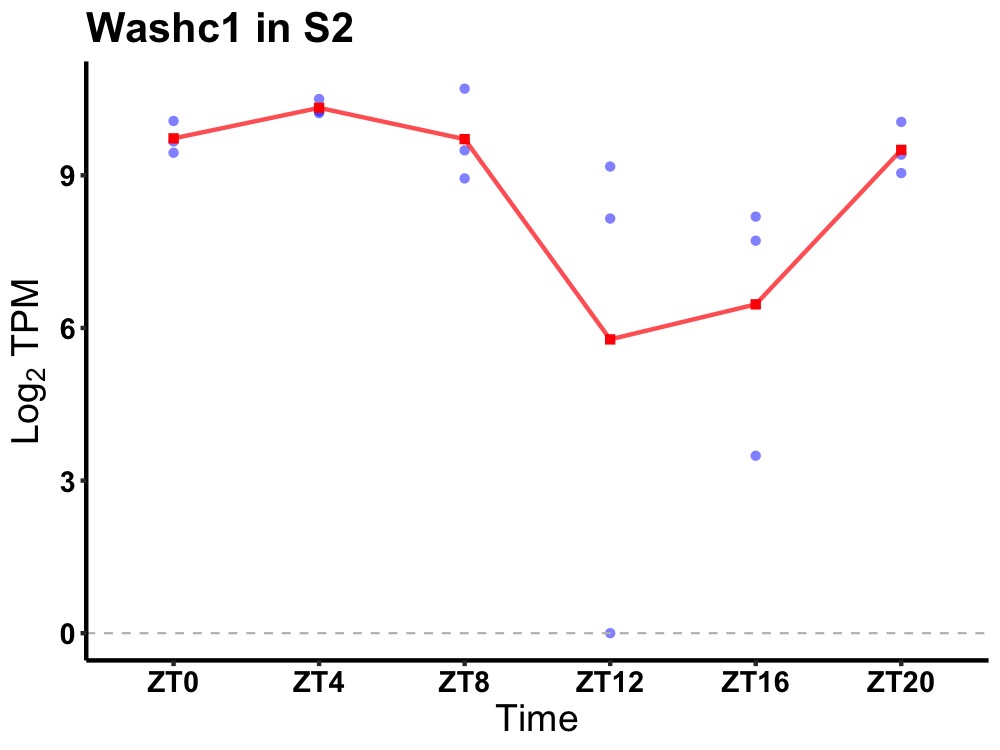 |
WASH complex subunit 1 | 0.001 | 24 | 4 | 1.04 |
| ENSMUSG00000028093 | Ndrg1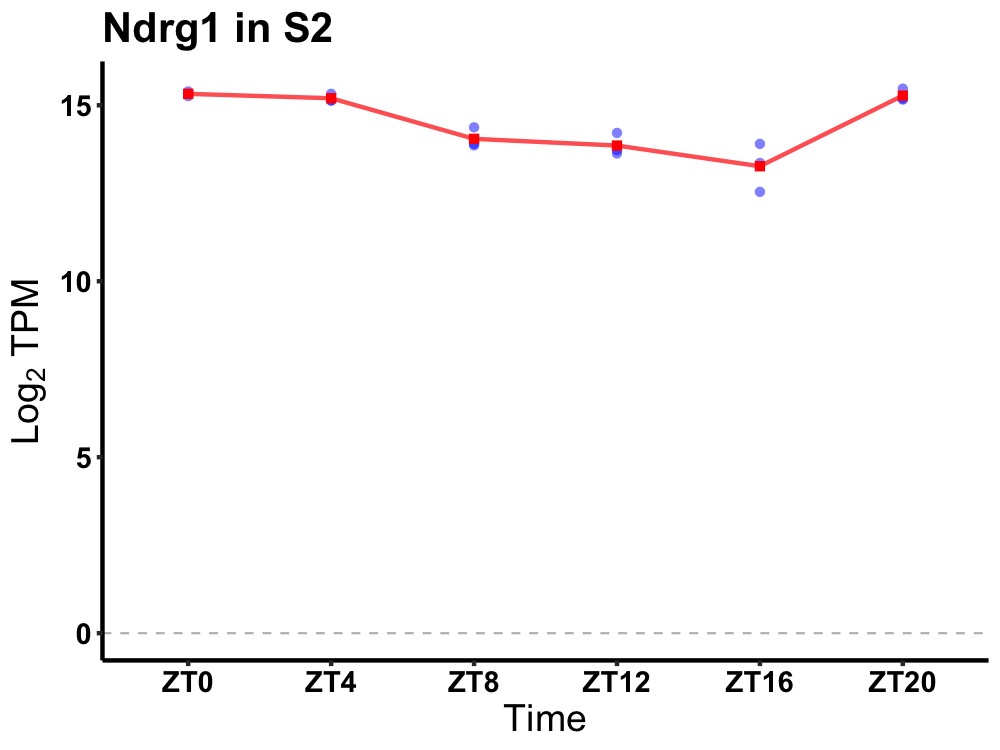 |
N-myc downstream regulated gene 1 | 0.005 | 24 | 2 | 1.04 |
| ENSMUSG00000018565 | Ccnl2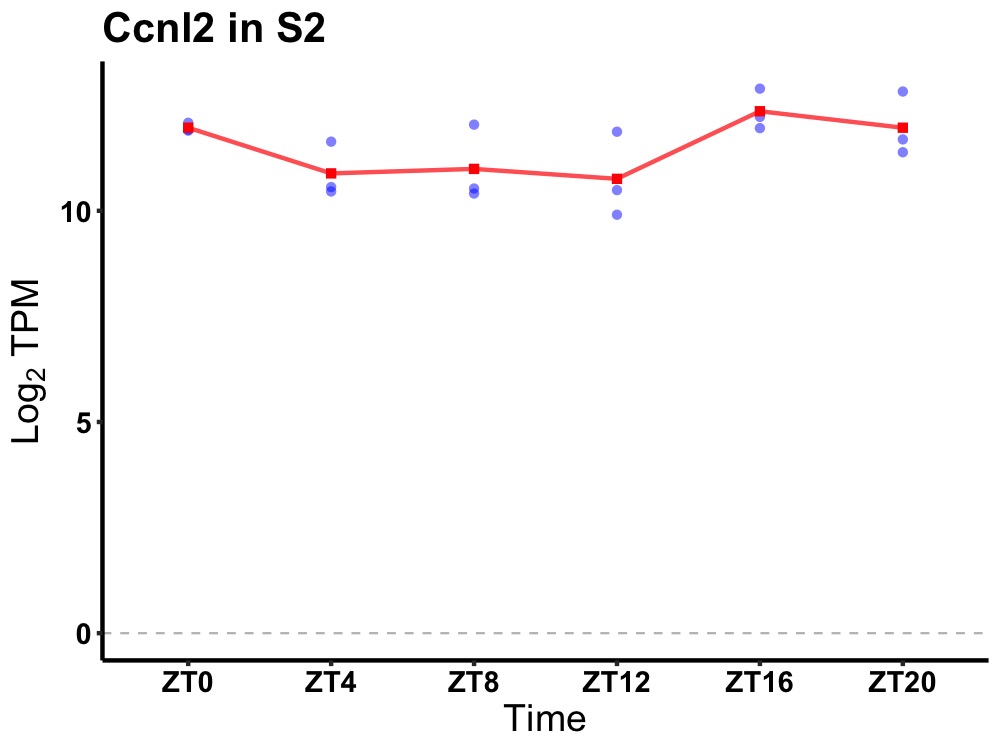 |
cyclin L2 | 0.049 | 20 | 18 | 1.03 |
| ENSMUSG00000096544 | Ddx3y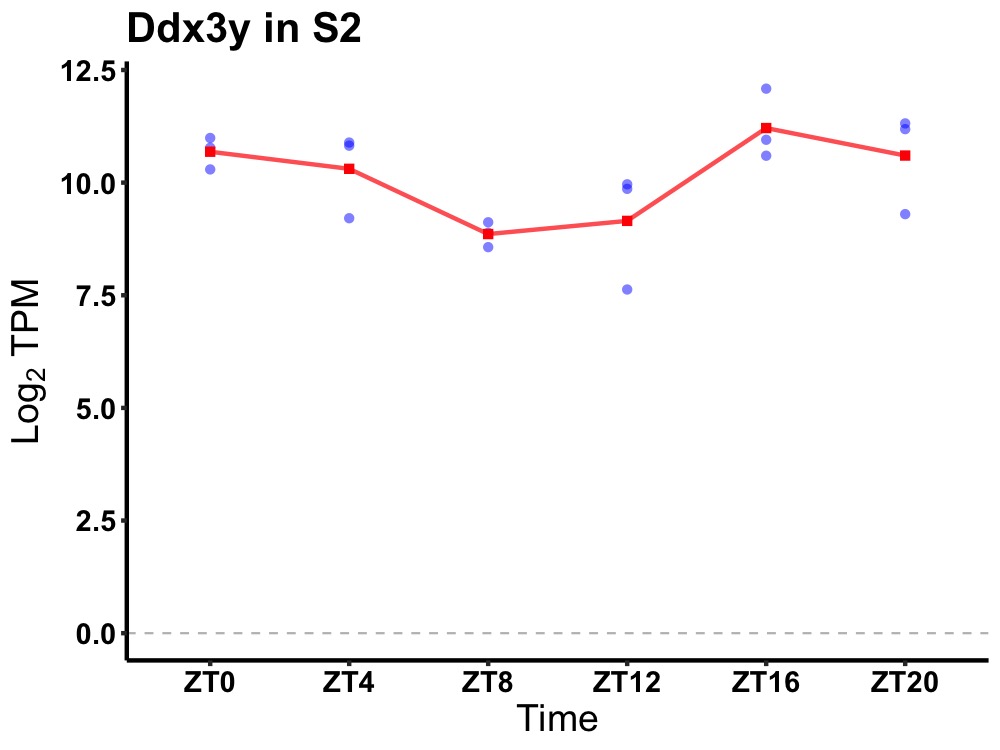 |
DEAD (Asp-Glu-Ala-Asp) box polypeptide 3, Y-linked | 0.049 | 24 | 22 | 1.03 |
| ENSMUSG00000036278 | Cnot10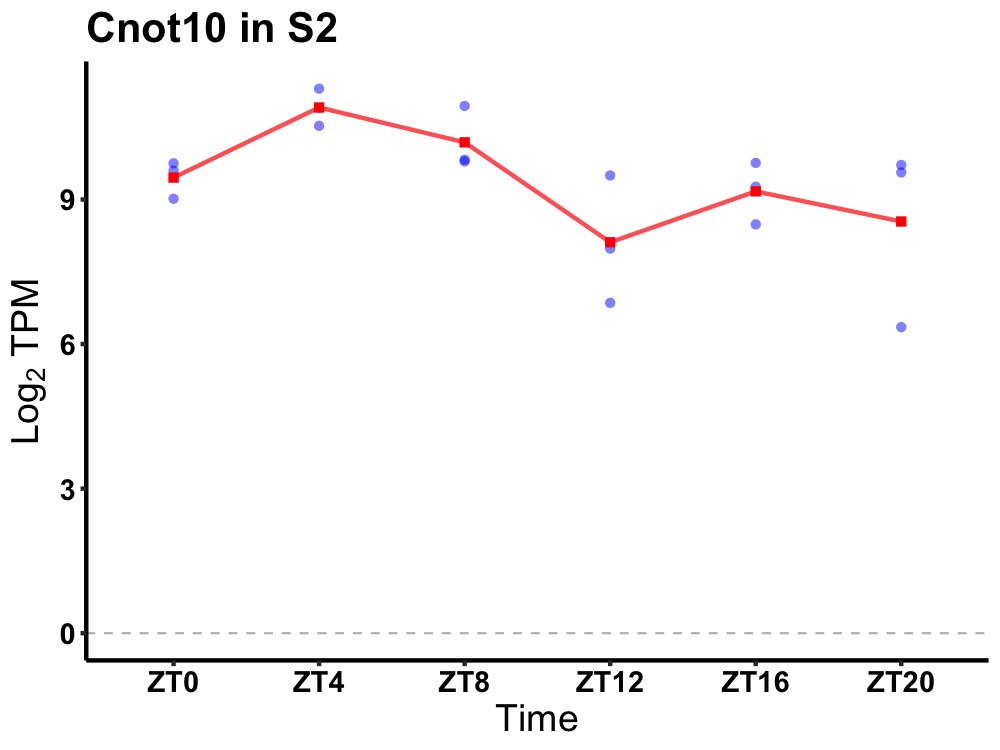 |
CCR4-NOT transcription complex, subunit 10 | 0.036 | 20 | 6 | 1.03 |
| ENSMUSG00000110935 | Sec23a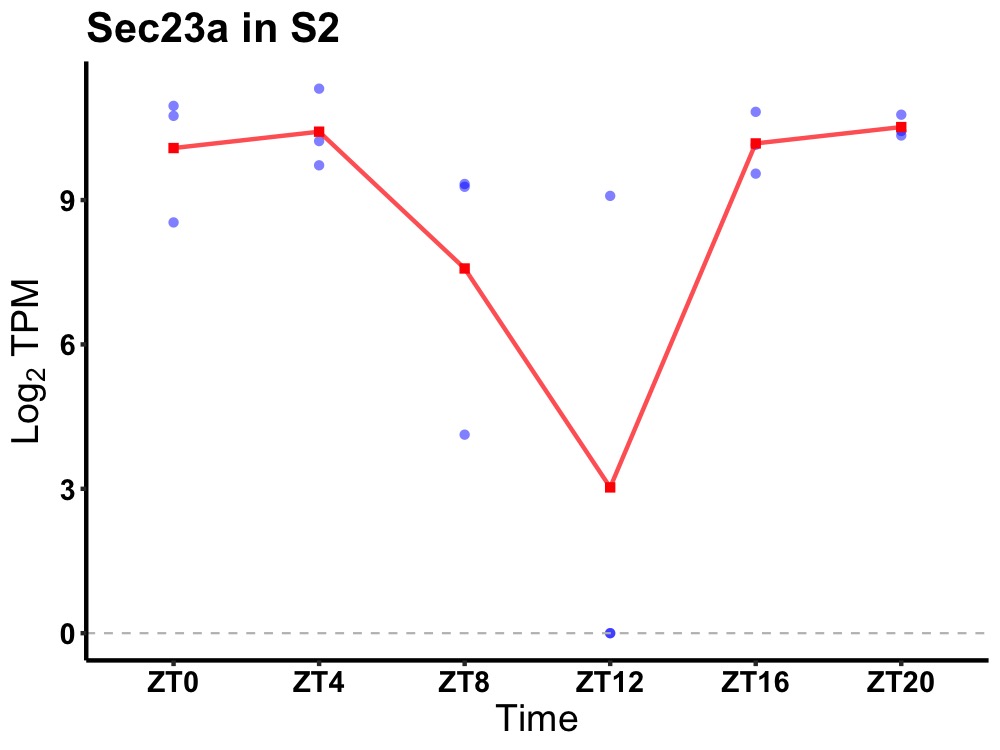 |
protein transport protein Sec23A | 0.019 | 24 | 0 | 1.02 |
| ENSMUSG00000108366 | Hus1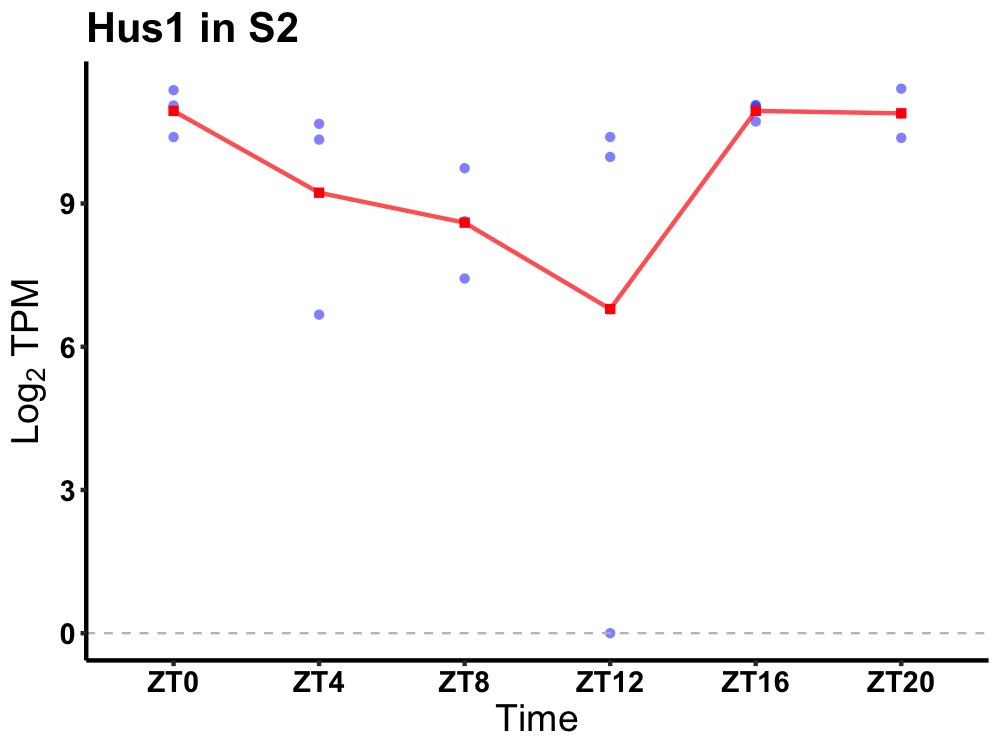 |
HUS1 checkpoint clamp component | 0.019 | 20 | 0 | 1.02 |
| ENSMUSG00000083834 | Gm12577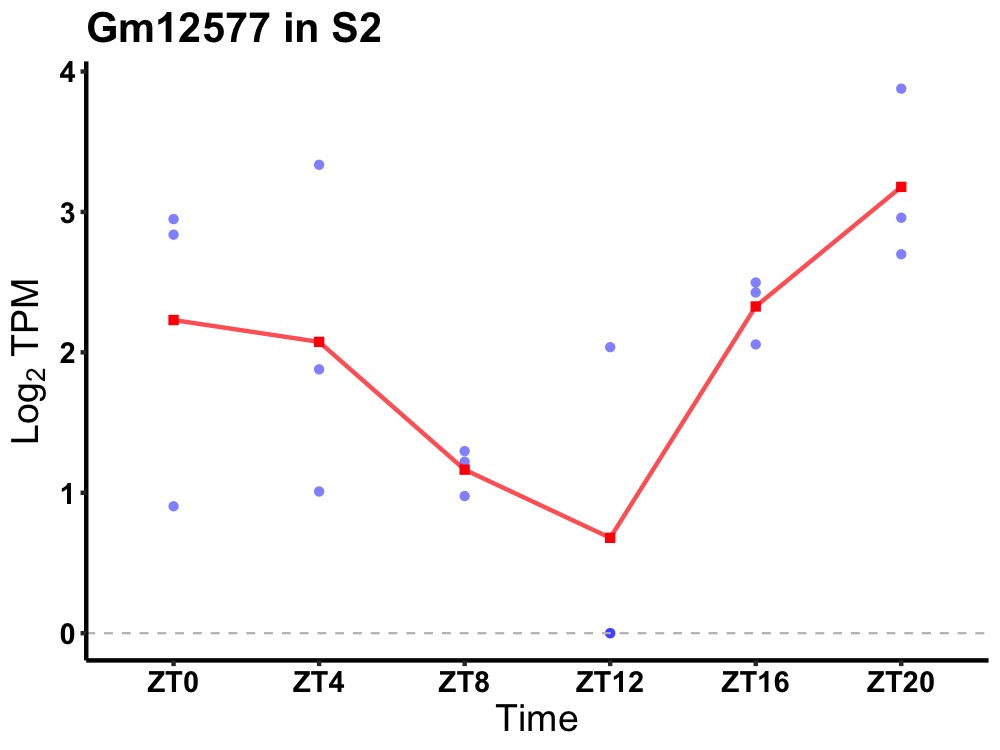 |
none | 0.049 | 24 | 22 | 1.00 |
| ENSMUSG00000083676 | Polr2b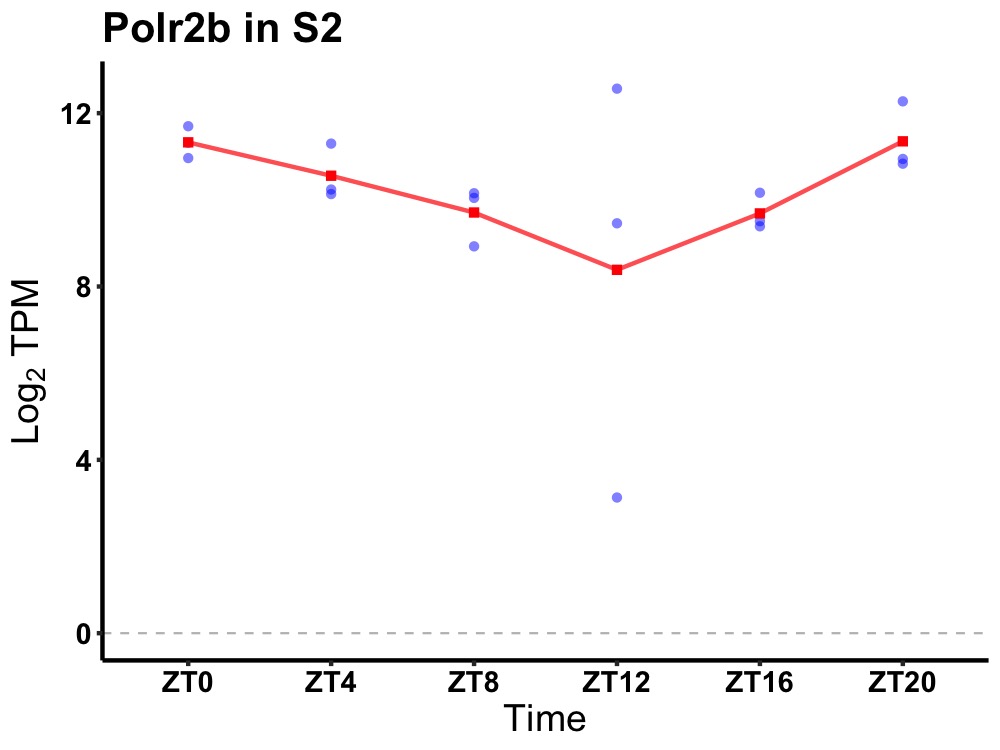 |
polymerase (RNA) II (DNA directed) polypeptide B | 0.049 | 24 | 0 | 1.00 |
| ENSMUSG00000014361 | 2210016L21Rik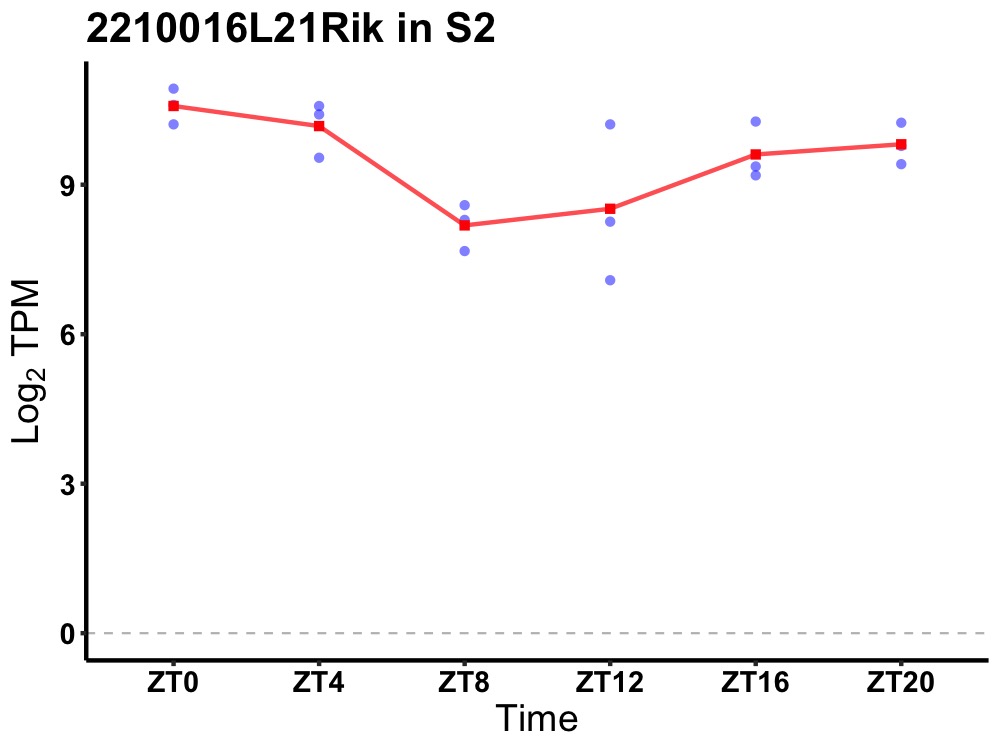 |
RIKEN cDNA 2210016L21 gene | 0.010 | 24 | 0 | 0.99 |
| ENSMUSG00000025371 | Itgb6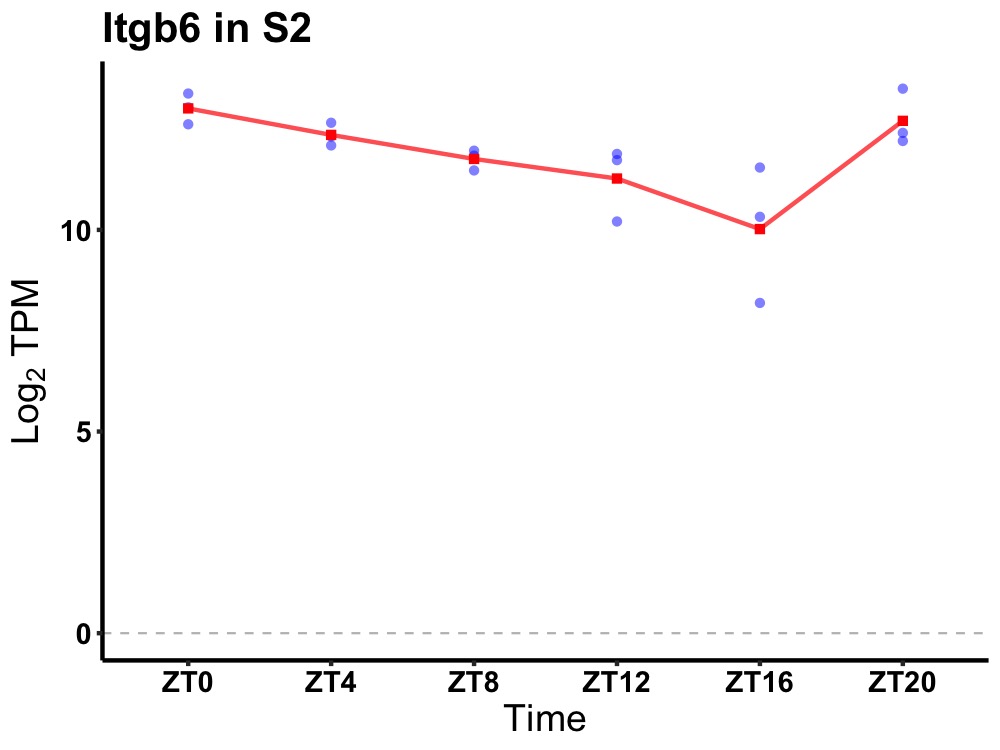 |
integrin beta 6 | 0.003 | 24 | 2 | 0.99 |
| ENSMUSG00000080715 | Ecd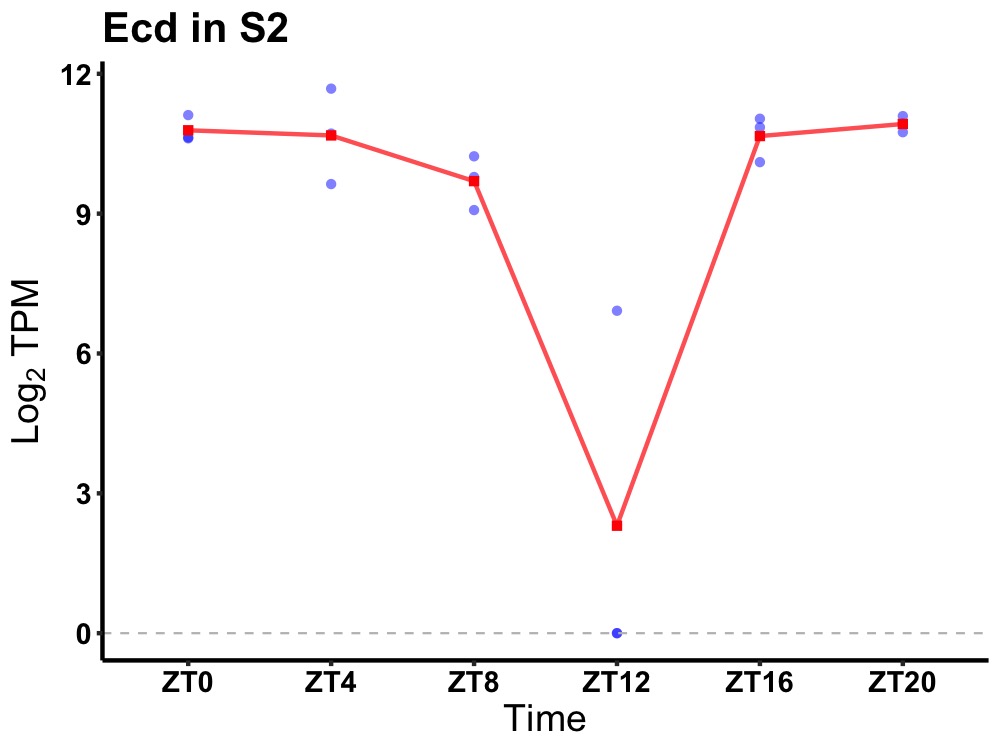 |
protein ecdysoneless homolog | 0.019 | 20 | 2 | 0.99 |
| ENSMUSG00000089694 | Nat8f7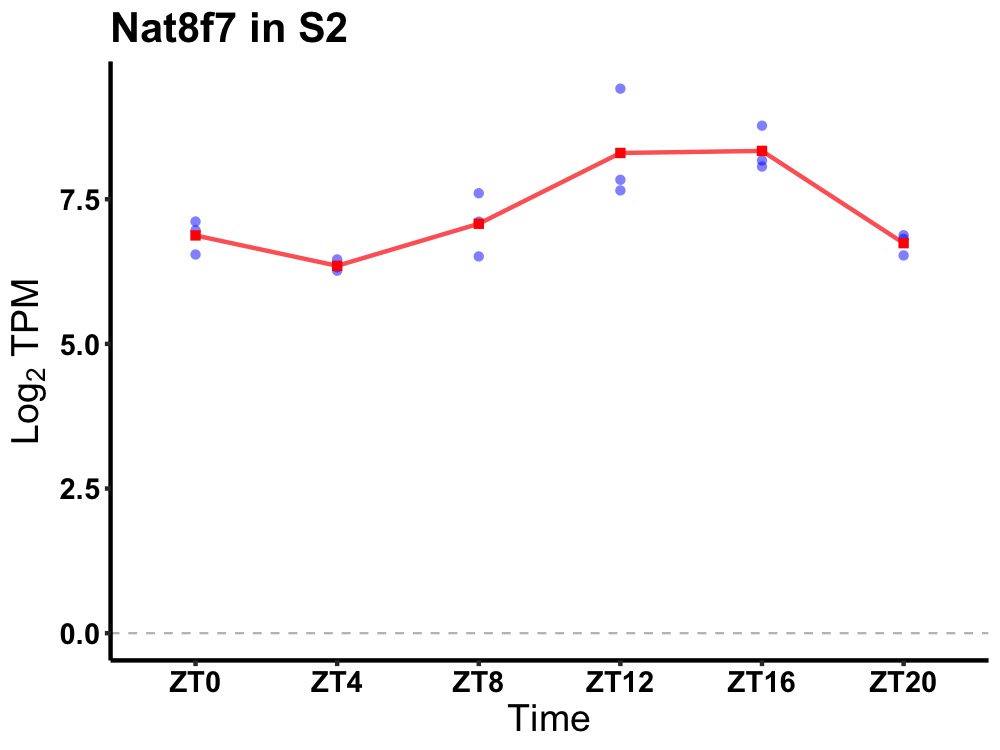 |
N-acetyltransferase 8 (GCN5-related) family member 7 | 0.000 | 20 | 14 | 0.98 |
| ENSMUSG00000109532 | Alcam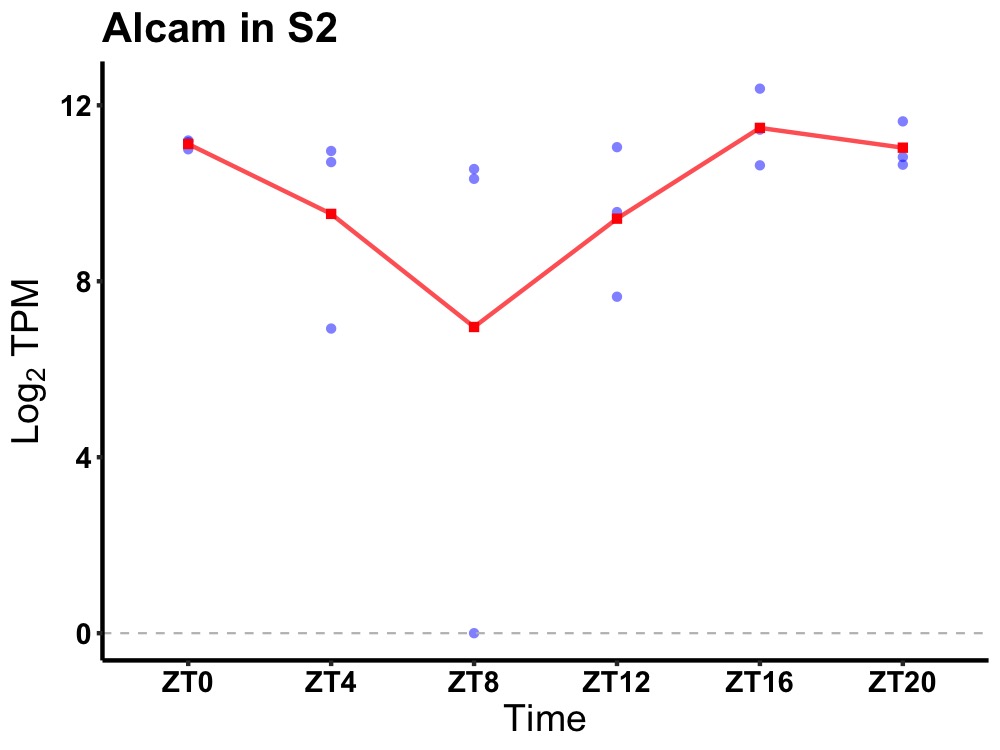 |
activated leukocyte cell adhesion molecule | 0.049 | 20 | 18 | 0.97 |
| ENSMUSG00000020644 | Rnf145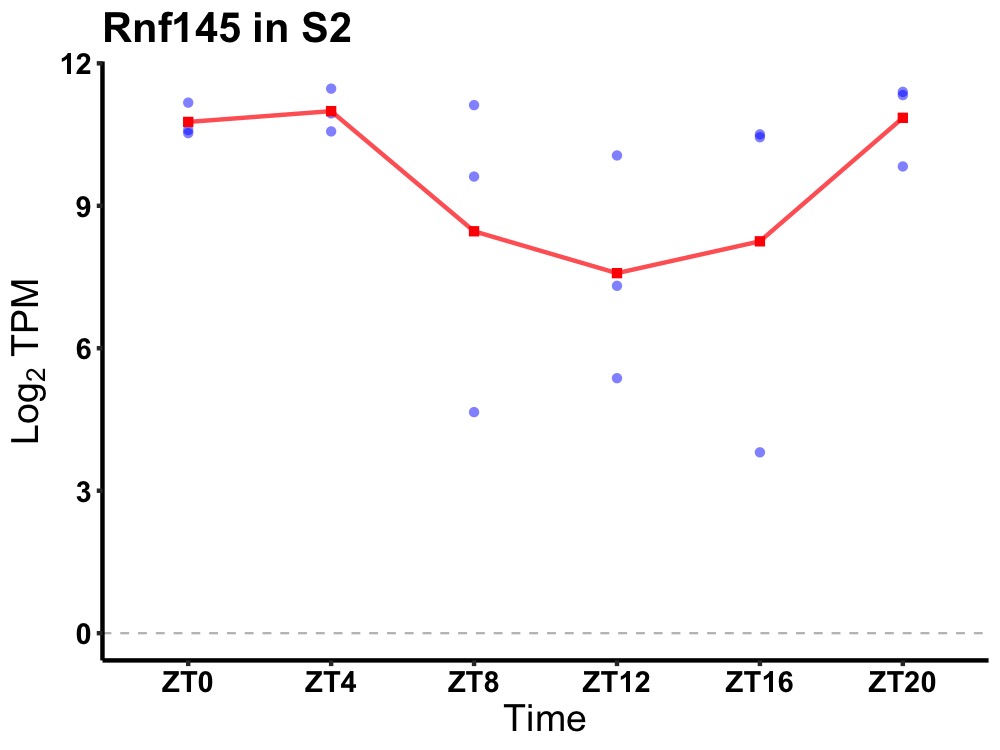 |
ring finger protein 145 | 0.049 | 20 | 4 | 0.96 |
| ENSMUSG00000069678 | Pcgf1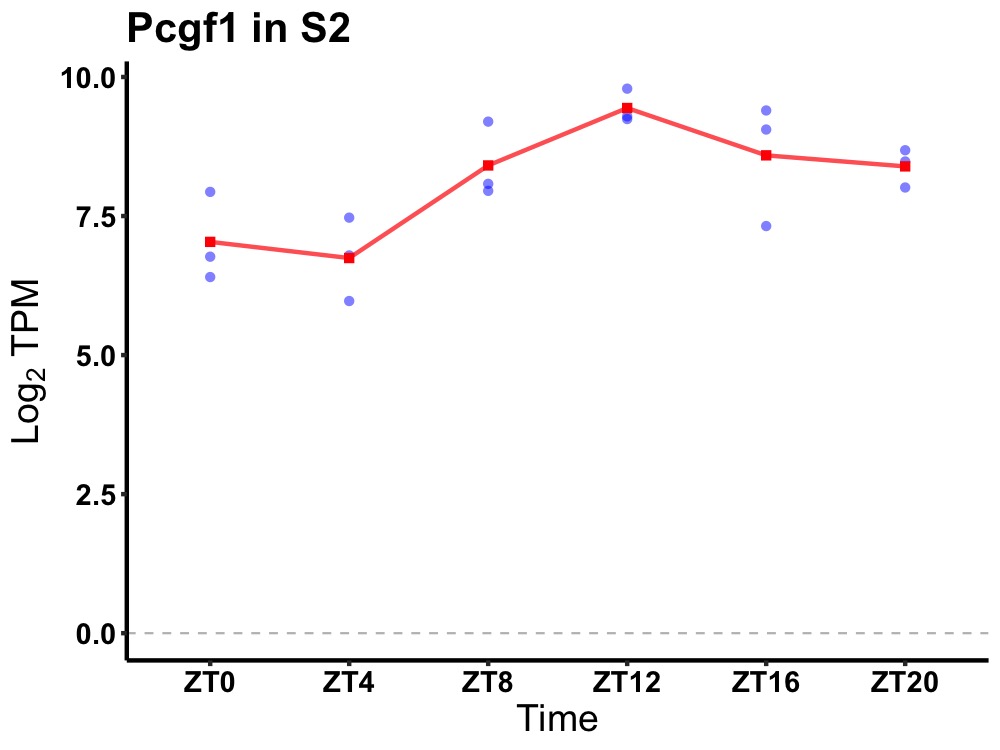 |
polycomb group ring finger 1 | 0.003 | 24 | 14 | 0.96 |
| ENSMUSG00000061488 | Dctn4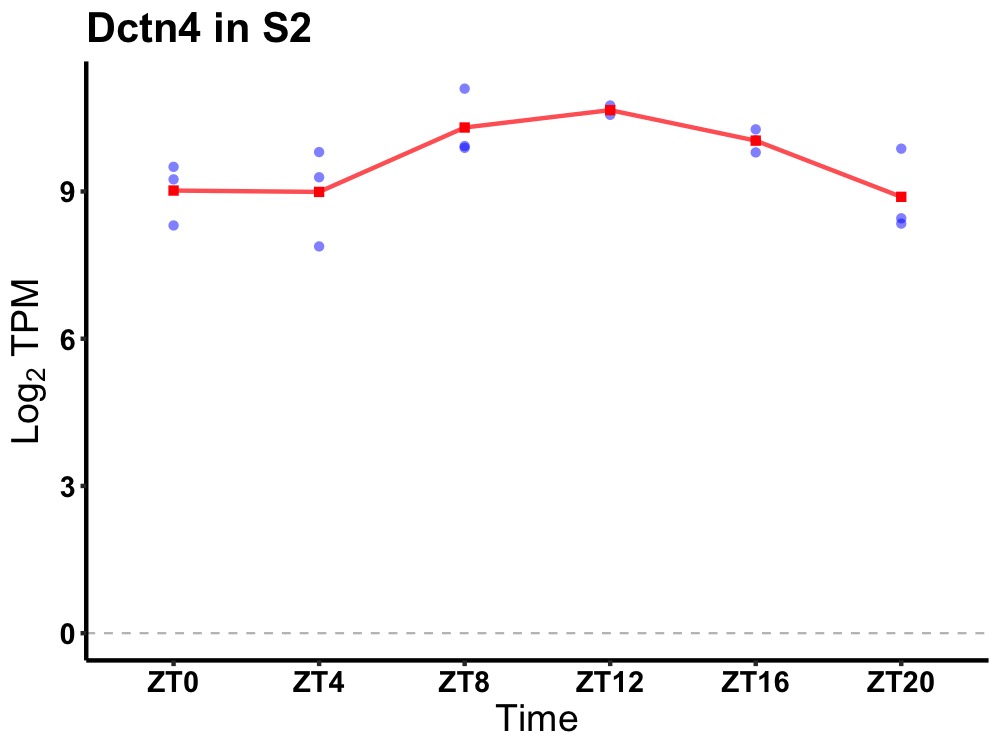 |
dynactin 4 | 0.005 | 24 | 12 | 0.95 |
| ENSMUSG00000110057 | Nuak2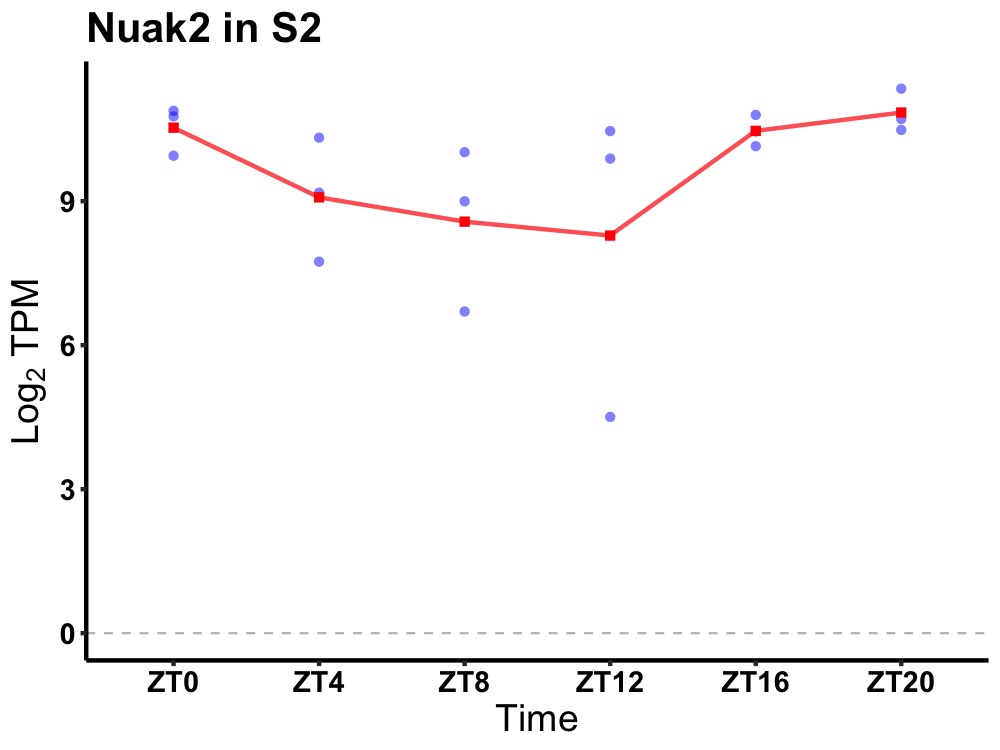 |
NUAK family, SNF1-like kinase, 2 | 0.036 | 24 | 22 | 0.94 |
| ENSMUSG00000013973 | Dedd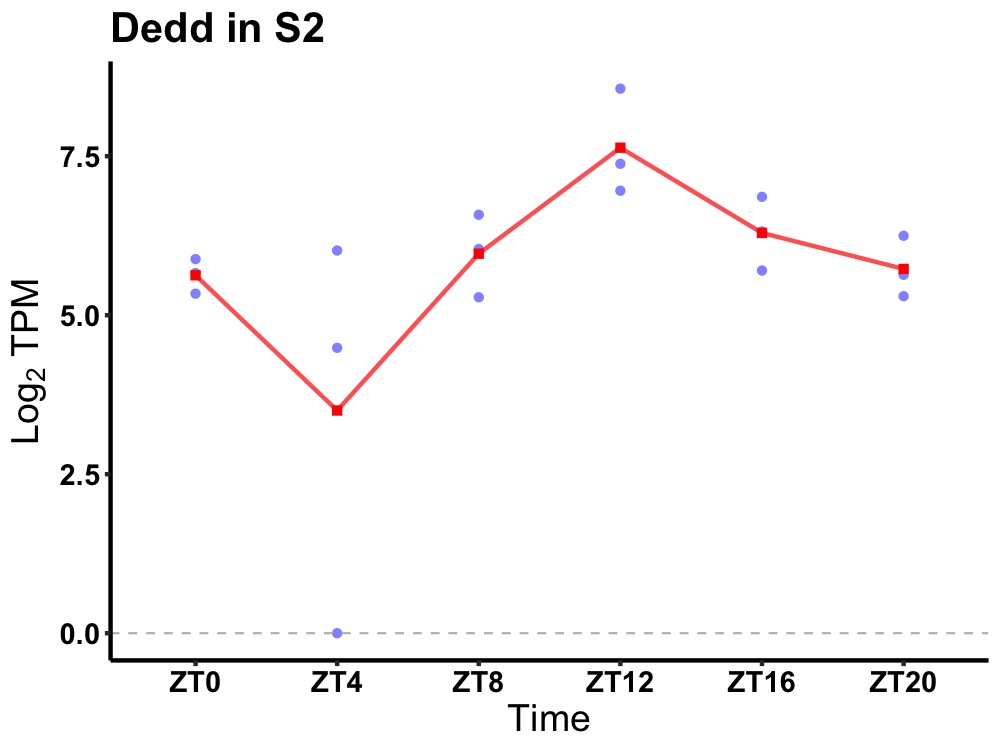 |
death effector domain-containing | 0.007 | 20 | 14 | 0.93 |
| ENSMUSG00000081614 | Gm9436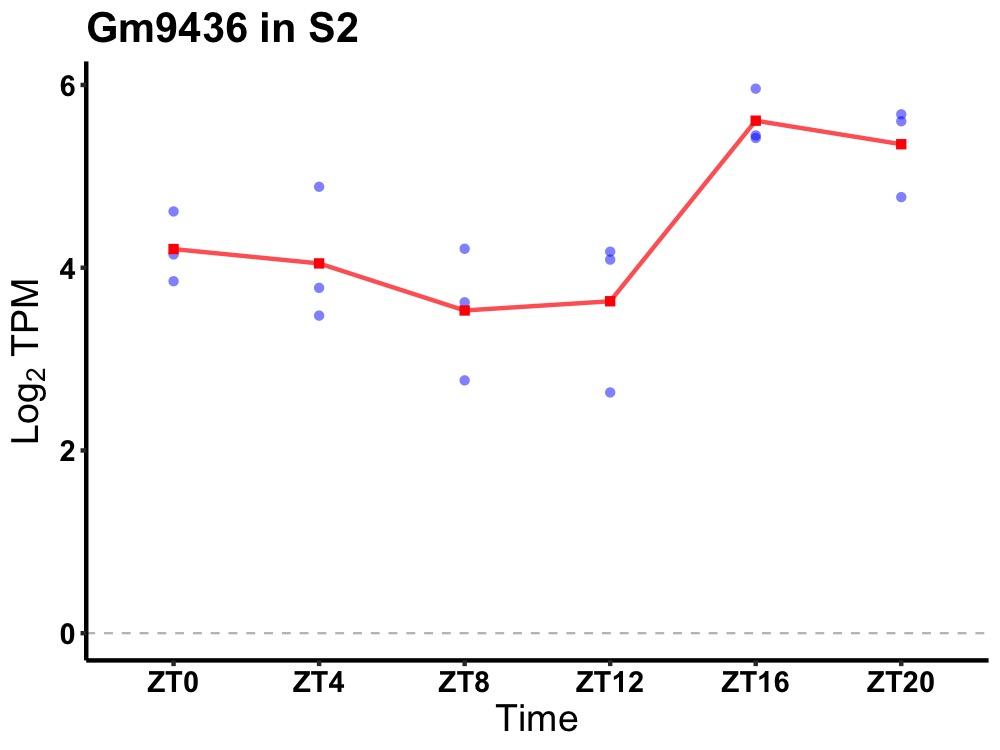 |
predicted gene 9436 | 0.027 | 24 | 20 | 0.93 |
| ENSMUSG00000083856 | Gm13886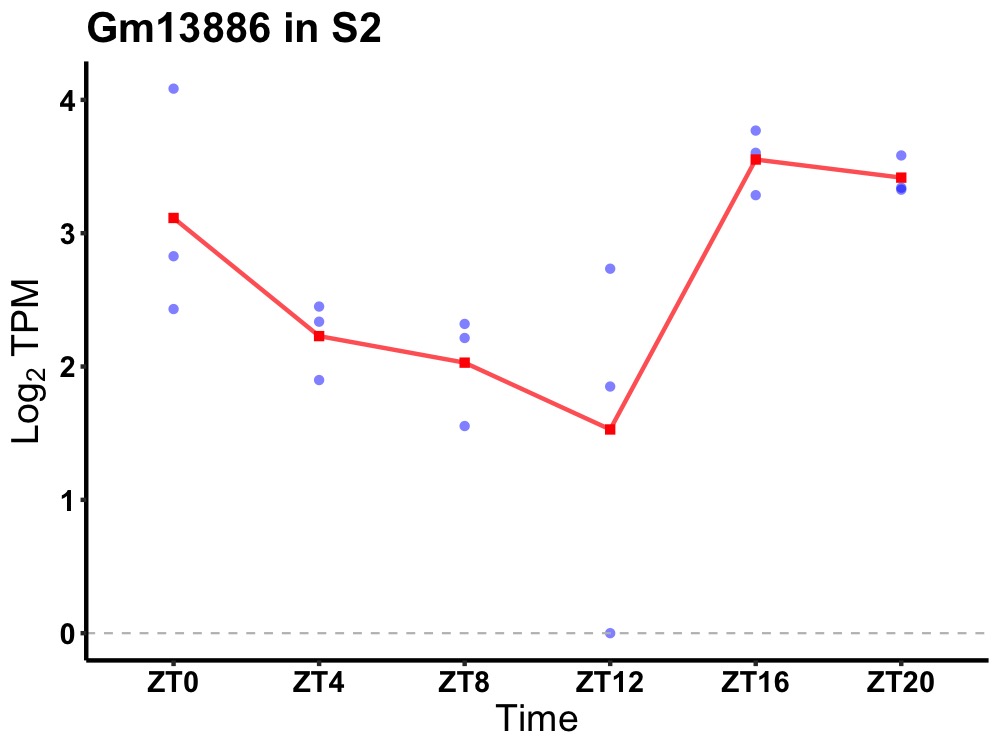 |
predicted gene 13886 | 0.027 | 24 | 22 | 0.93 |
| ENSMUSG00000106454 | Gm17775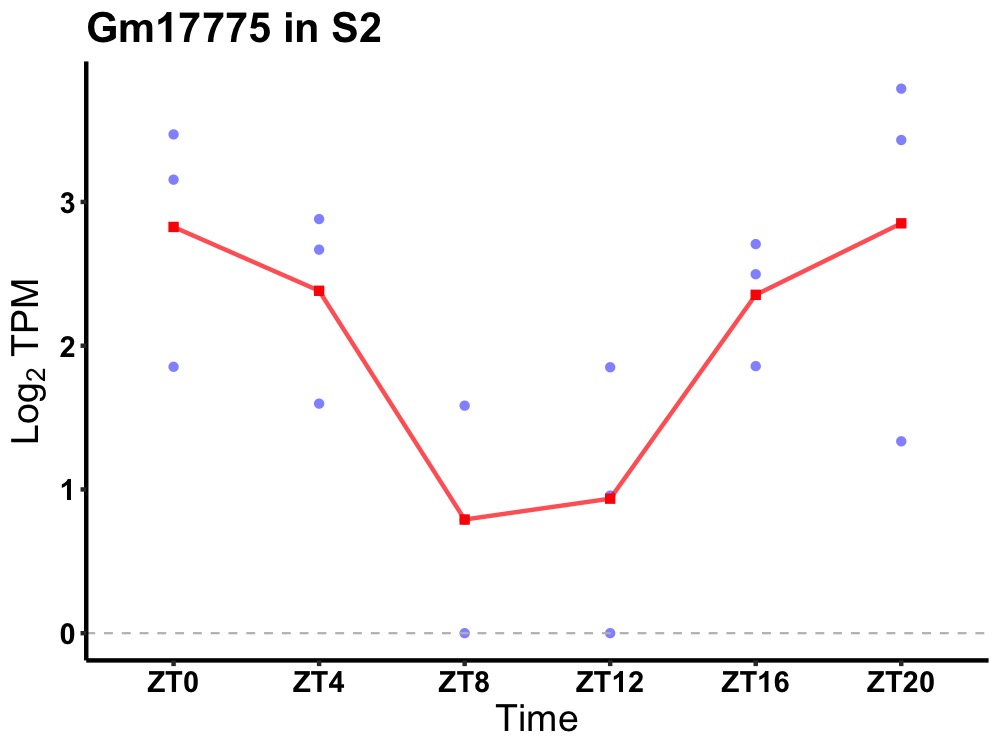 |
none | 0.023 | 24 | 22 | 0.92 |
| ENSMUSG00000084347 | Akt2-ps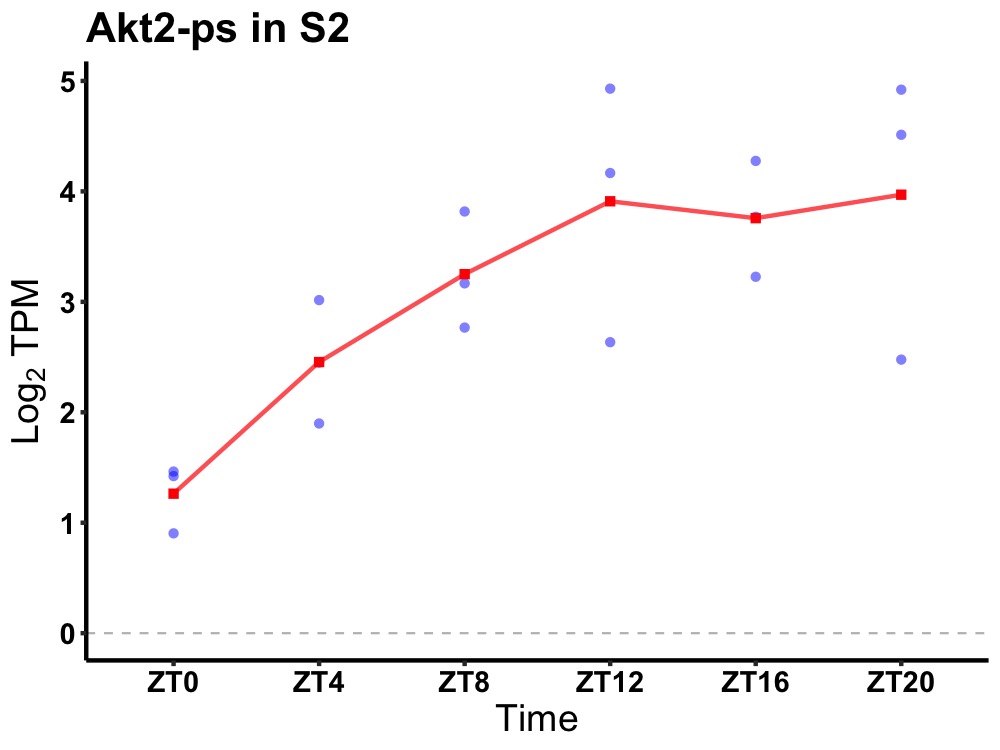 |
thymoma viral proto-oncogene 2, pseudogene | 0.036 | 24 | 14 | 0.91 |
| ENSMUSG00000014905 | Acmsd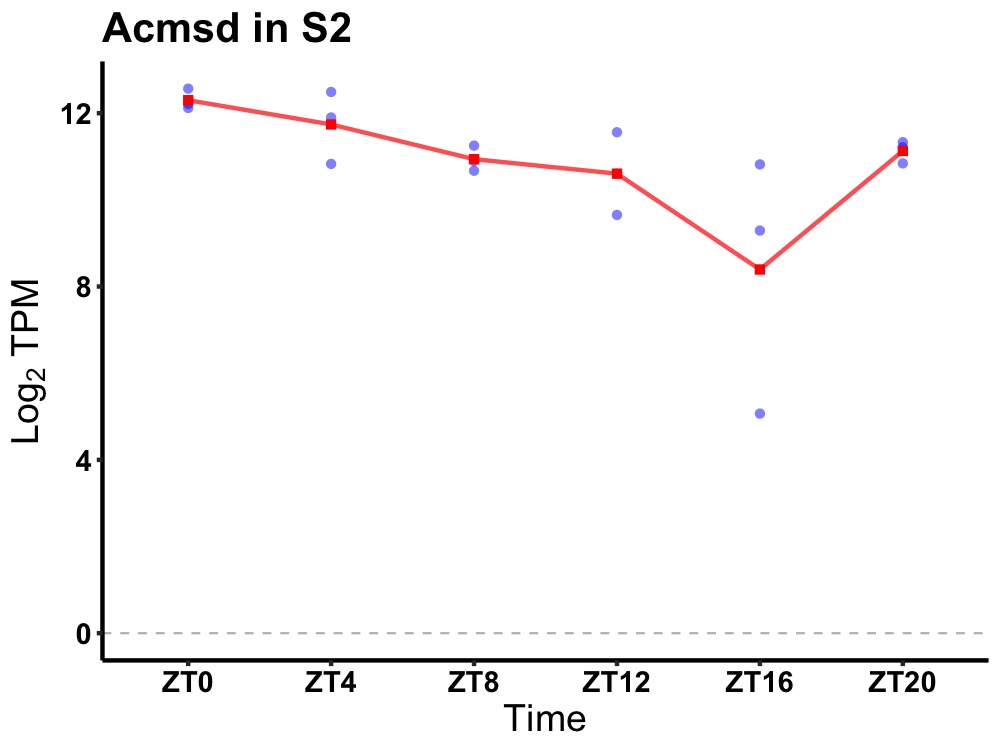 |
amino carboxymuconate semialdehyde decarboxylase | 0.014 | 24 | 2 | 0.90 |
| ENSMUSG00000021748 | Sel1l3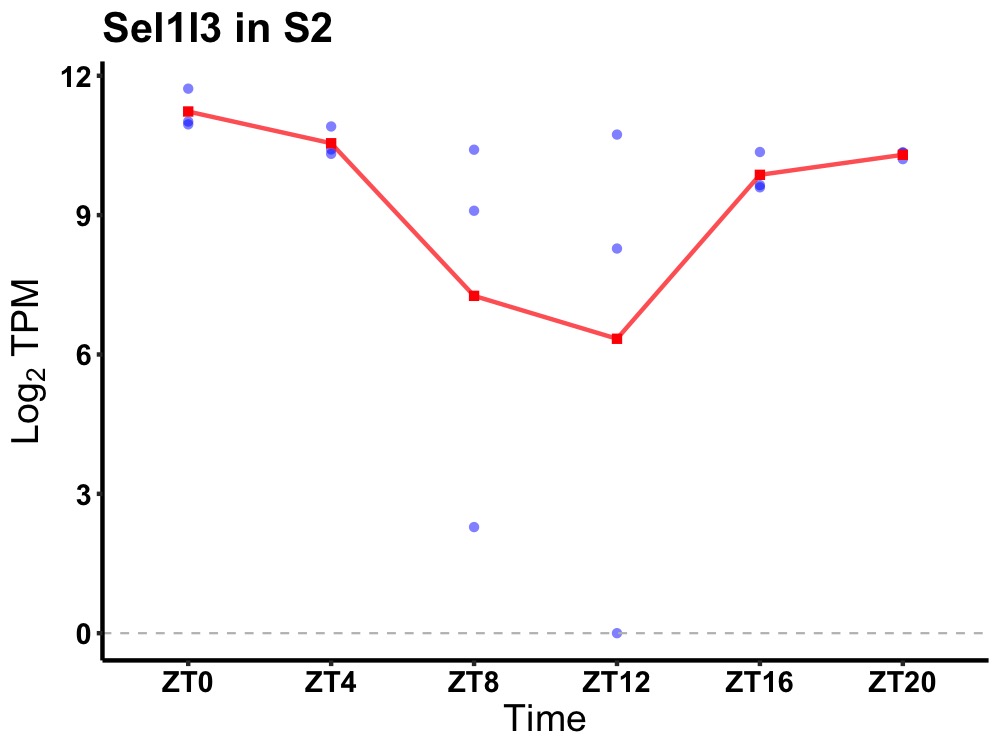 |
sel-1 suppressor of lin-12-like 3 (C. elegans) | 0.036 | 24 | 2 | 0.89 |
| ENSMUSG00000083392 | Gm6335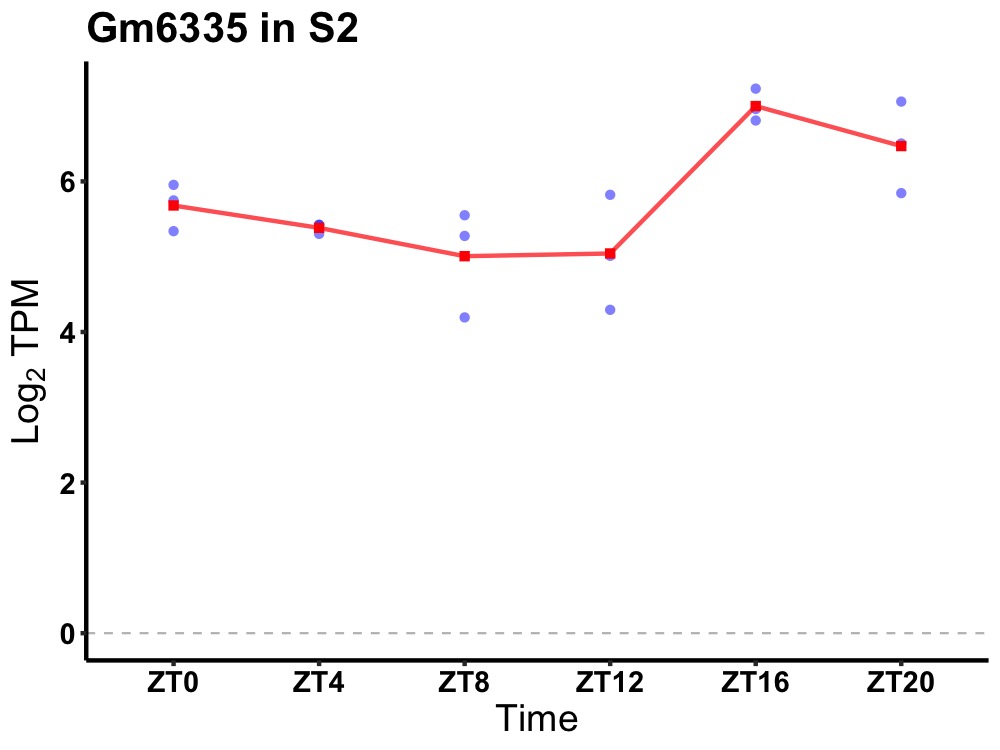 |
predicted gene 6335 | 0.019 | 24 | 20 | 0.89 |
| ENSMUSG00000024807 | Cnot6l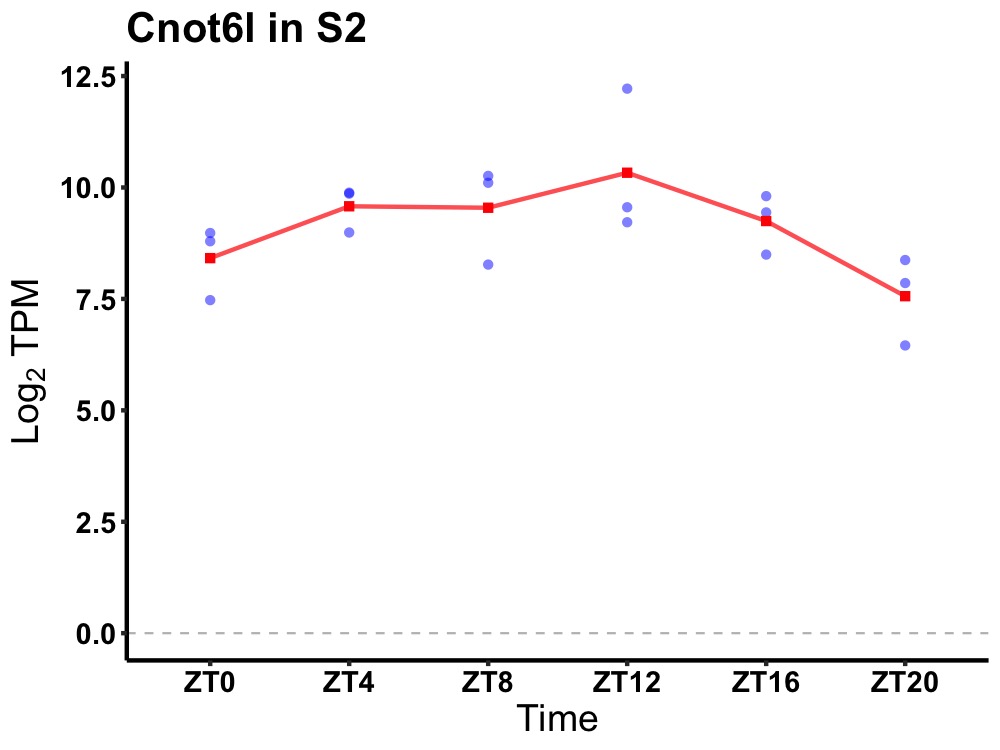 |
CCR4-NOT transcription complex, subunit 6-like | 0.027 | 24 | 10 | 0.88 |
| ENSMUSG00000032422 | Jpt2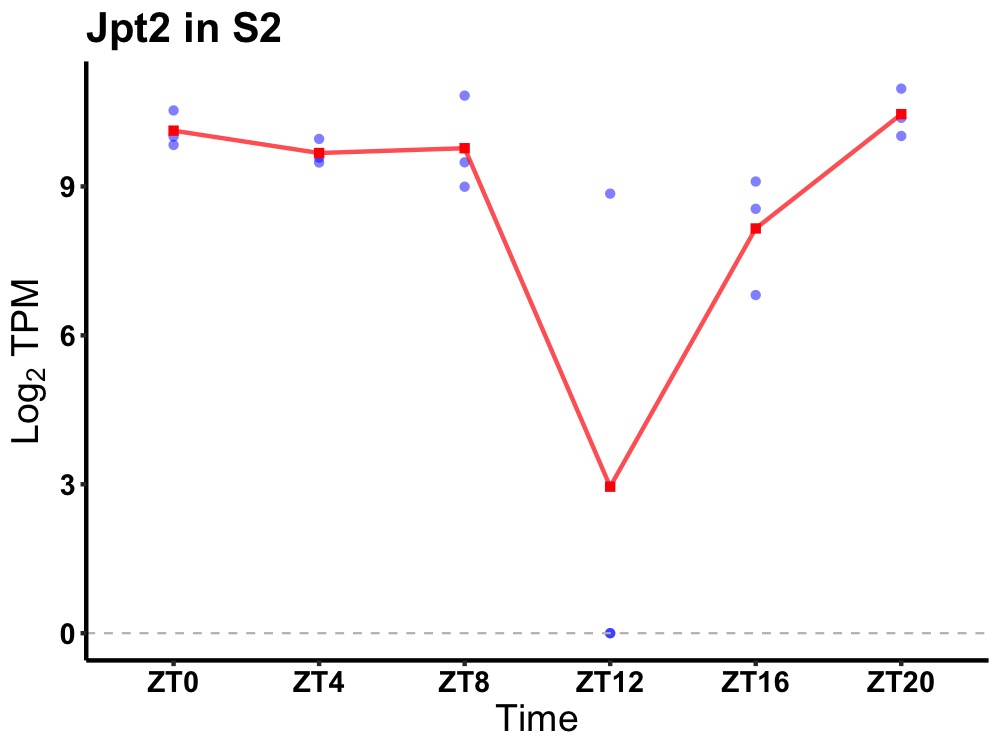 |
jupiter microtubule associated homolog 2 | 0.005 | 20 | 2 | 0.88 |
| ENSMUSG00000080808 | Gm12608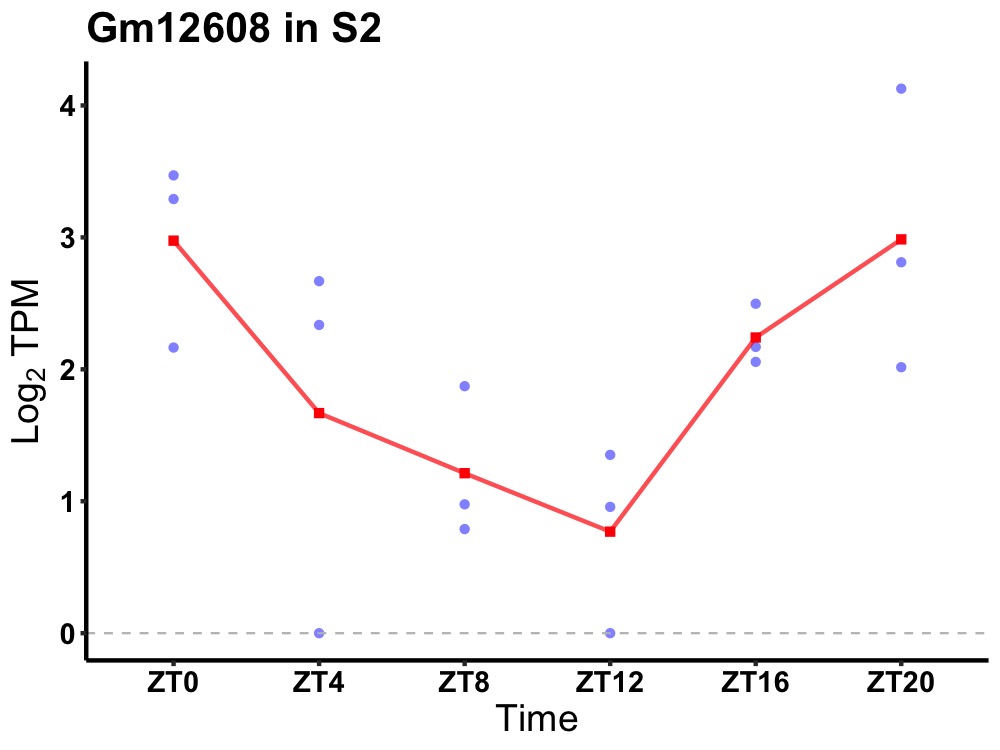 |
none | 0.016 | 24 | 0 | 0.87 |
| ENSMUSG00000059796 | Tardbp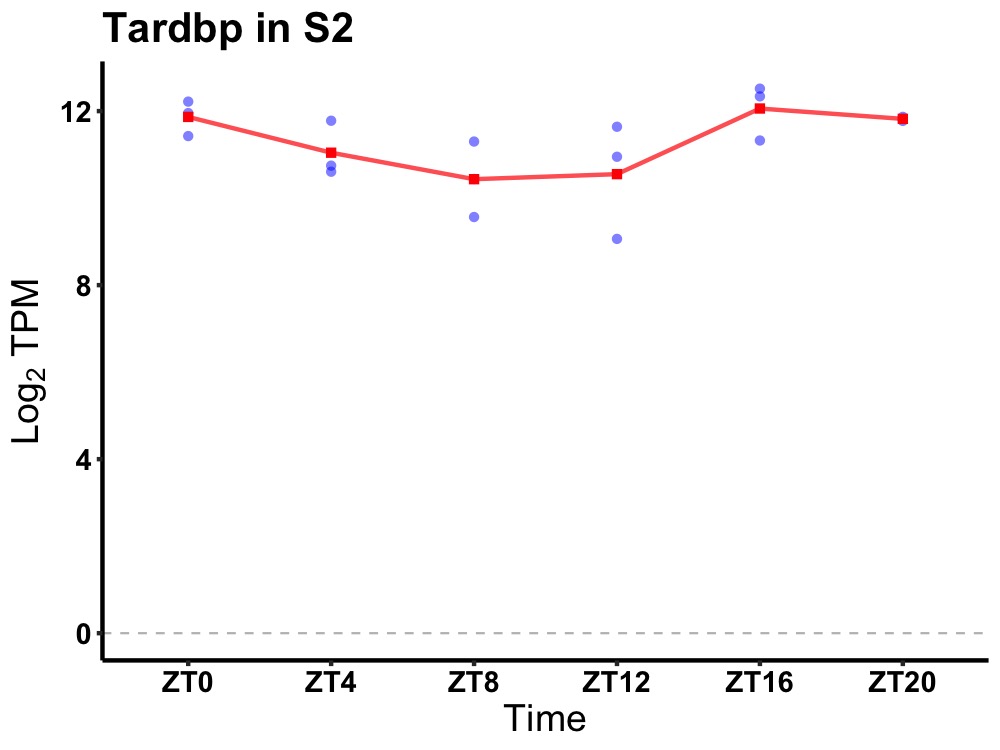 |
TAR DNA binding protein | 0.014 | 20 | 18 | 0.87 |
| ENSMUSG00000107907 | Gm8574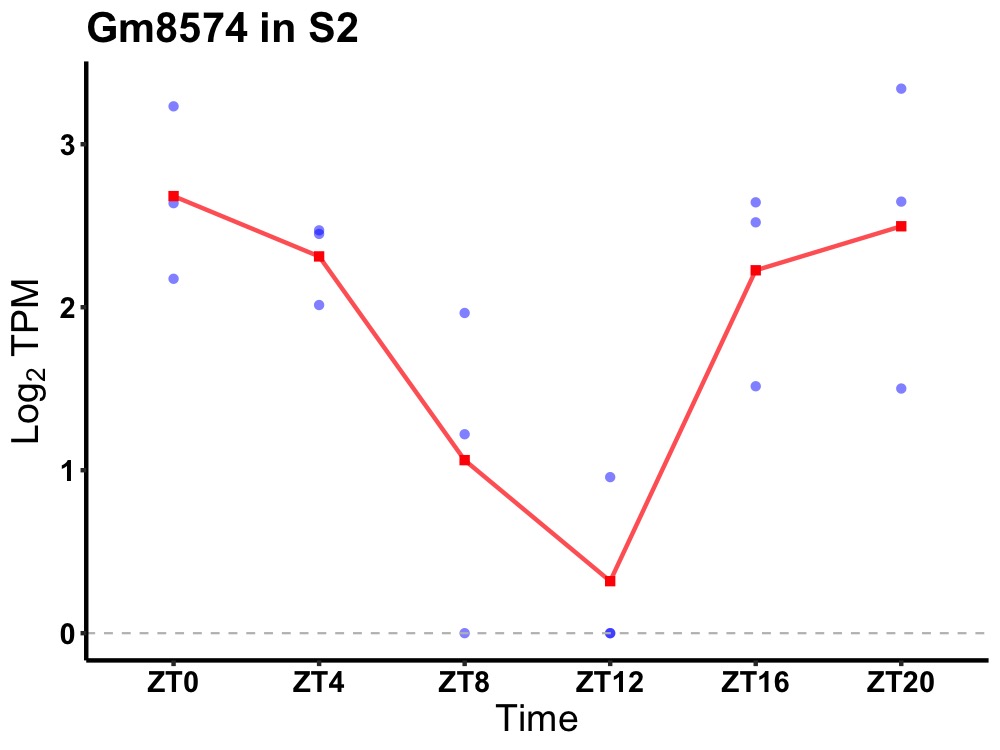 |
none | 0.014 | 20 | 2 | 0.86 |
| ENSMUSG00000083705 | Mafg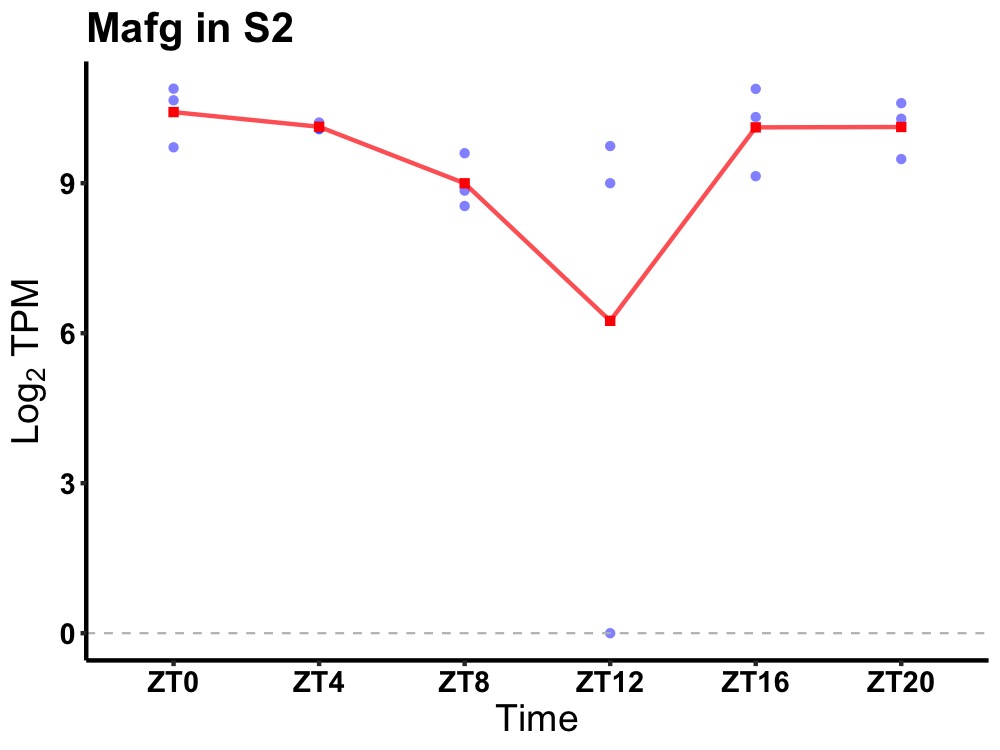 |
v-maf musculoaponeurotic fibrosarcoma oncogene family, protein G (avian) | 0.036 | 20 | 0 | 0.86 |
| ENSMUSG00000020381 | Metap1d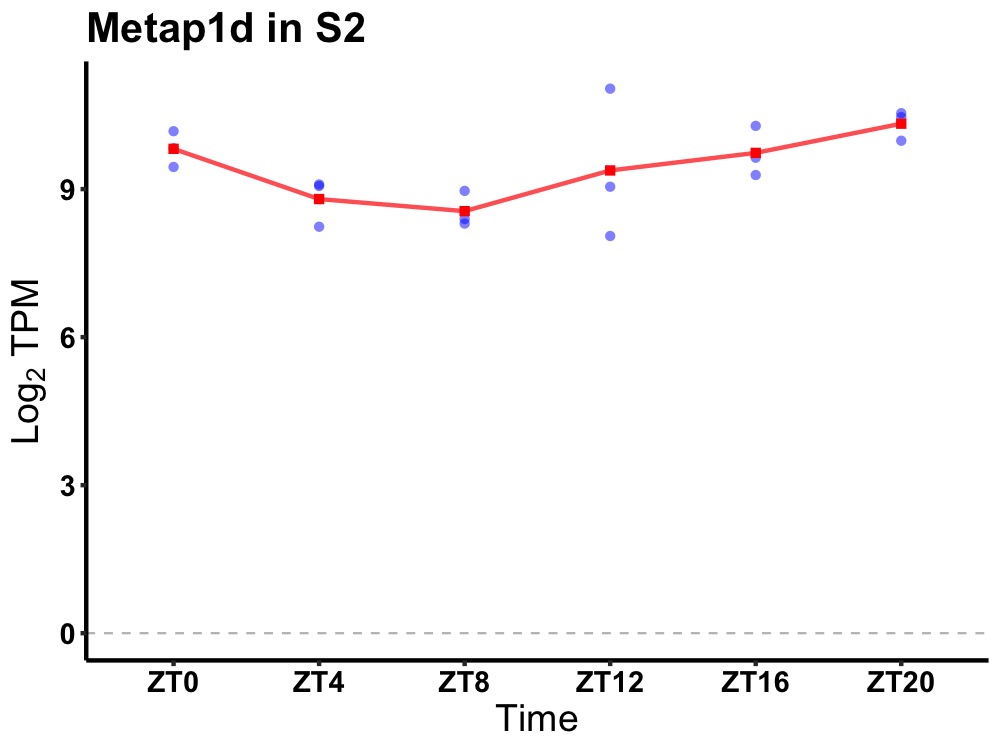 |
methionyl aminopeptidase type 1D (mitochondrial) | 0.014 | 24 | 22 | 0.86 |
| ENSMUSG00000106755 | Zc3h14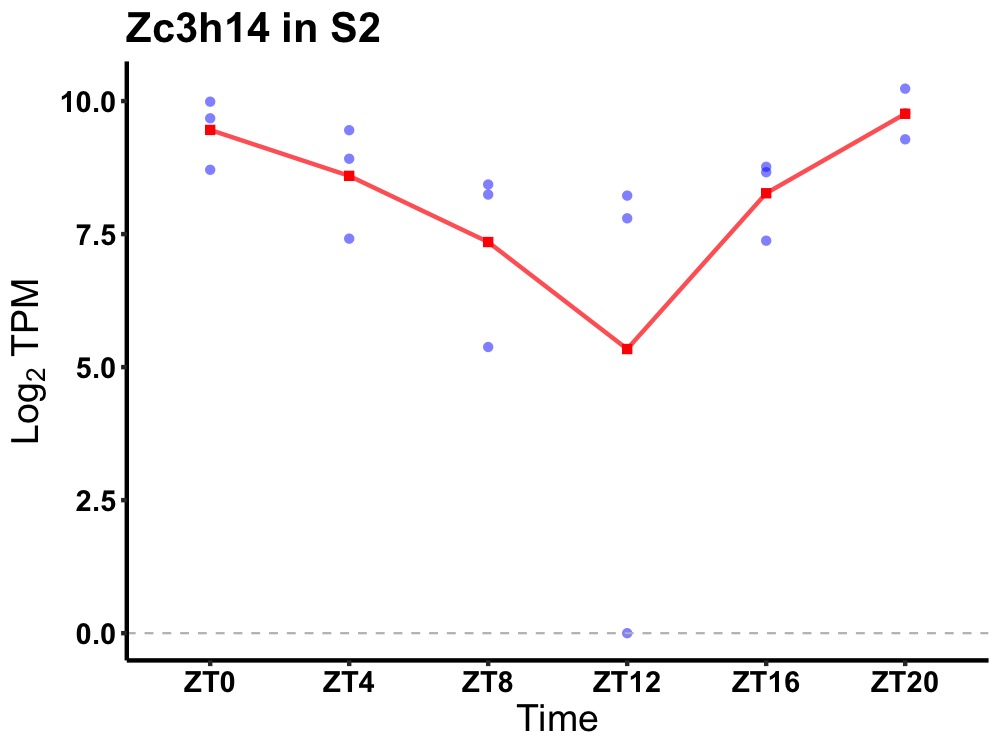 |
zinc finger CCCH type containing 14 | 0.001 | 20 | 2 | 0.86 |
| ENSMUSG00000023795 | Pisd-ps2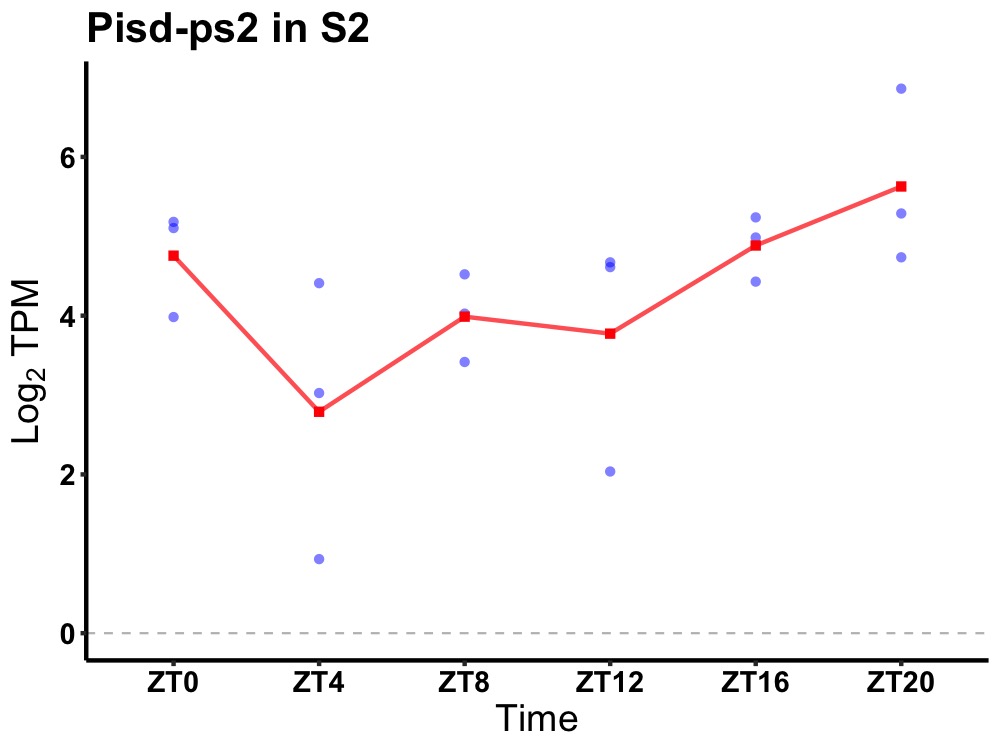 |
phosphatidylserine decarboxylase, pseudogene 2 | 0.036 | 24 | 20 | 0.86 |
| ENSMUSG00000007338 | Cggbp1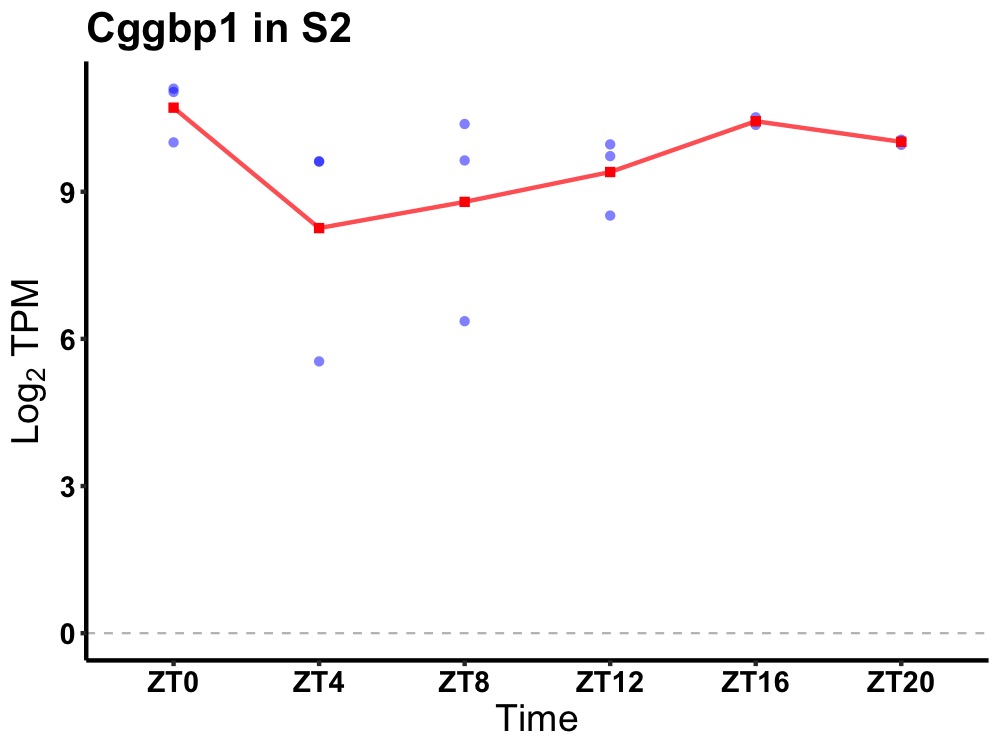 |
CGG triplet repeat binding protein 1 | 0.027 | 20 | 18 | 0.85 |
| ENSMUSG00000031505 | Ndst1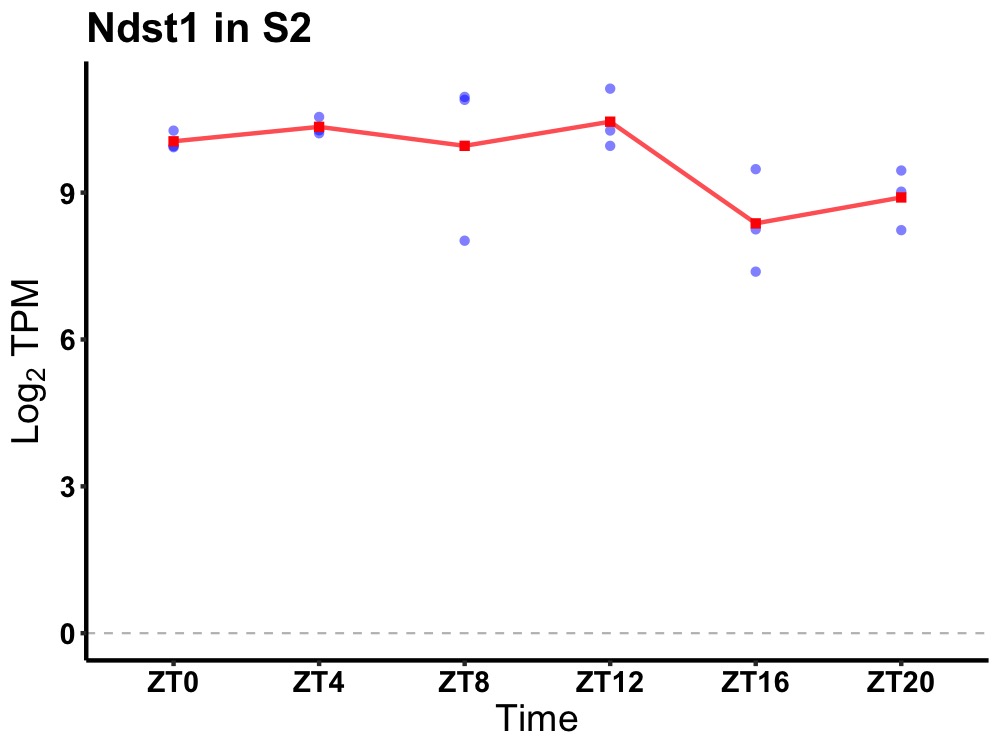 |
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 1 | 0.027 | 24 | 8 | 0.84 |
| ENSMUSG00000047409 | Ddc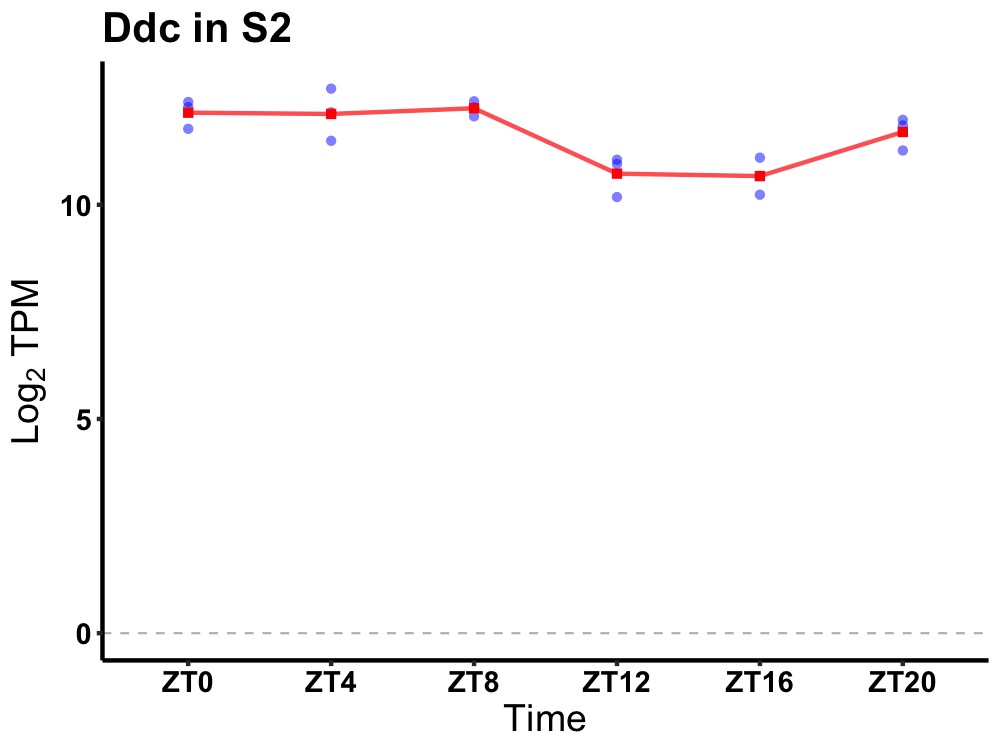 |
dopa decarboxylase | 0.010 | 24 | 4 | 0.83 |
| ENSMUSG00000026966 | Luc7l2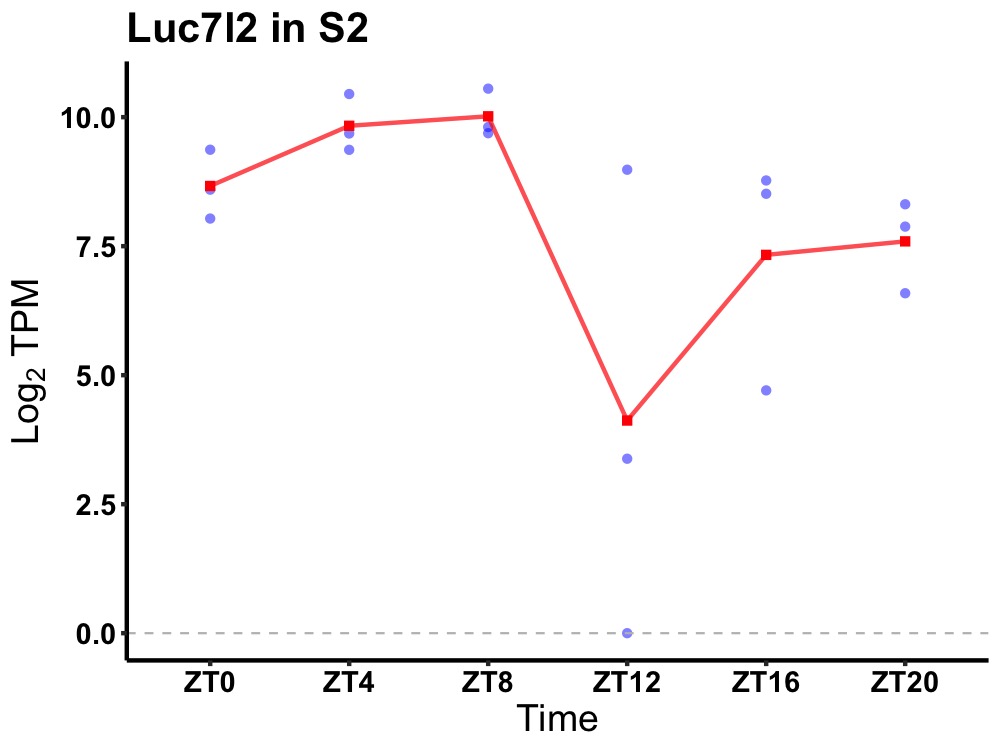 |
LUC7-like 2 (S. cerevisiae) | 0.049 | 24 | 8 | 0.83 |
| ENSMUSG00000023074 | Brap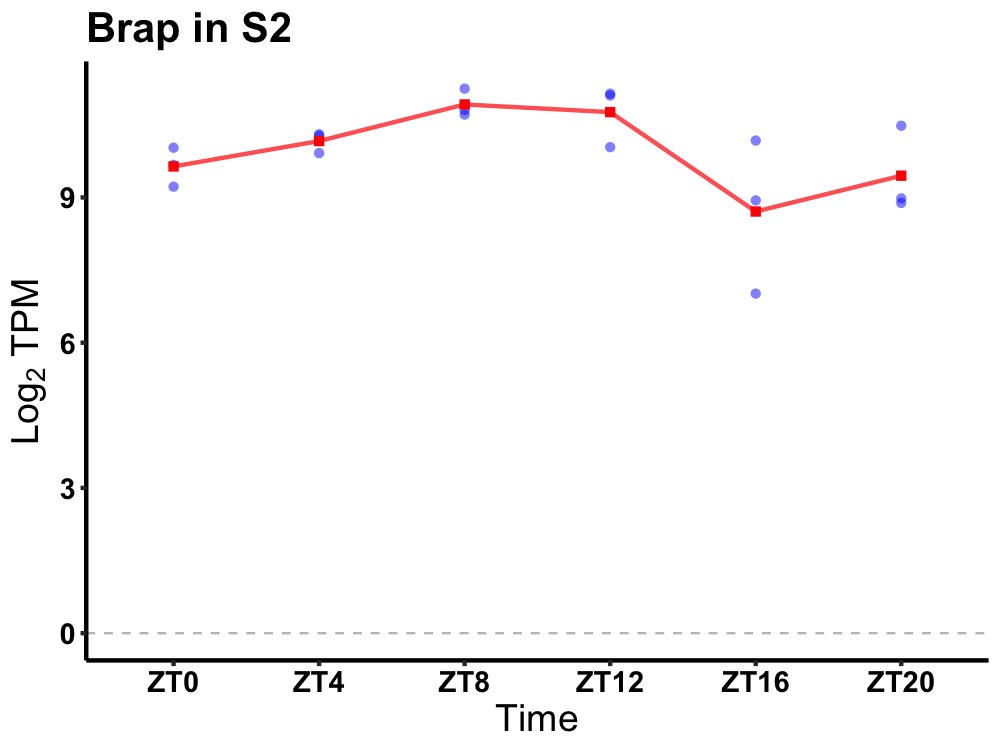 |
BRCA1 associated protein | 0.036 | 24 | 10 | 0.82 |
| ENSMUSG00000028649 | Gpkow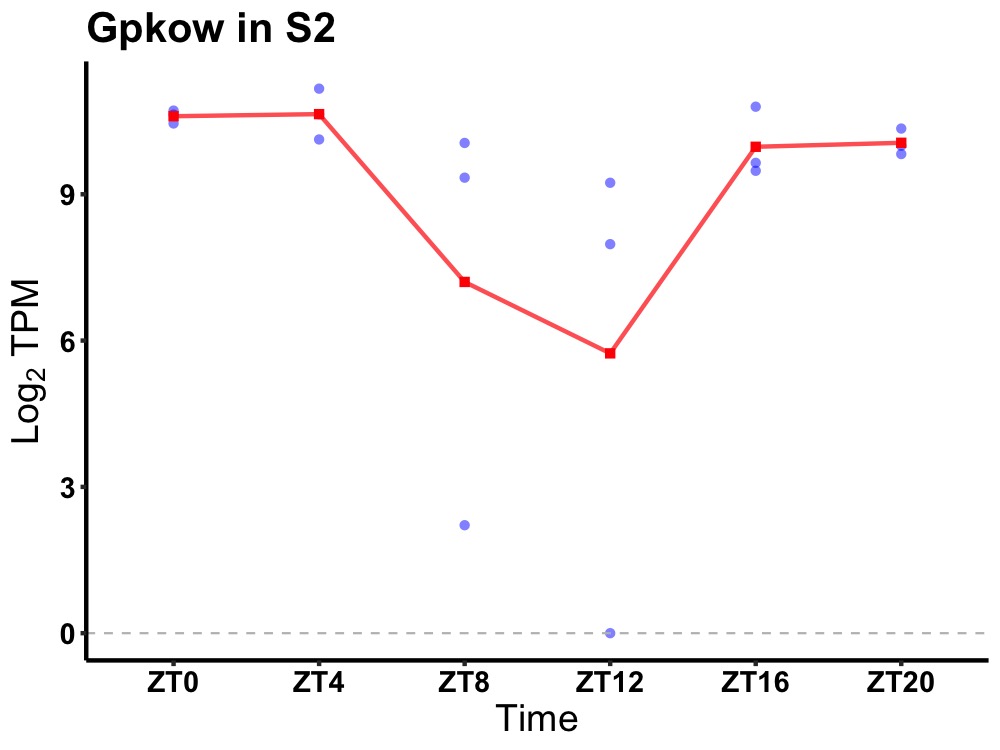 |
G patch domain and KOW motifs | 0.014 | 24 | 2 | 0.81 |
| ENSMUSG00000050199 | Sgk1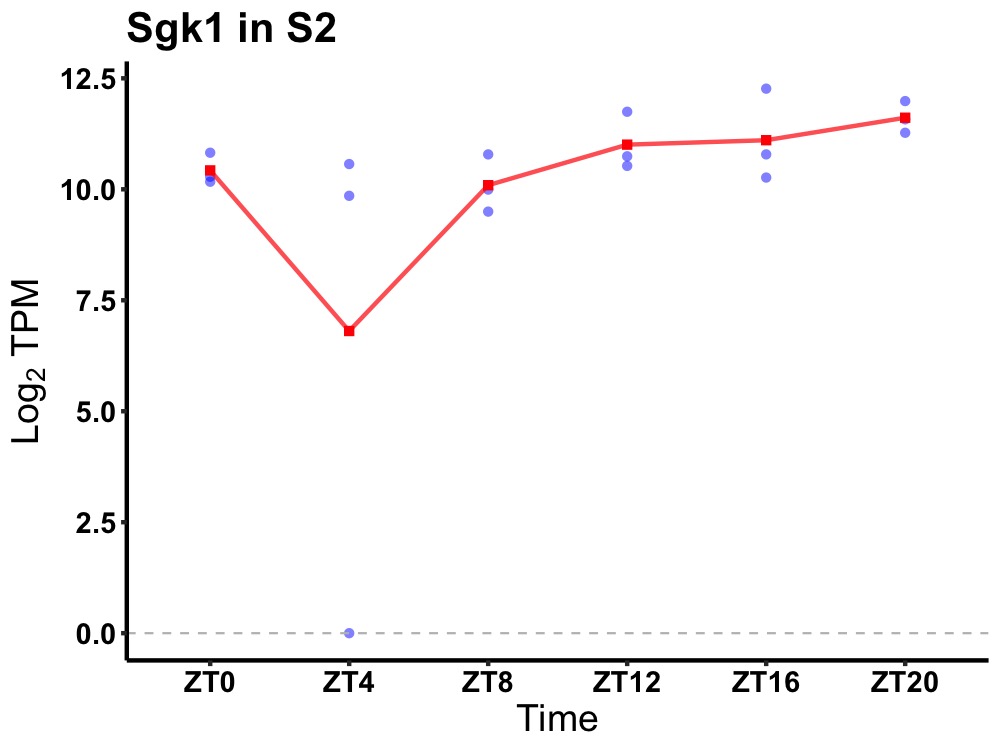 |
serum/glucocorticoid regulated kinase 1 | 0.049 | 24 | 18 | 0.81 |
| ENSMUSG00000050628 | Ubald2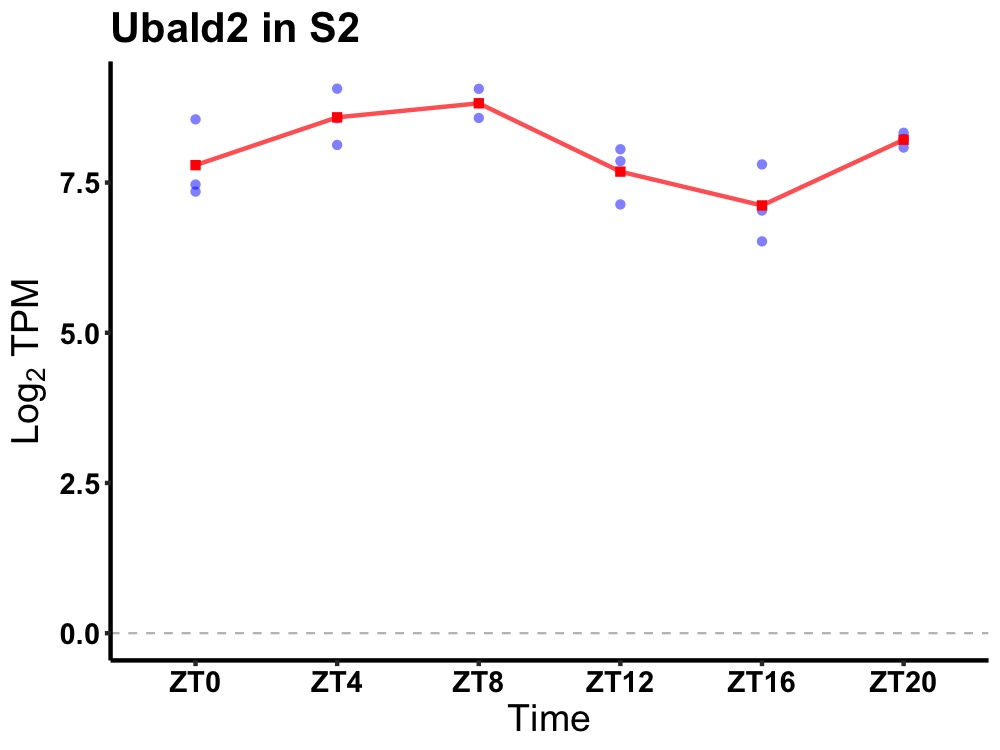 |
UBA-like domain containing 2 | 0.001 | 20 | 6 | 0.81 |
| ENSMUSG00000071640 | Stxbp3-ps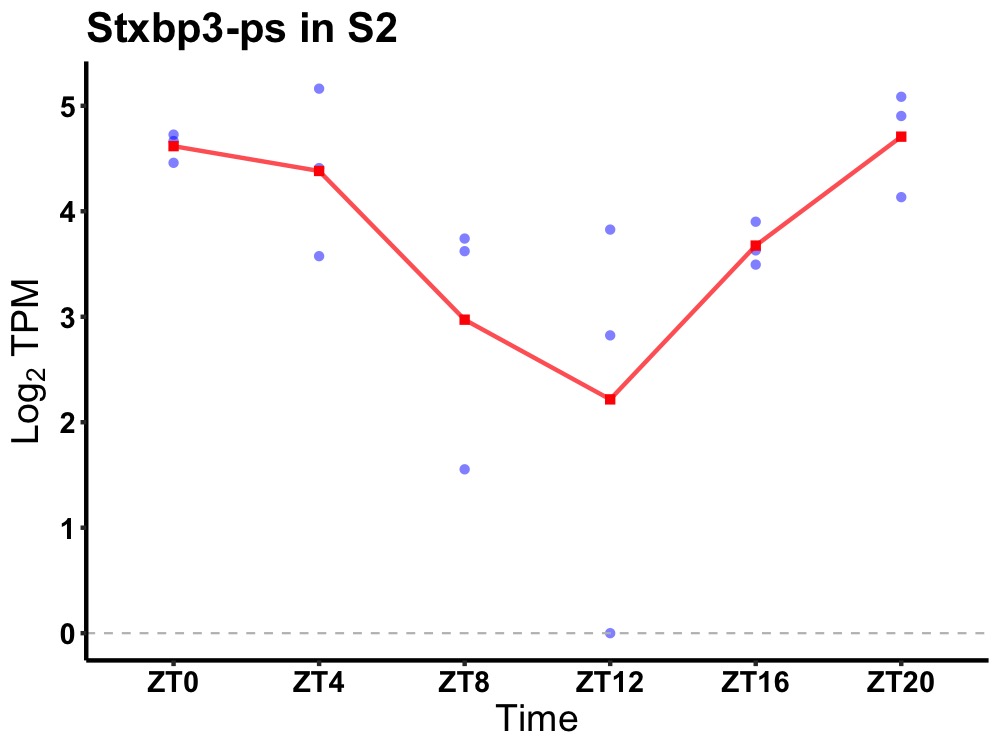 |
syntaxin-binding protein 3, pseudogene | 0.010 | 20 | 2 | 0.80 |
| ENSMUSG00000111877 | Khdc4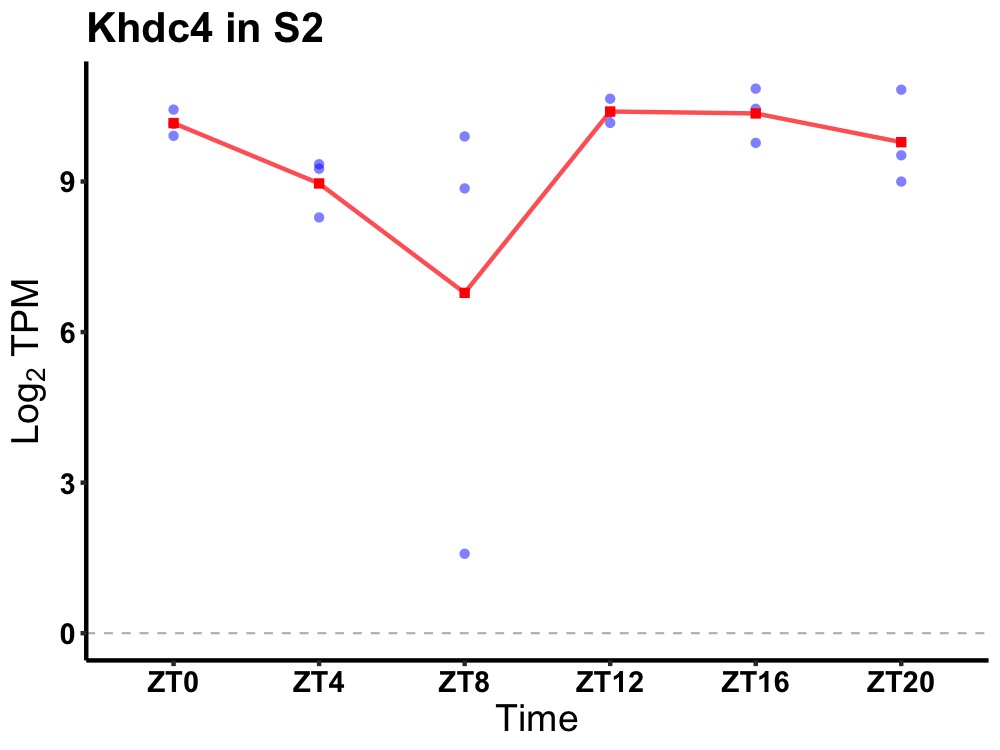 |
KH domain containing 4, pre-mRNA splicing factor | 0.027 | 20 | 16 | 0.80 |
| ENSMUSG00000076135 | Clk1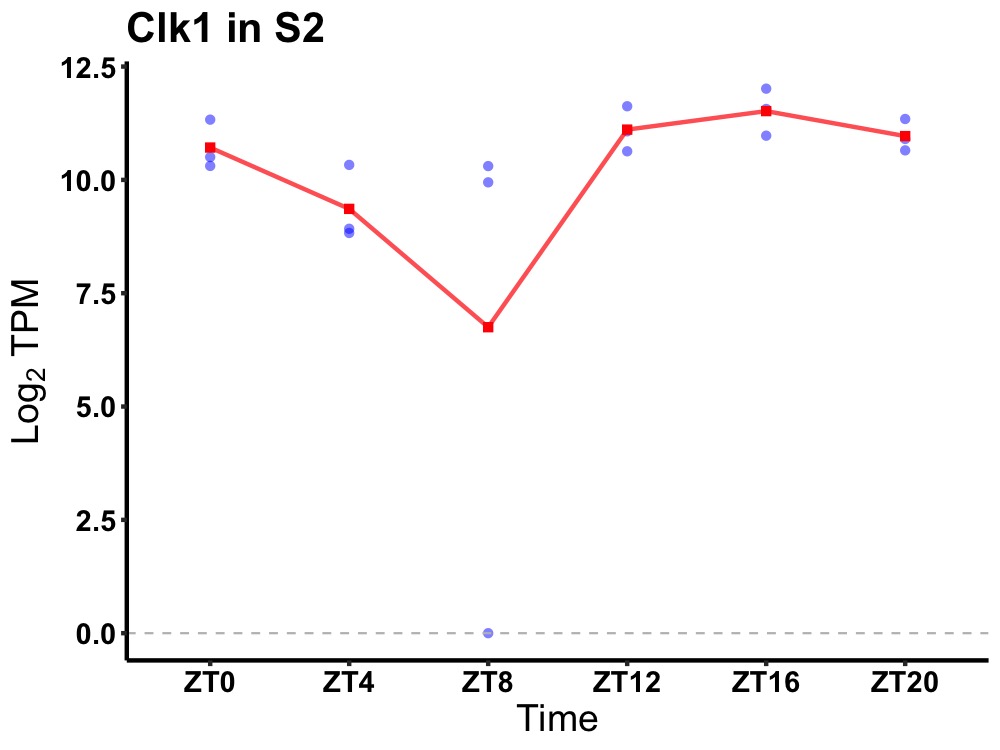 |
CDC-like kinase 1 | 0.002 | 24 | 18 | 0.80 |
| ENSMUSG00000028221 | Farsa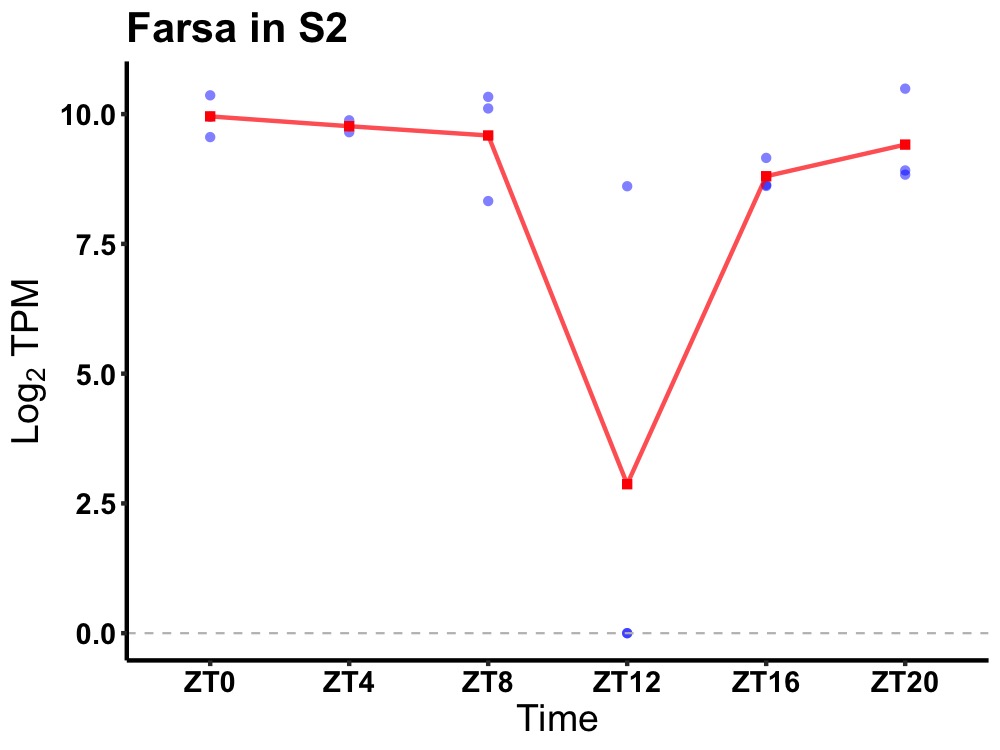 |
phenylalanine--tRNA ligase alpha subunit | 0.049 | 24 | 2 | 0.80 |
| ENSMUSG00000112017 | Gm7842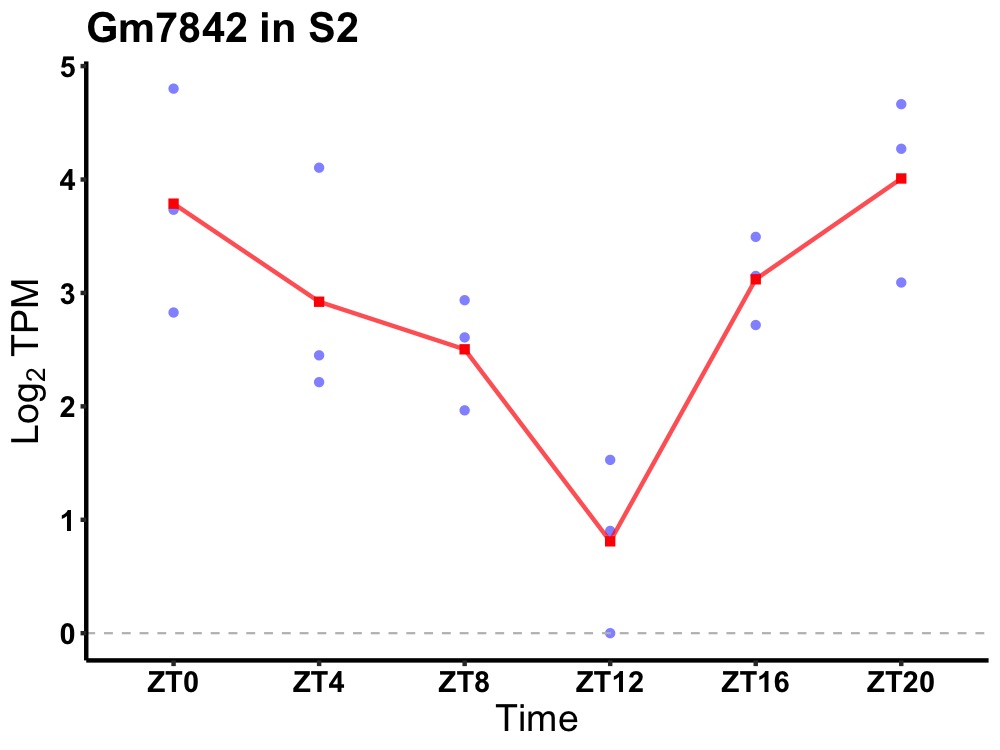 |
predicted gene 7842 | 0.007 | 20 | 2 | 0.80 |
| ENSMUSG00000114167 | Gm7002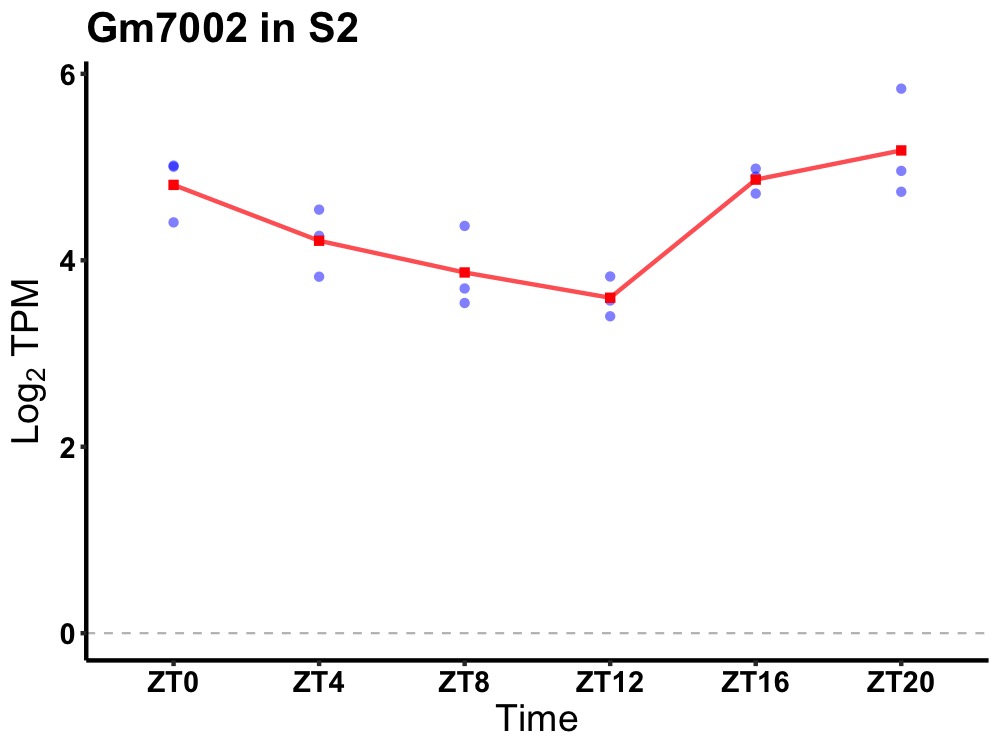 |
predicted gene 7002 | 0.002 | 24 | 22 | 0.79 |
| ENSMUSG00000091498 | Uggt1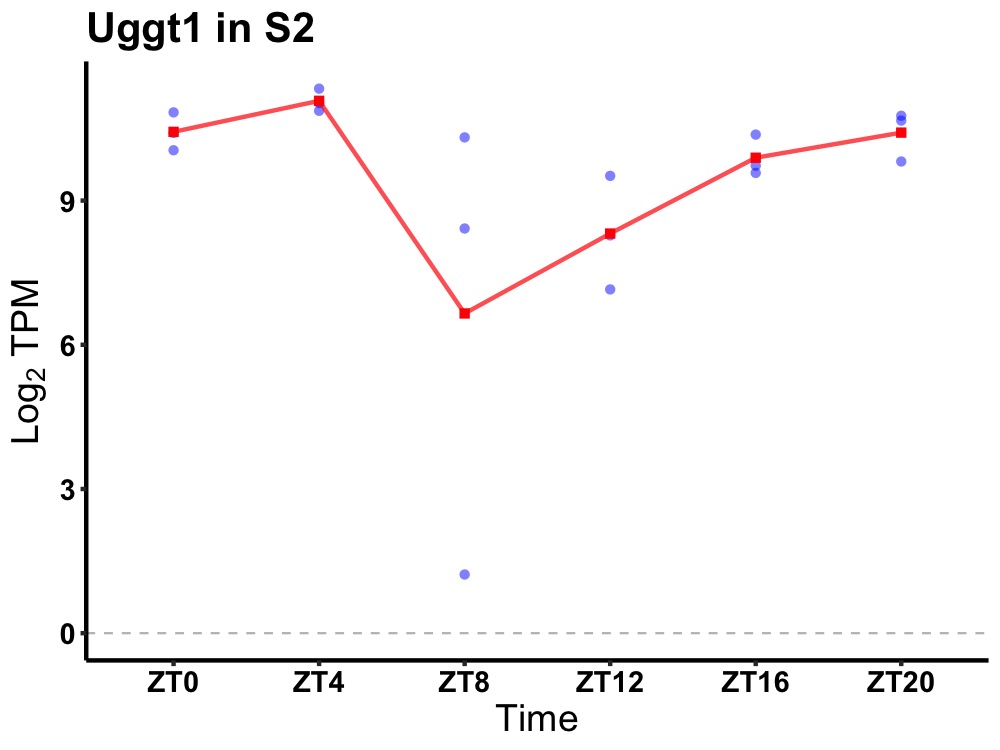 |
UDP-glucose glycoprotein glucosyltransferase 1 | 0.002 | 20 | 4 | 0.78 |
| ENSMUSG00000114034 | BC005537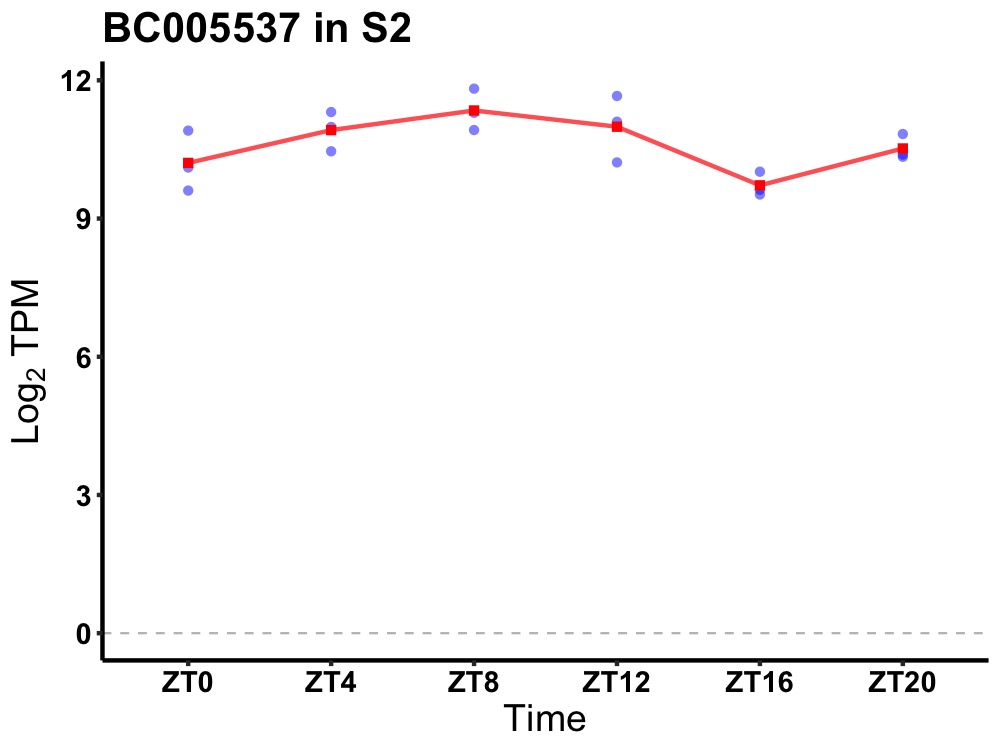 |
cDNA sequence BC005537 | 0.005 | 20 | 8 | 0.78 |
| ENSMUSG00000027751 | Abhd17c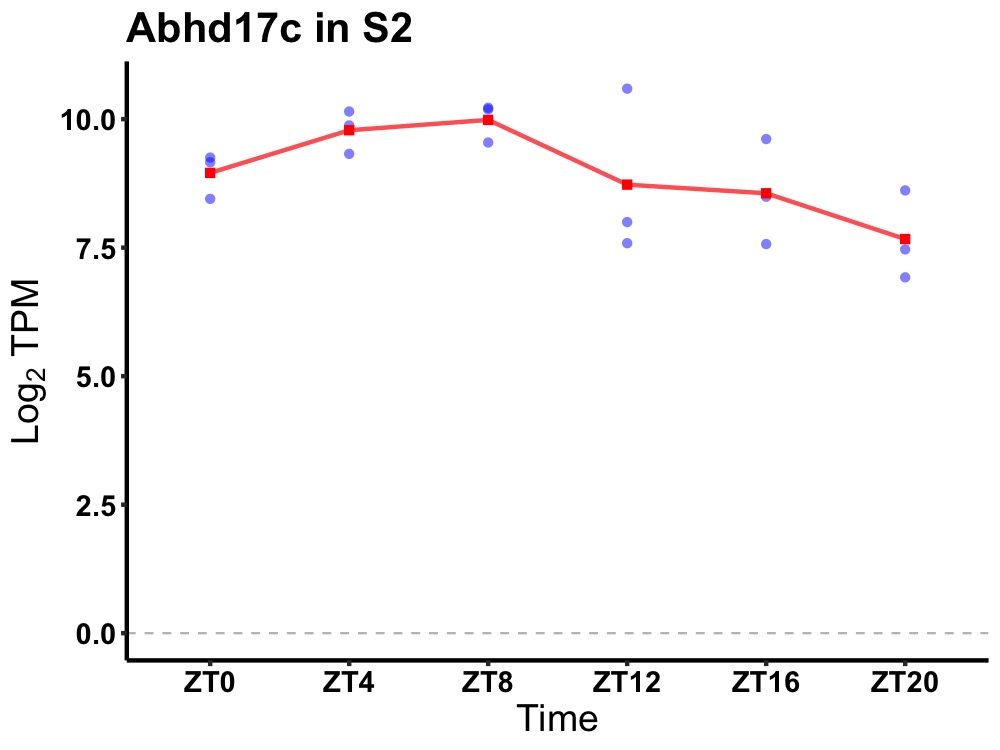 |
abhydrolase domain containing 17C | 0.049 | 24 | 8 | 0.78 |
| ENSMUSG00000000385 | Zdhhc21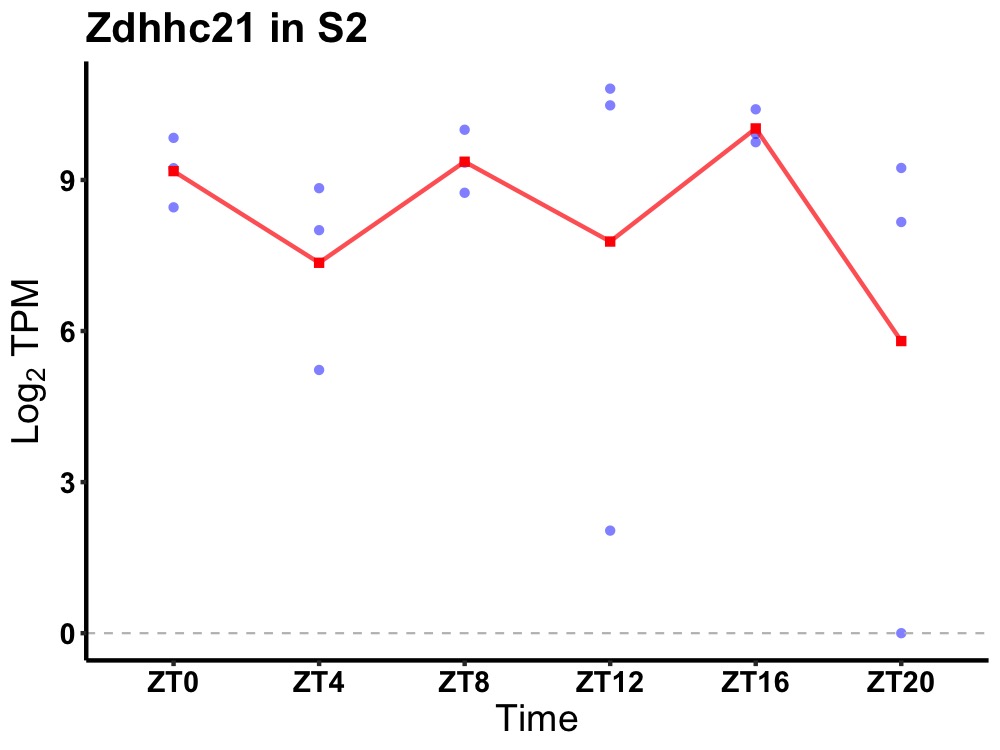 |
zinc finger, DHHC domain containing 21 | 0.049 | 20 | 14 | 0.77 |
| ENSMUSG00000074516 | Apob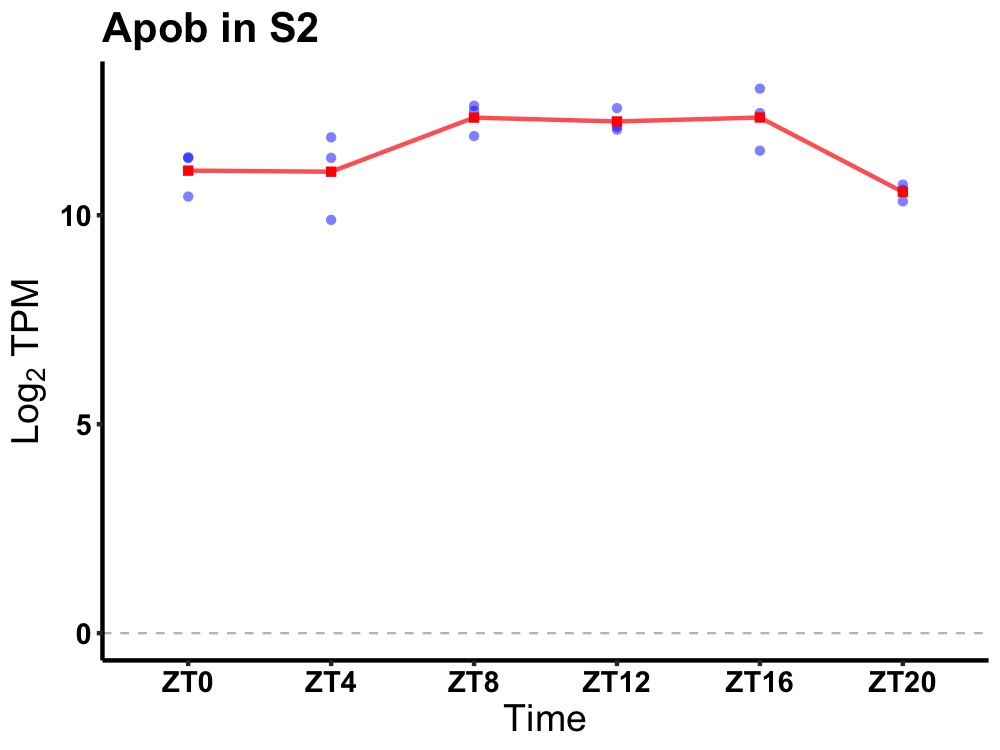 |
apolipoprotein B | 0.010 | 20 | 12 | 0.76 |
| ENSMUSG00000115916 | Gm8702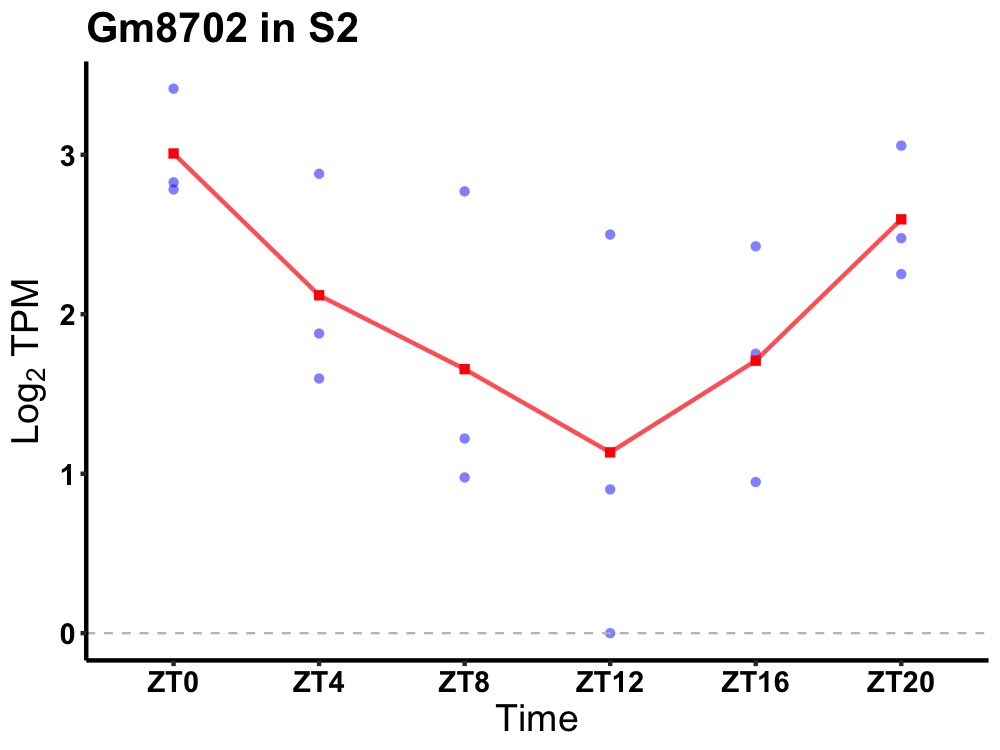 |
predicted gene 8702 | 0.036 | 24 | 0 | 0.76 |
| ENSMUSG00000112341 | Gm4796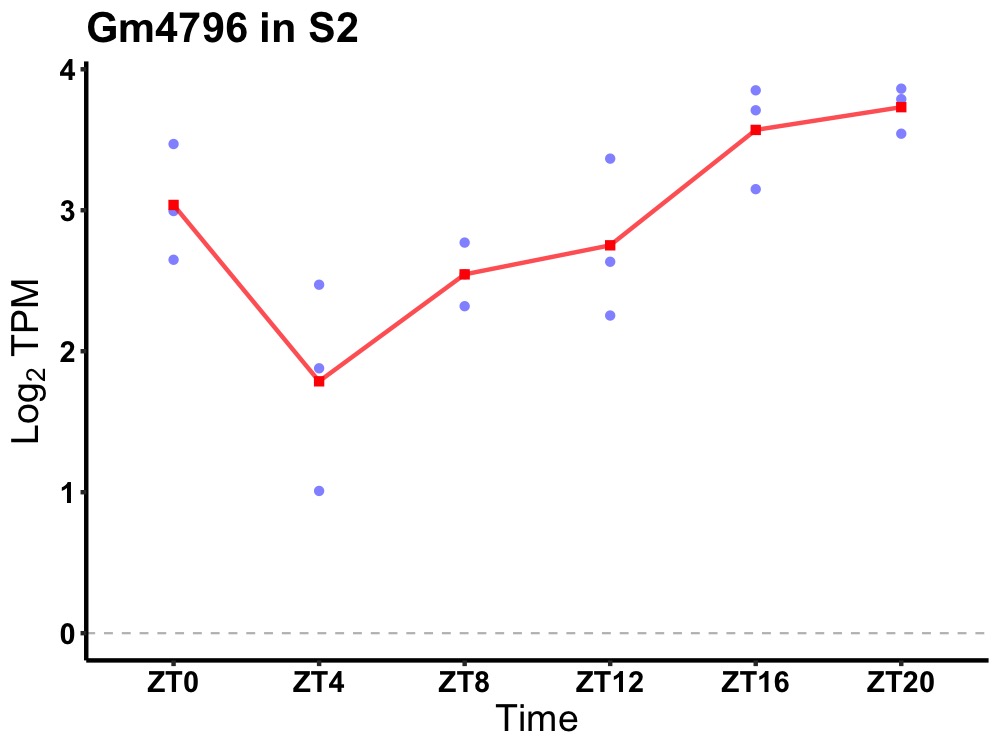 |
predicted gene 4796 | 0.003 | 24 | 20 | 0.76 |
| ENSMUSG00000059461 | Npnt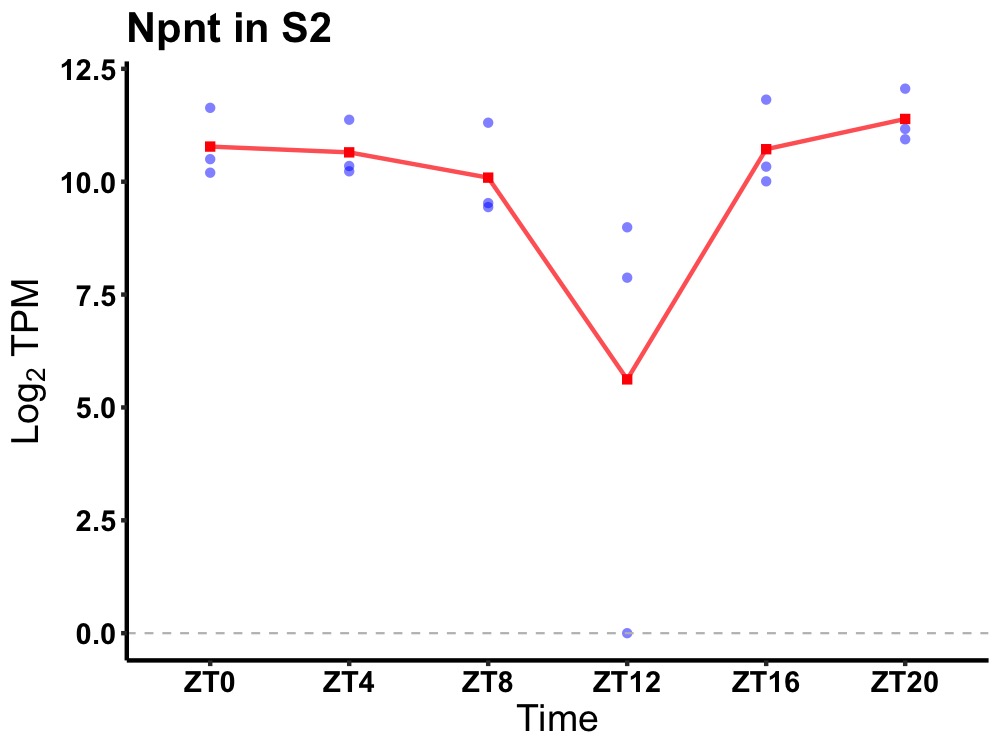 |
nephronectin | 0.019 | 20 | 2 | 0.75 |
| ENSMUSG00000047843 | Bri3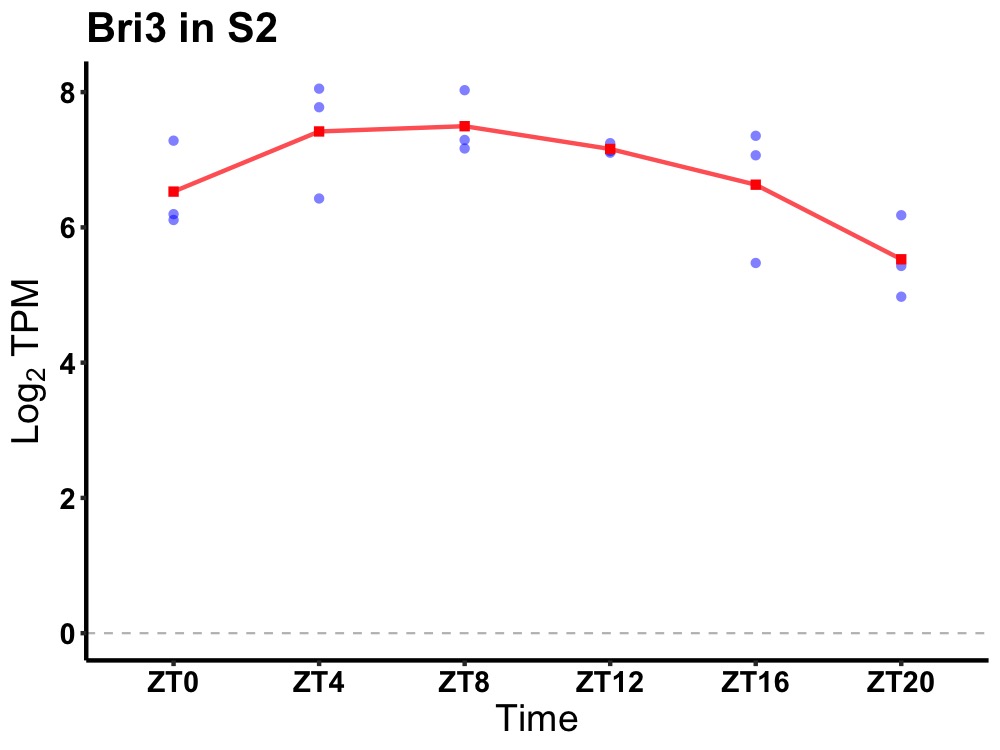 |
brain protein I3 | 0.036 | 24 | 8 | 0.74 |
| ENSMUSG00000039684 | Gm5422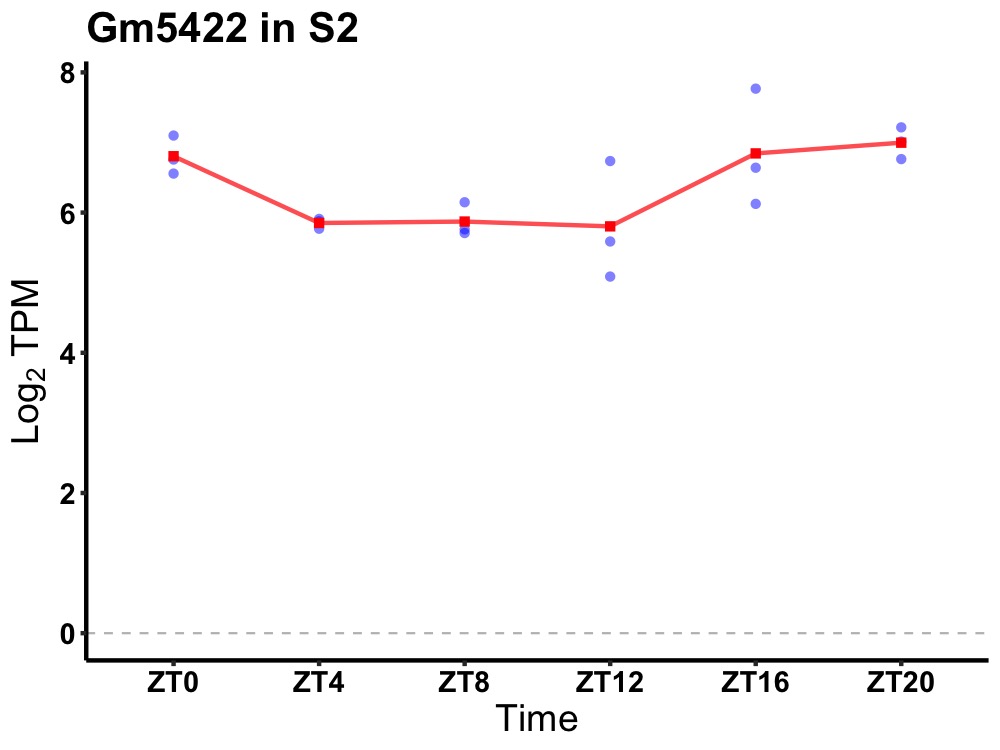 |
predicted pseudogene 5422 | 0.014 | 24 | 22 | 0.74 |
| ENSMUSG00000100615 | Gm5511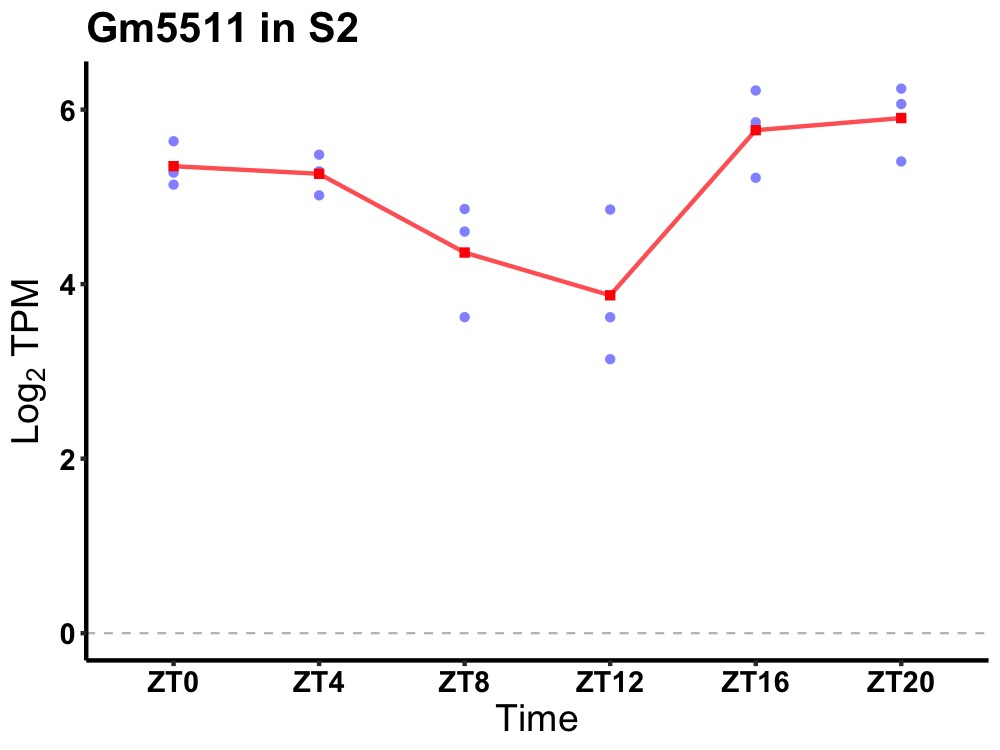 |
predicted gene 5511 | 0.014 | 24 | 22 | 0.74 |
| ENSMUSG00000024538 | Lin52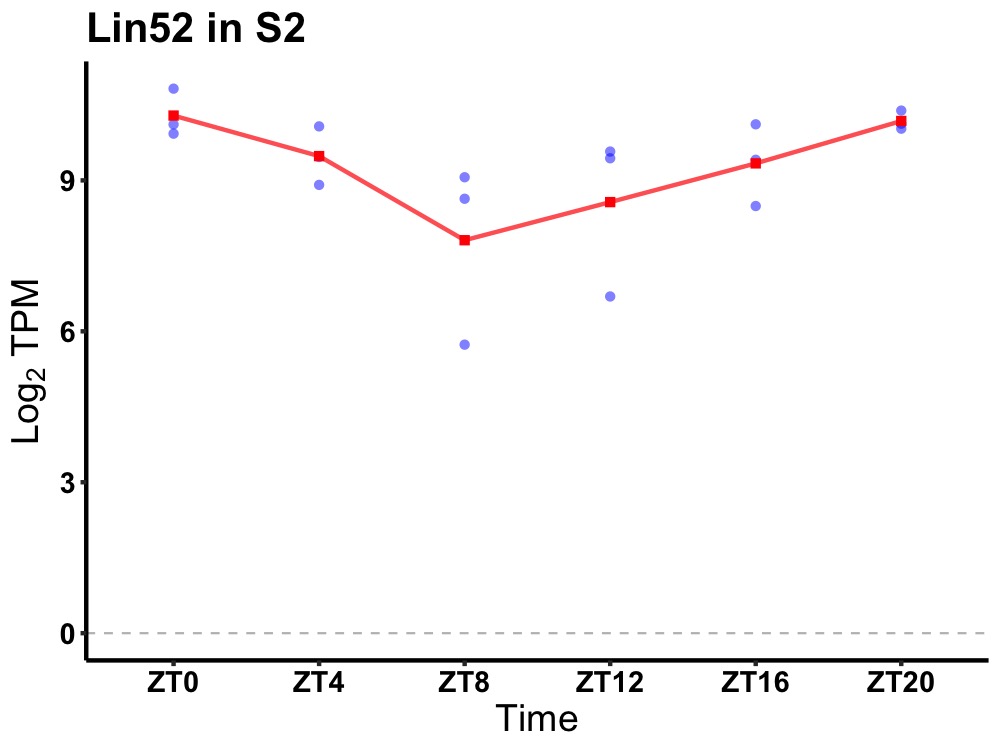 |
lin-52 homolog (C. elegans) | 0.019 | 24 | 22 | 0.74 |
| ENSMUSG00000113706 | Ddi2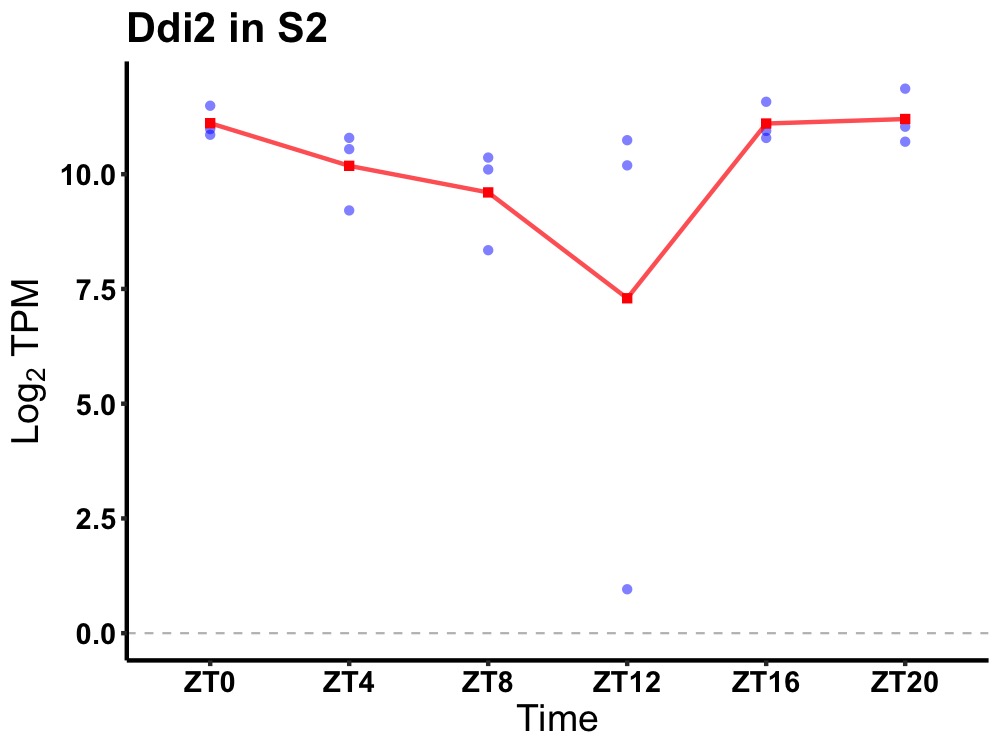 |
DNA-damage inducible protein 2 | 0.010 | 24 | 22 | 0.73 |
| ENSMUSG00000091697 | Arsg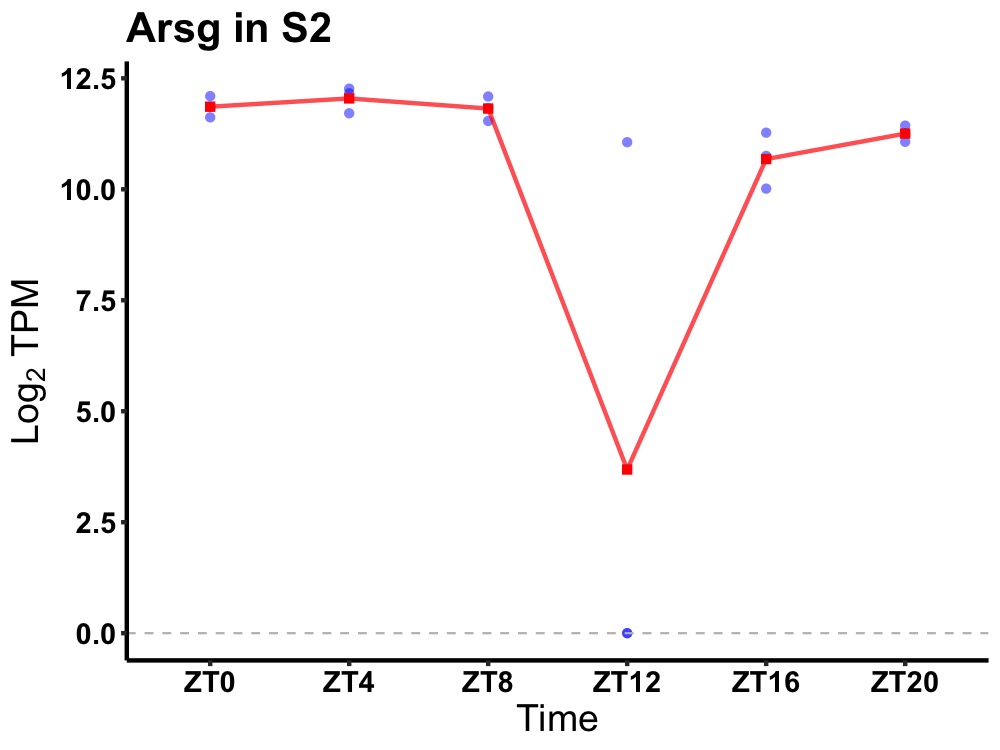 |
arylsulfatase G | 0.000 | 24 | 4 | 0.73 |
| ENSMUSG00000056999 | Fubp1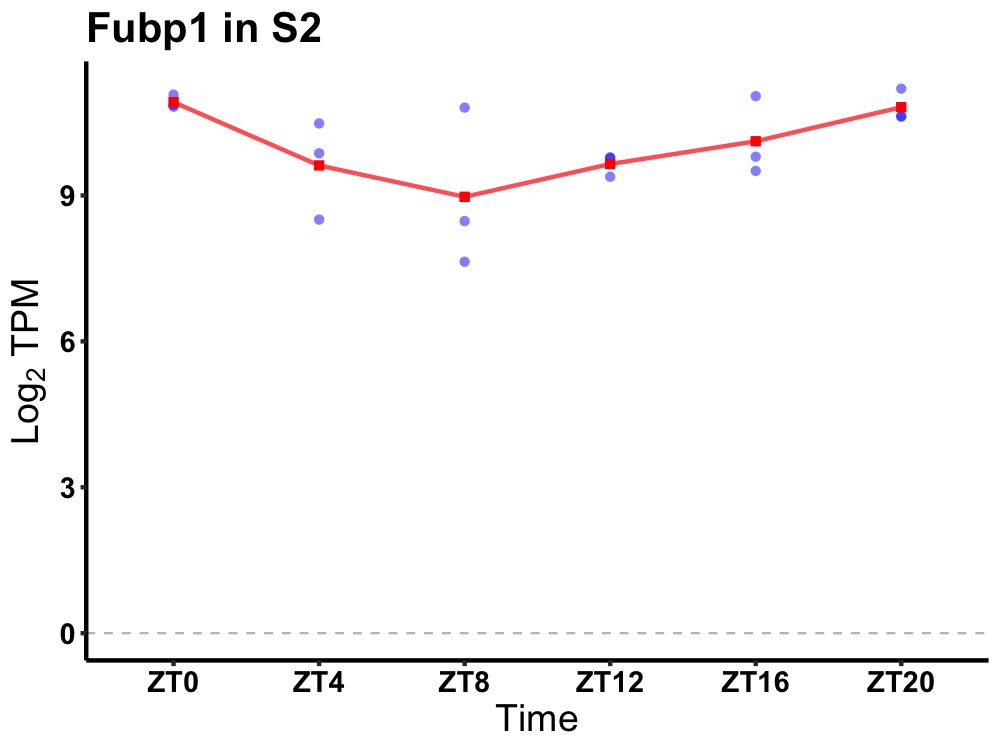 |
far upstream element (FUSE) binding protein 1 | 0.019 | 20 | 0 | 0.72 |
| ENSMUSG00000024826 | Slc16a12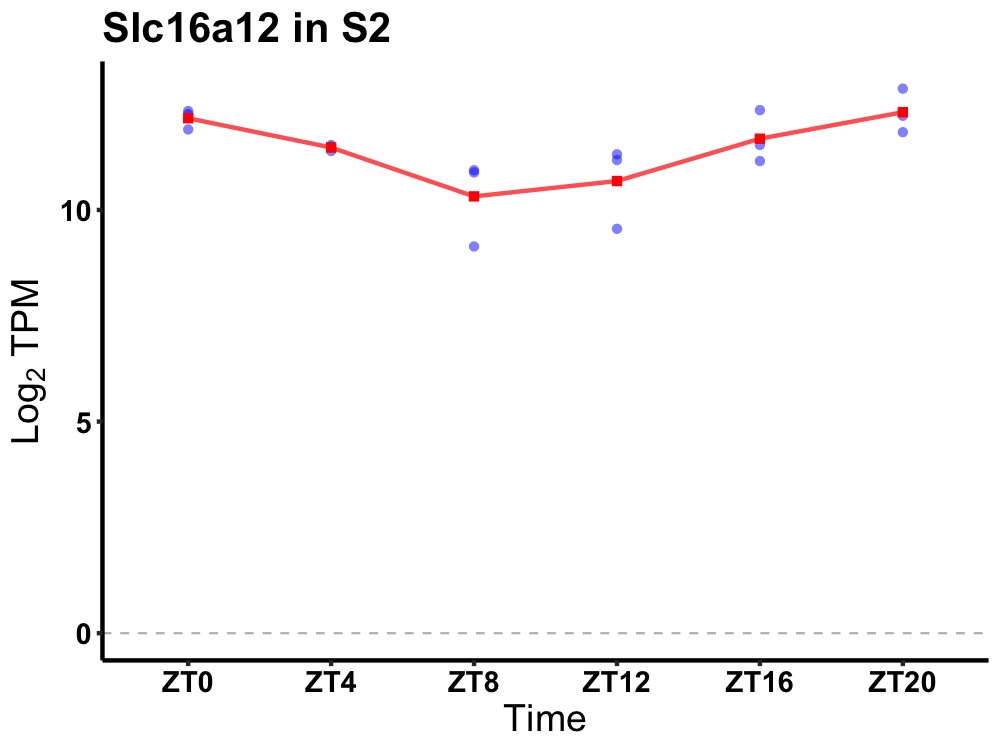 |
solute carrier family 16 (monocarboxylic acid transporters), member 12 | 0.000 | 20 | 0 | 0.72 |
| ENSMUSG00000084329 | Gm6733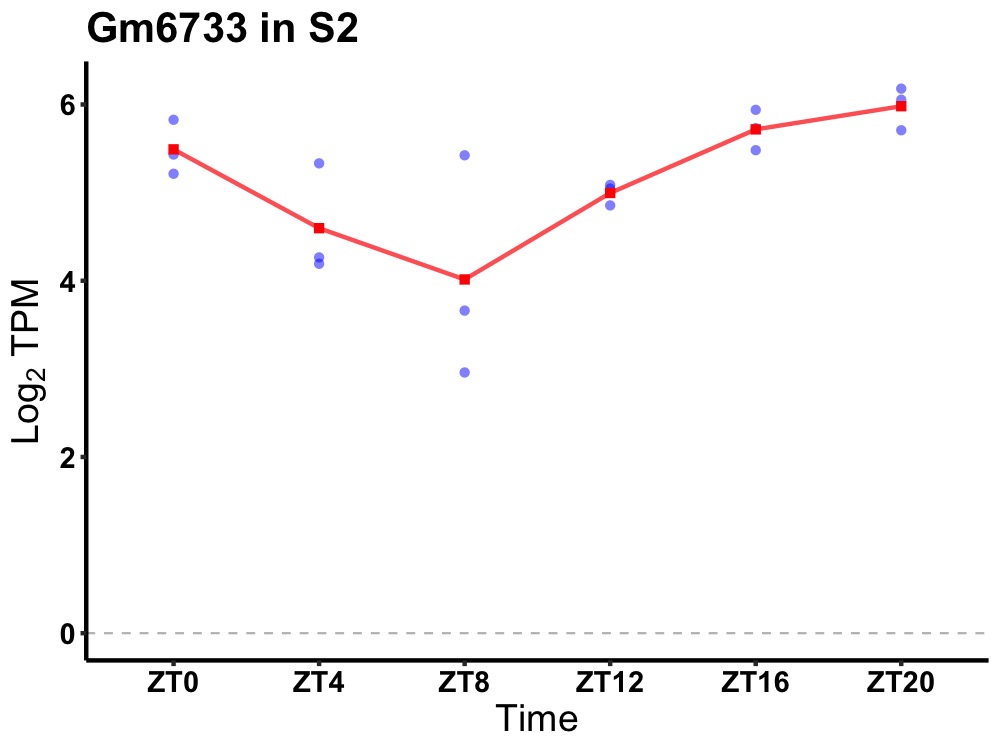 |
predicted gene 6733 | 0.000 | 24 | 20 | 0.72 |
| ENSMUSG00000115730 | Gm7459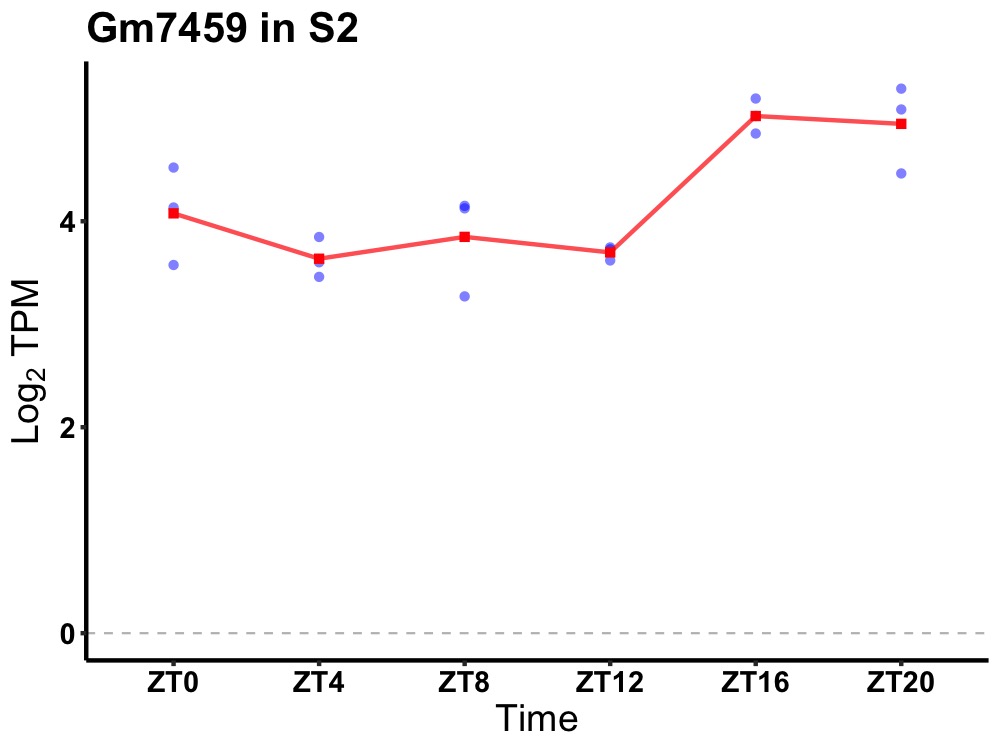 |
predicted gene 7459 | 0.036 | 24 | 20 | 0.71 |
| ENSMUSG00000066072 | Nat8f6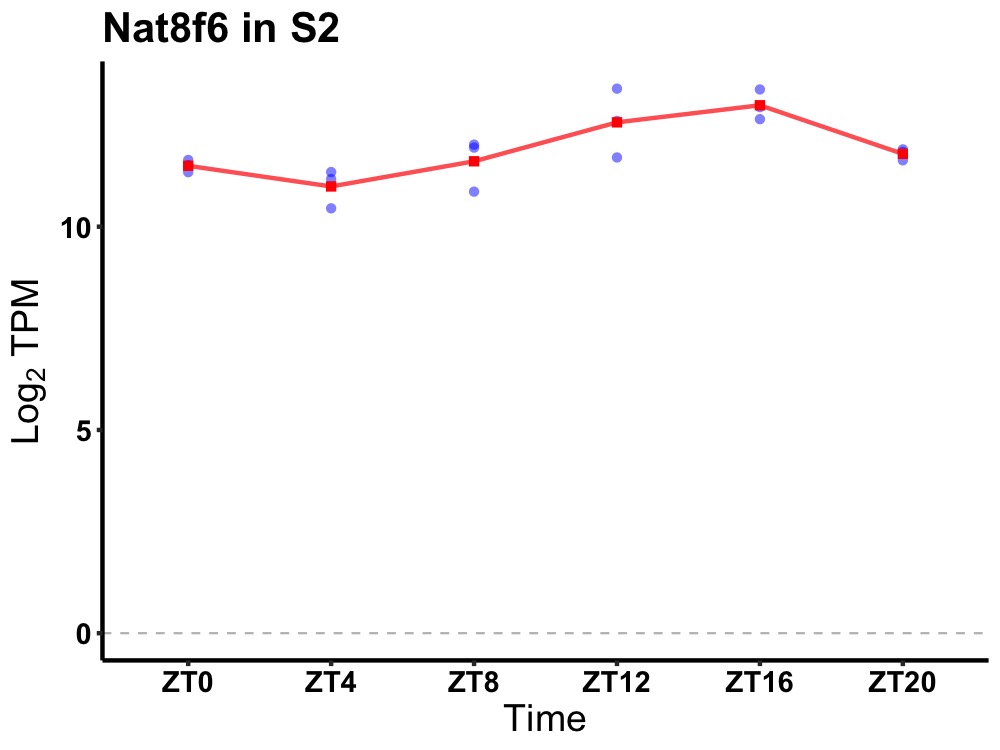 |
N-acetyltransferase 8 (GCN5-related) family member 6 | 0.002 | 24 | 16 | 0.71 |
| ENSMUSG00000114558 | Gm9570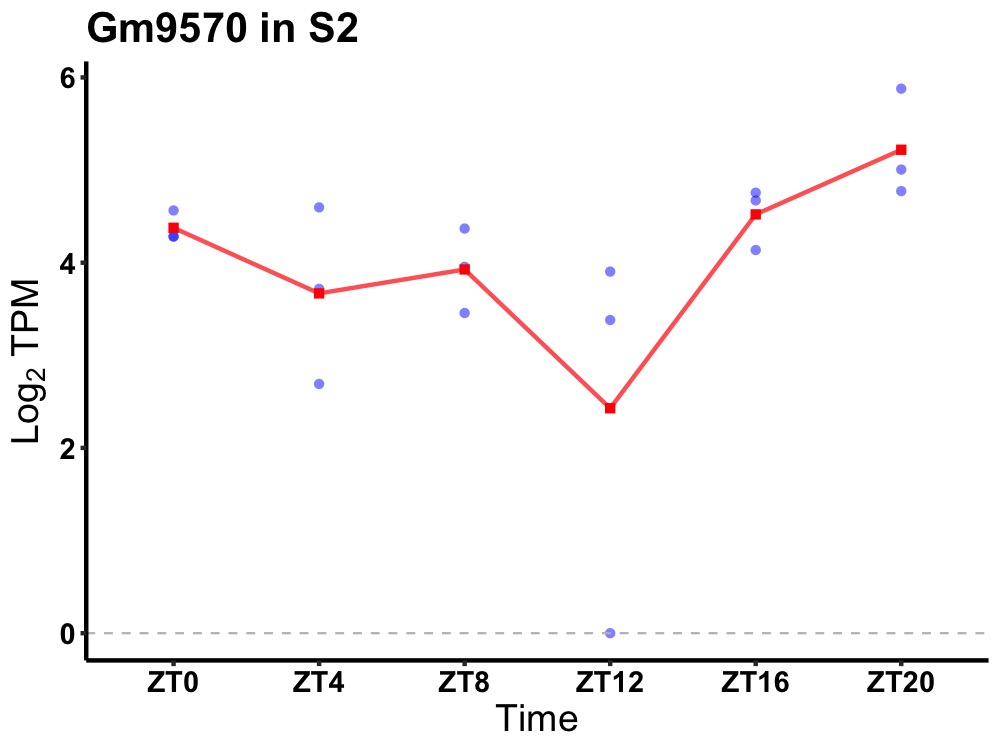 |
predicted gene 9570 | 0.049 | 24 | 22 | 0.70 |
| ENSMUSG00000081021 | Il13ra1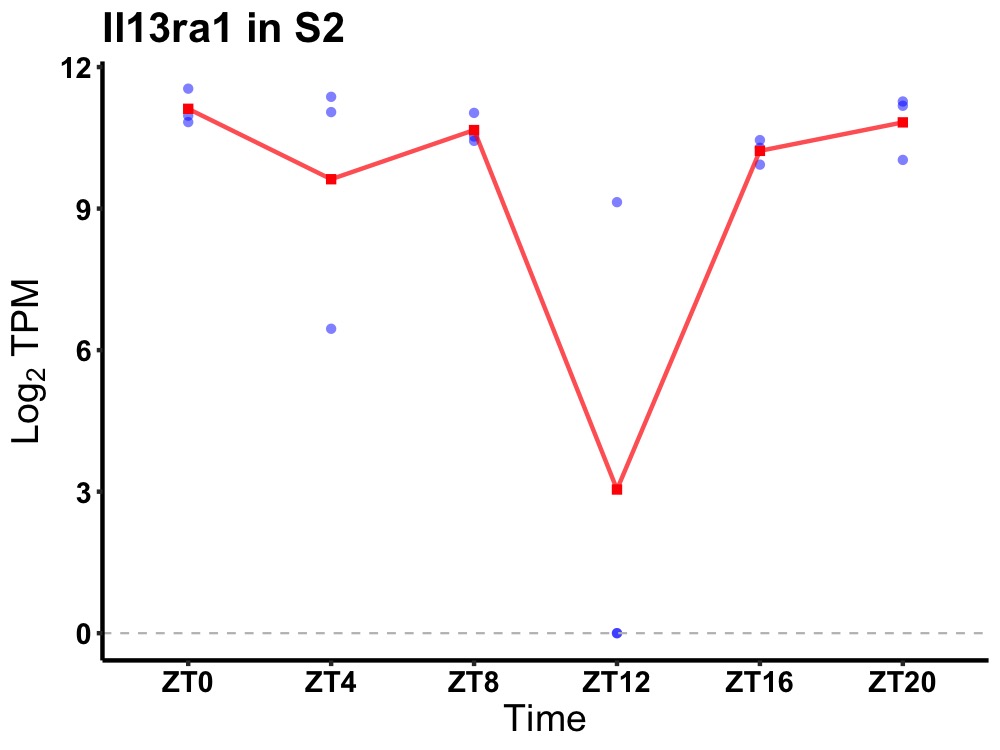 |
interleukin-13 receptor subunit alpha-1 | 0.036 | 24 | 2 | 0.69 |
| ENSMUSG00000026281 | Osgin1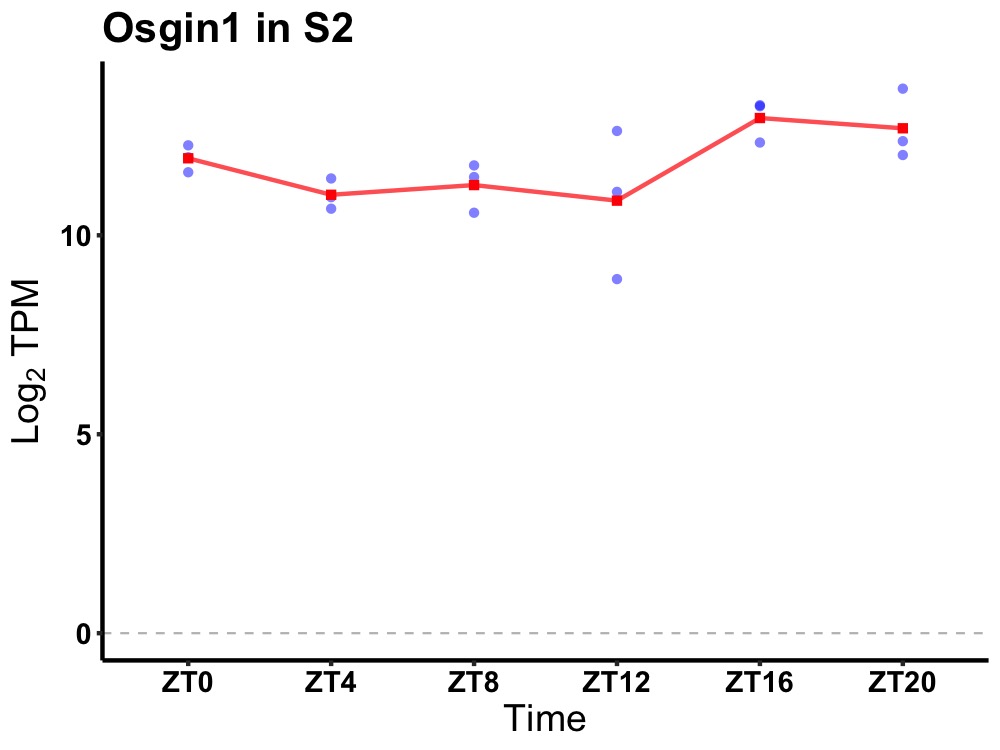 |
oxidative stress induced growth inhibitor 1 | 0.027 | 24 | 18 | 0.69 |
| ENSMUSG00000070348 | Prnp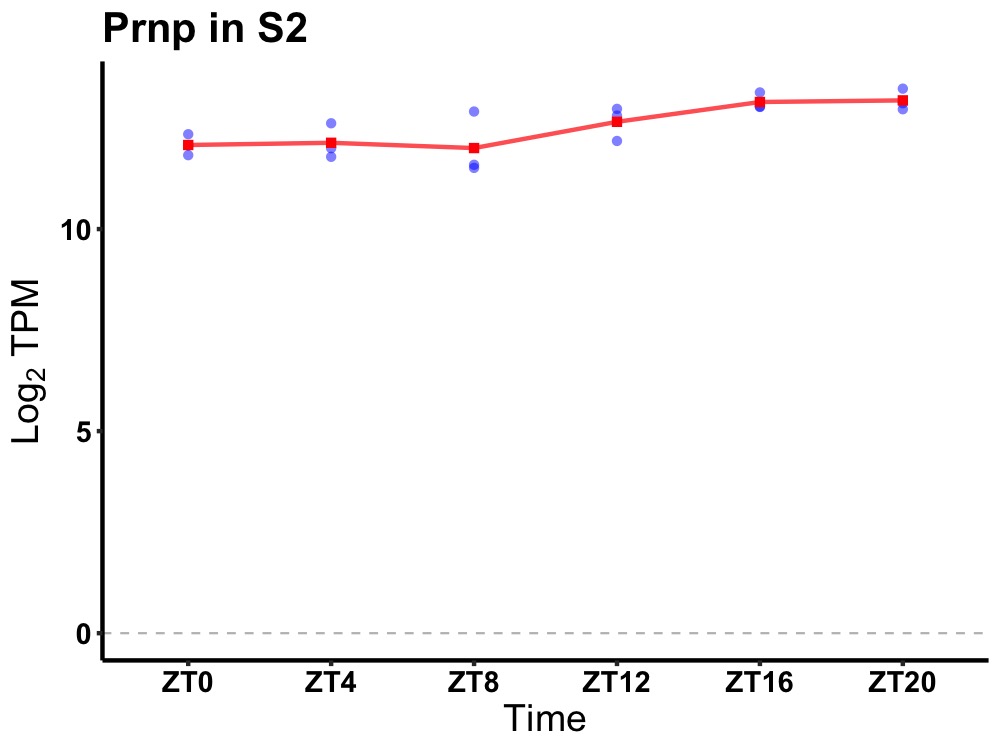 |
prion protein | 0.007 | 24 | 18 | 0.69 |
| ENSMUSG00000080775 | Gm6368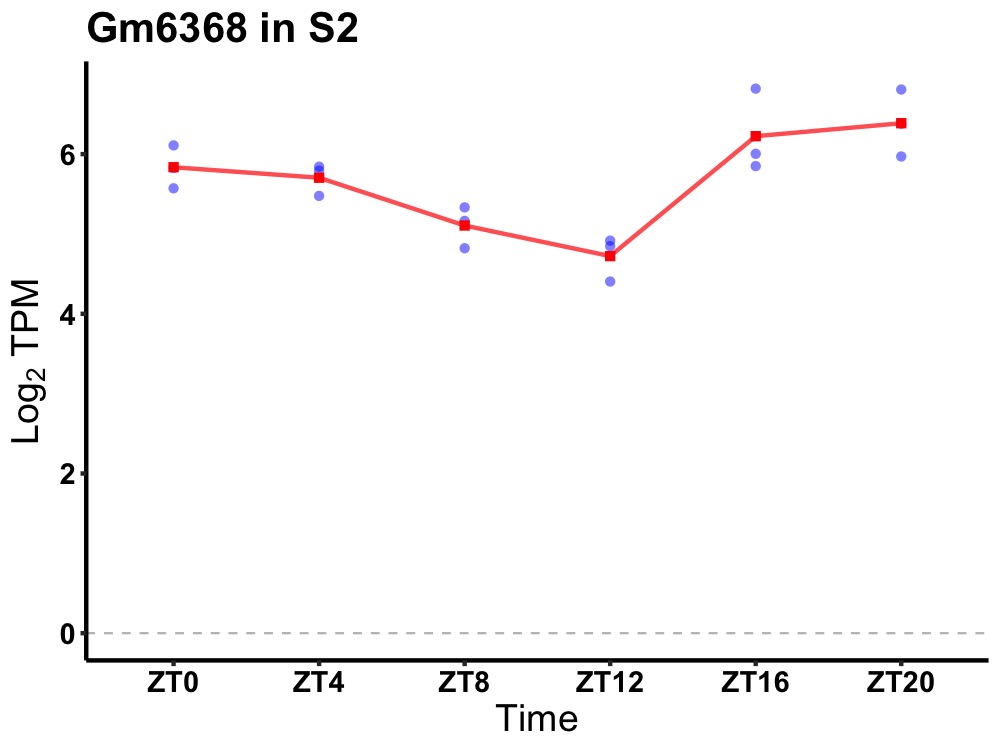 |
predicted gene 6368 | 0.005 | 24 | 22 | 0.69 |
| ENSMUSG00000083627 | Gm12502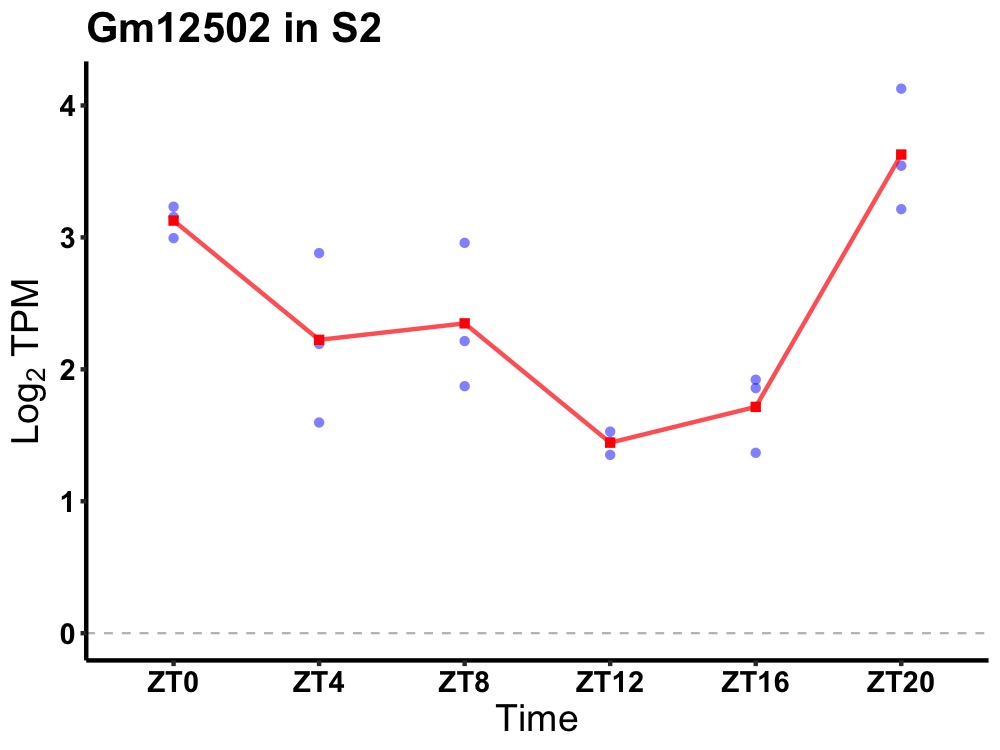 |
predicted gene 12502 | 0.001 | 20 | 2 | 0.69 |
| ENSMUSG00000080021 | Slc35d2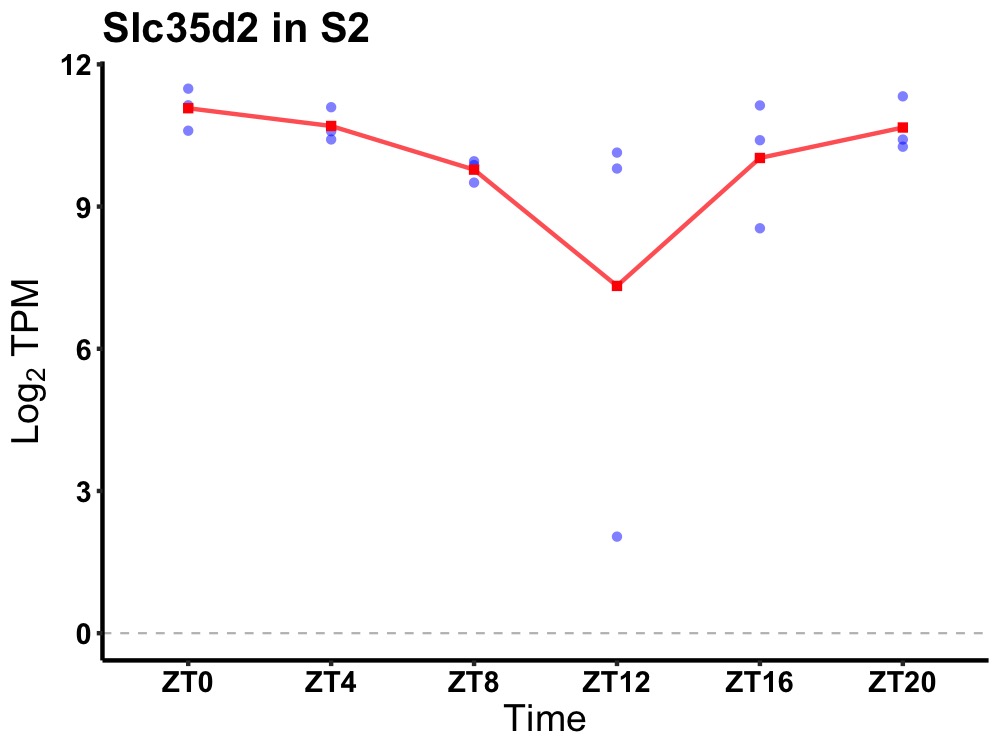 |
solute carrier family 35, member D2 | 0.014 | 24 | 0 | 0.69 |
| ENSMUSG00000082908 | Cdc37l1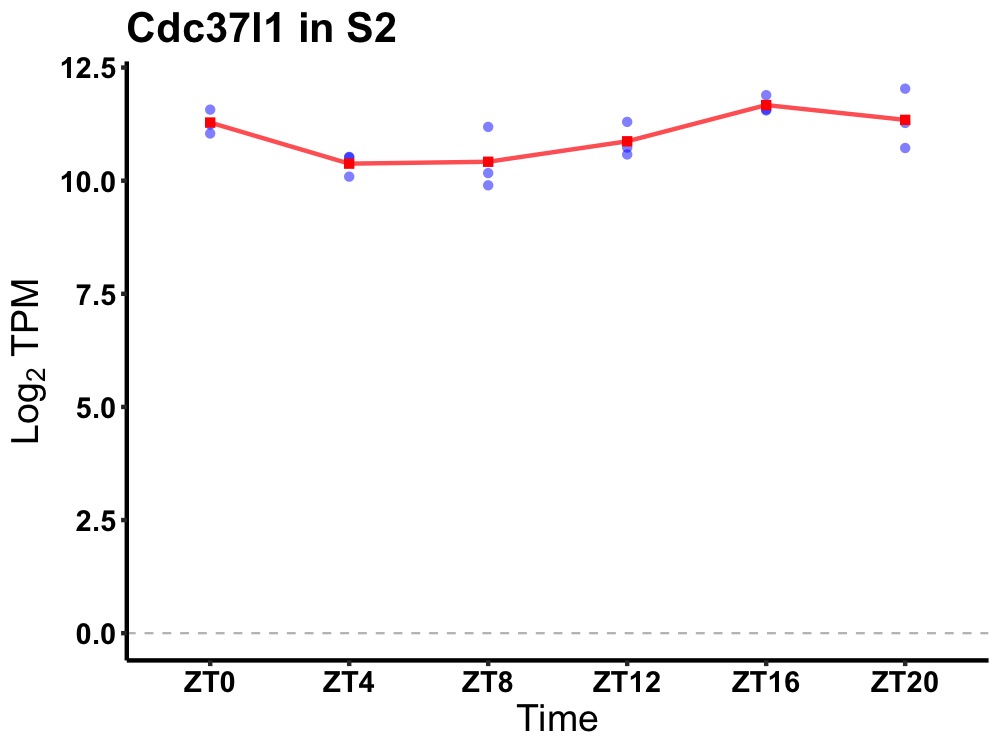 |
cell division cycle 37-like 1 | 0.005 | 20 | 18 | 0.69 |
| ENSMUSG00000020261 | Nbn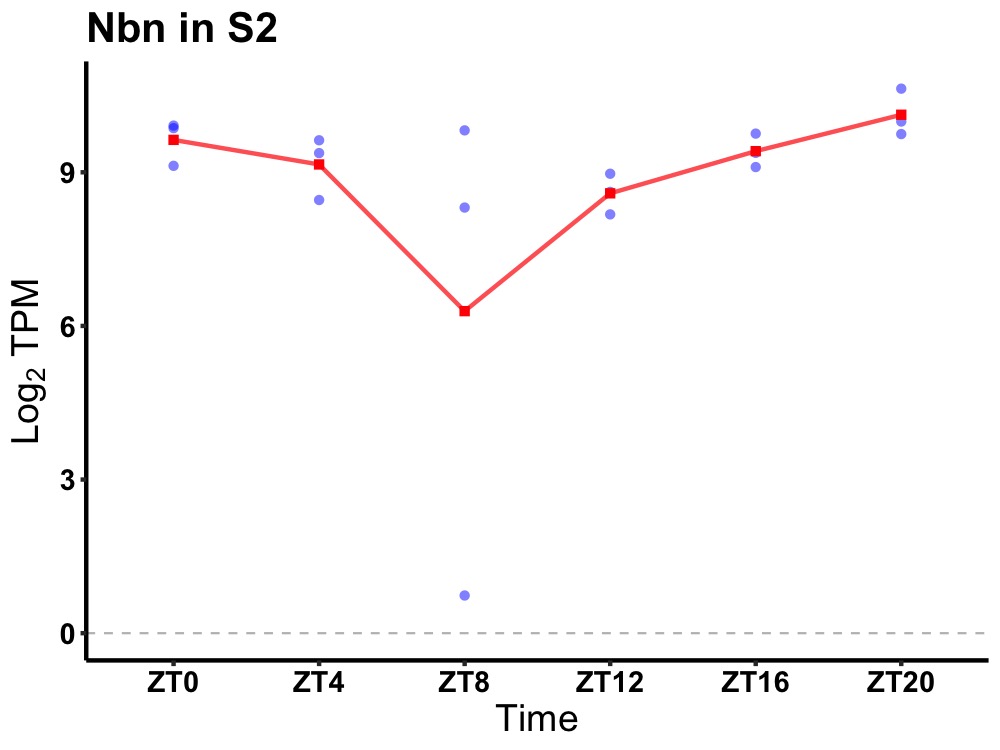 |
nibrin | 0.010 | 24 | 22 | 0.67 |
| ENSMUSG00000111446 | Rorc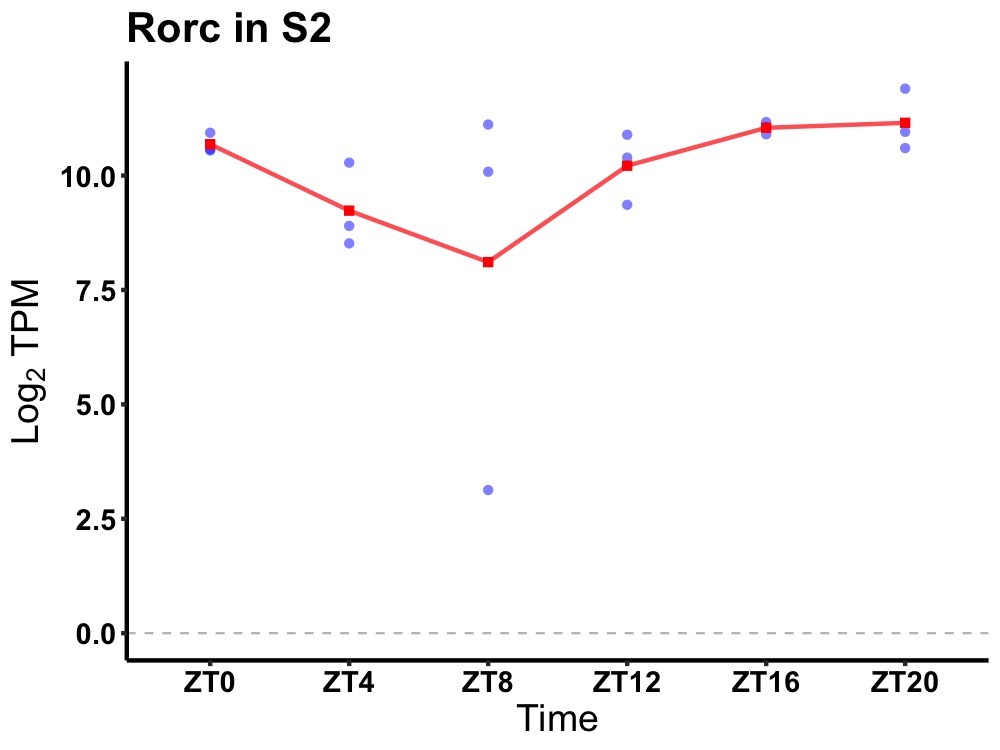 |
RAR-related orphan receptor gamma | 0.027 | 24 | 20 | 0.67 |
| ENSMUSG00000002076 | Cdk4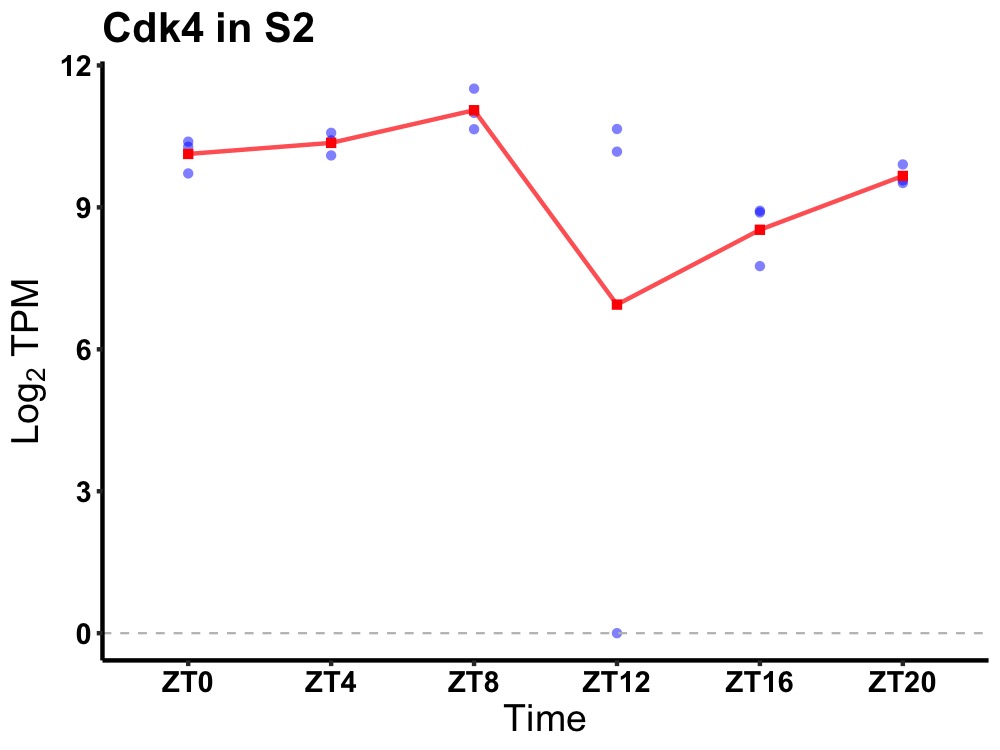 |
cyclin-dependent kinase 4 | 0.019 | 20 | 8 | 0.67 |
| ENSMUSG00000029804 | Asb9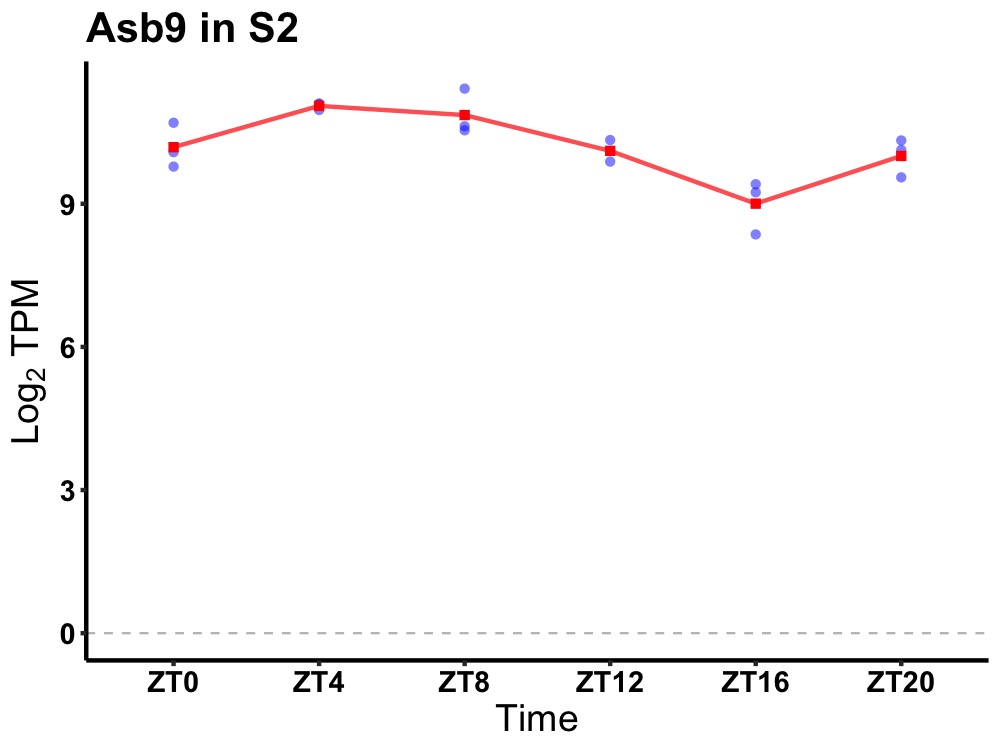 |
ankyrin repeat and SOCS box-containing 9 | 0.001 | 24 | 6 | 0.67 |
| ENSMUSG00000026701 | Agtr1a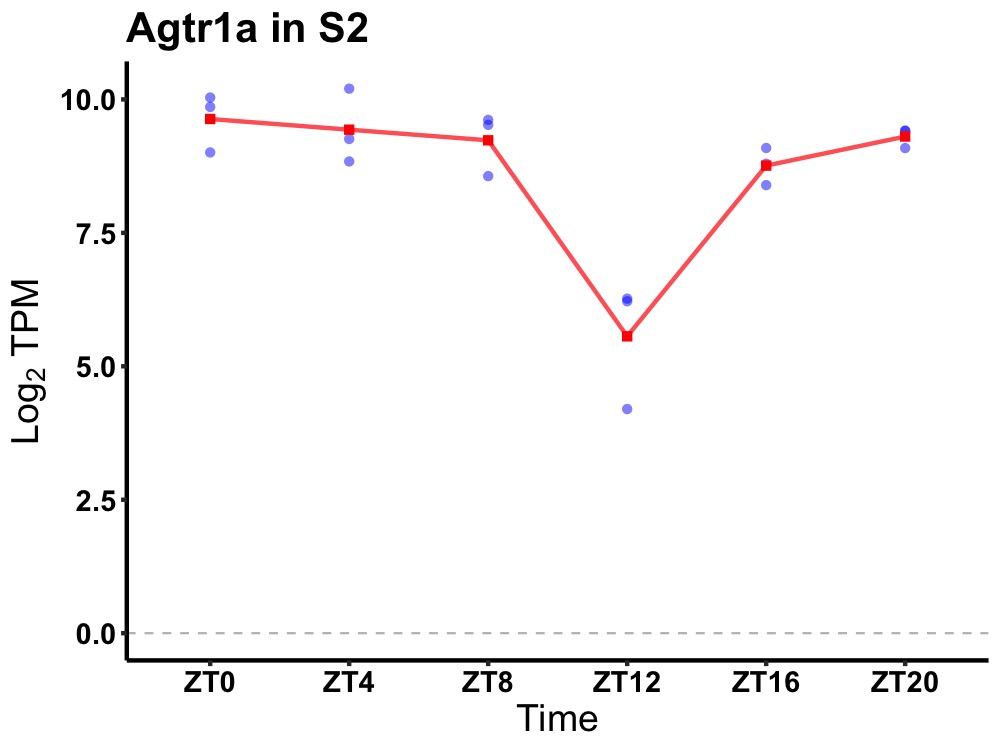 |
angiotensin II receptor, type 1a | 0.049 | 24 | 2 | 0.67 |
| ENSMUSG00000031090 | Bfar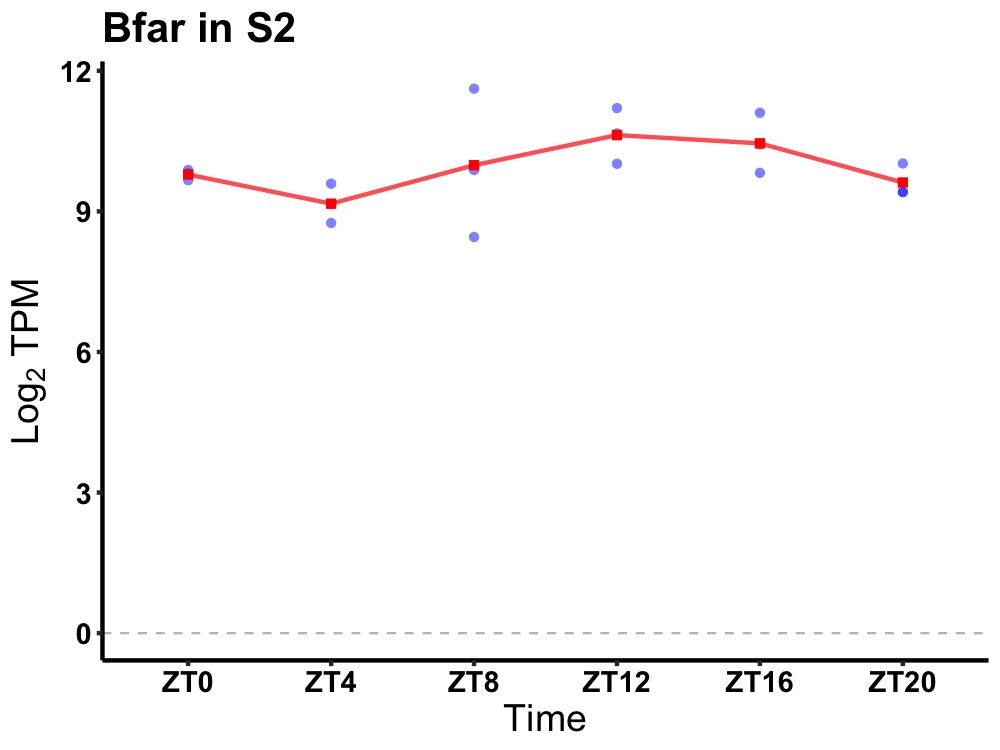 |
bifunctional apoptosis regulator | 0.036 | 20 | 14 | 0.67 |
| ENSMUSG00000085793 | Stard7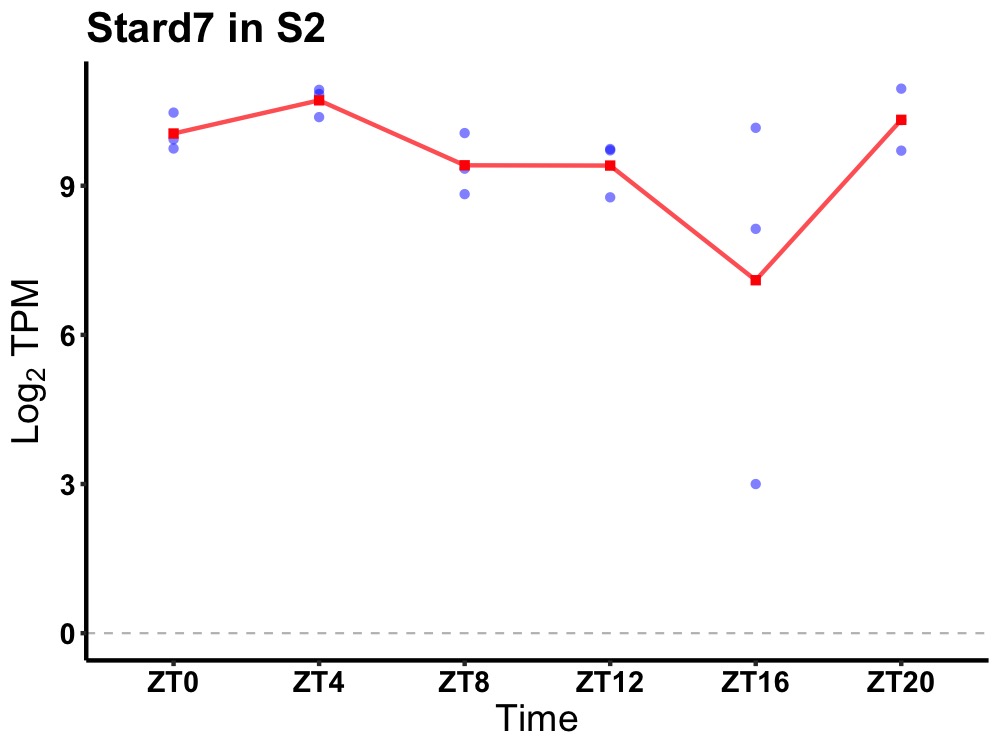 |
START domain containing 7 | 0.049 | 20 | 4 | 0.66 |
| ENSMUSG00000064370 | Wdr74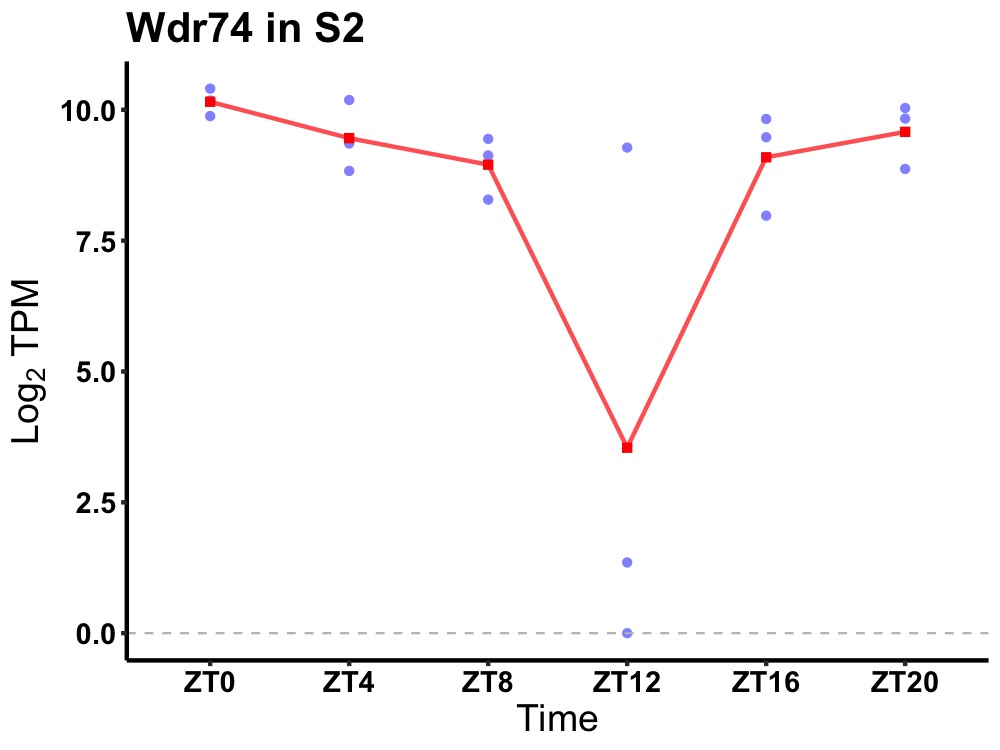 |
WD repeat domain 74 | 0.019 | 24 | 0 | 0.66 |
| ENSMUSG00000030096 | Dtd1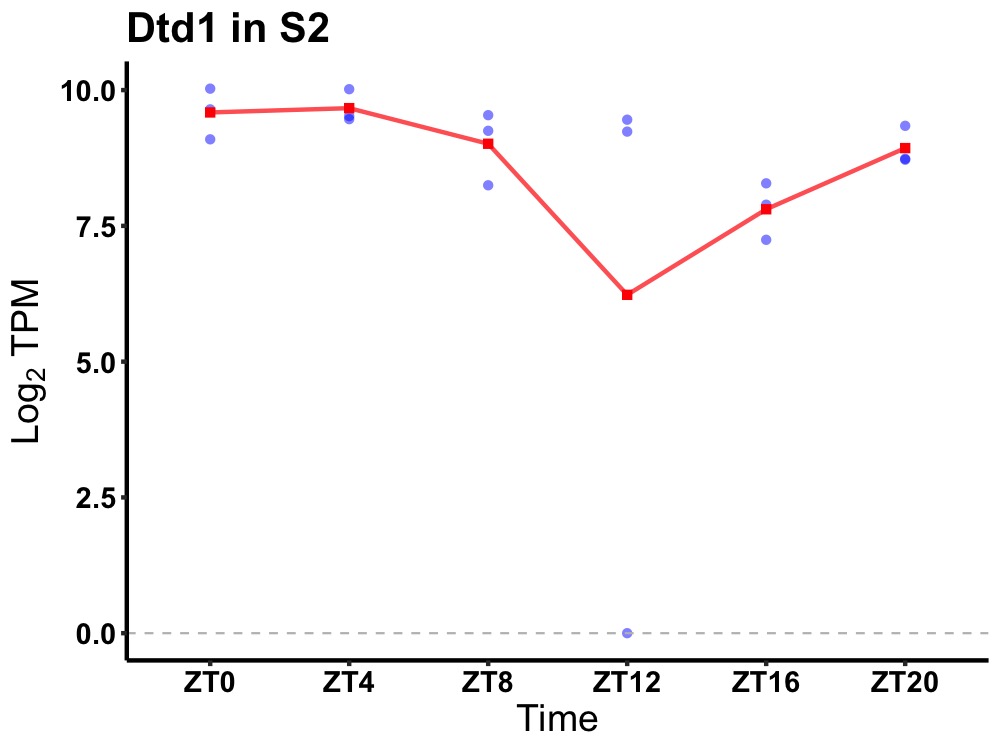 |
D-tyrosyl-tRNA deacylase 1 | 0.036 | 24 | 4 | 0.66 |
| ENSMUSG00000103137 | Gm37981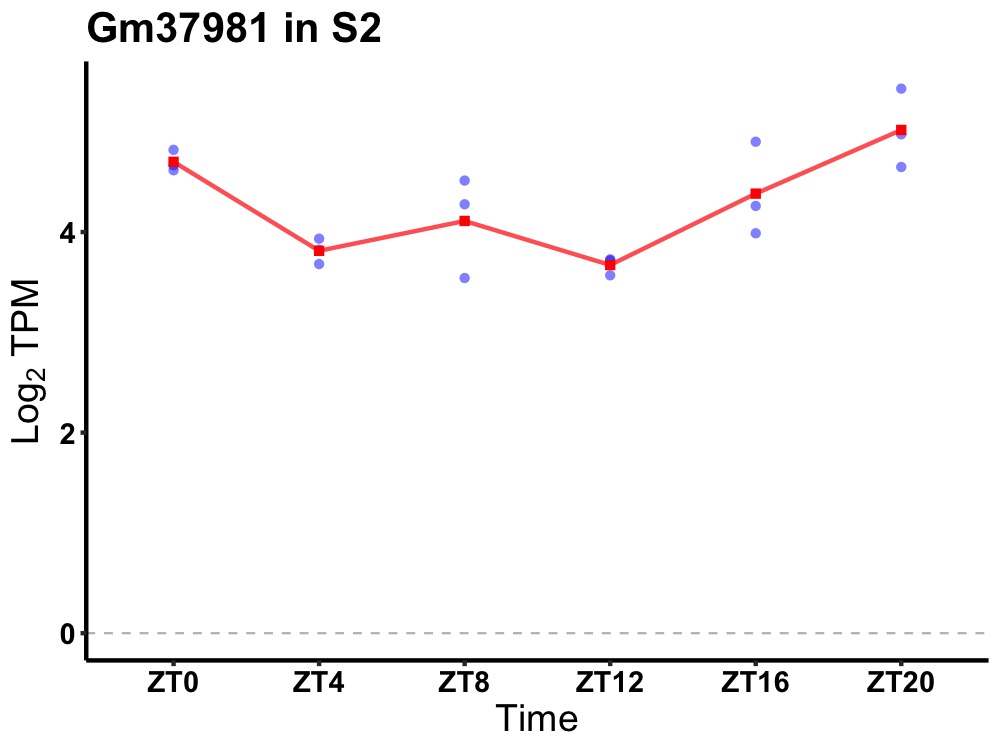 |
predicted gene, 37981 | 0.010 | 24 | 22 | 0.65 |
| ENSMUSG00000026034 | Nr1h4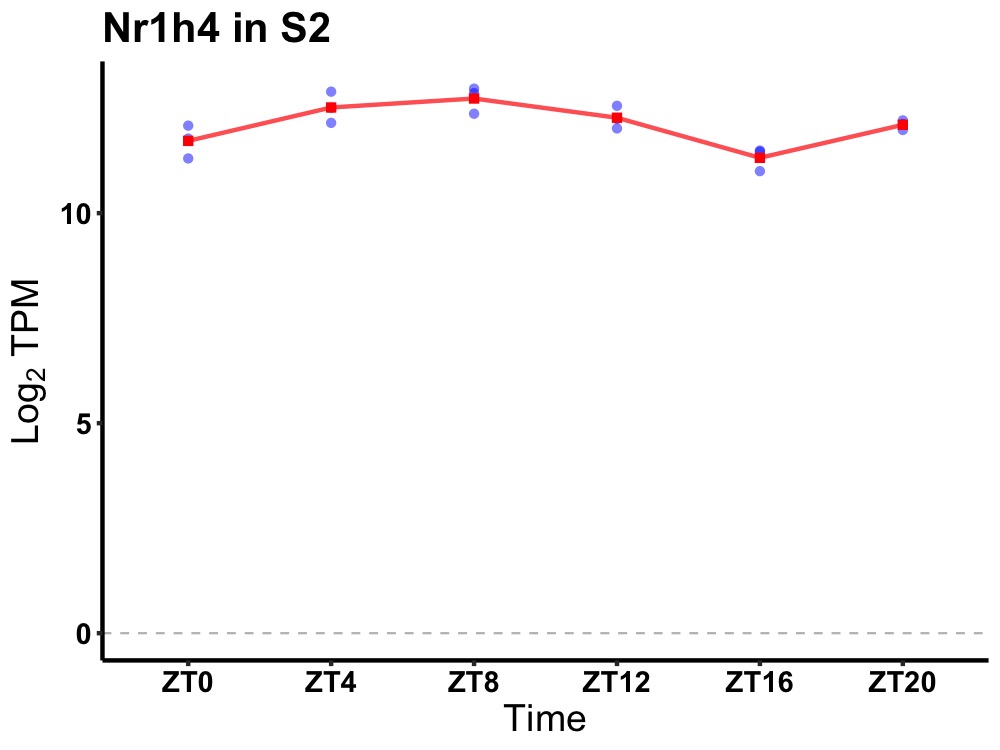 |
nuclear receptor subfamily 1, group H, member 4 | 0.000 | 20 | 8 | 0.65 |
| ENSMUSG00000083305 | Gm13315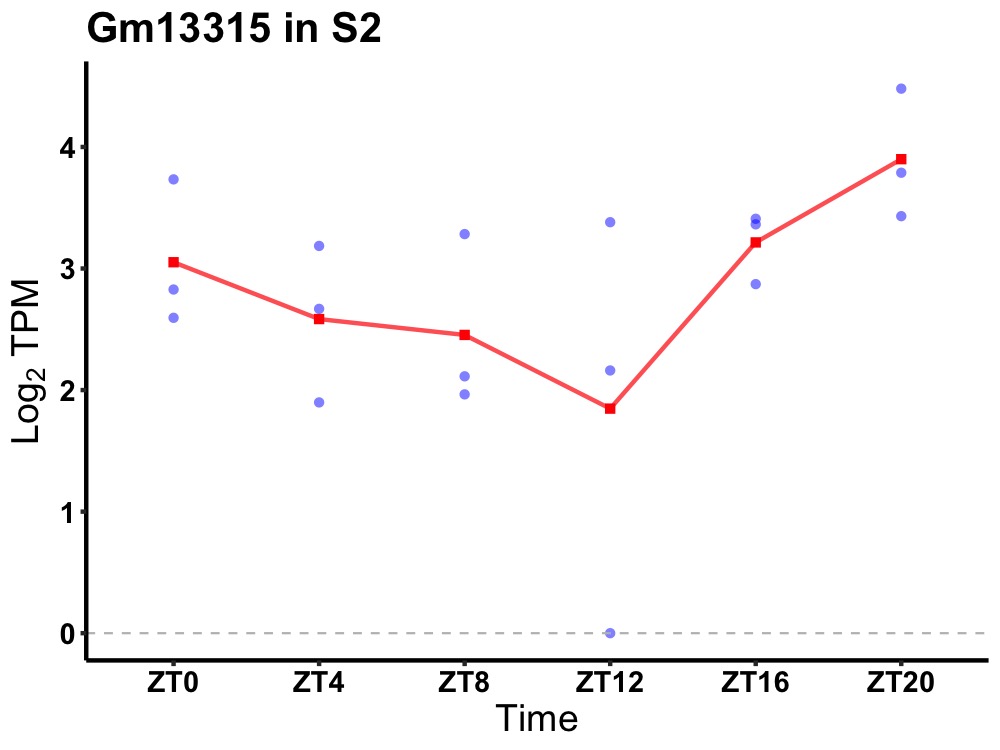 |
predicted gene 13315 | 0.049 | 24 | 20 | 0.65 |
| ENSMUSG00000021905 | Etfrf1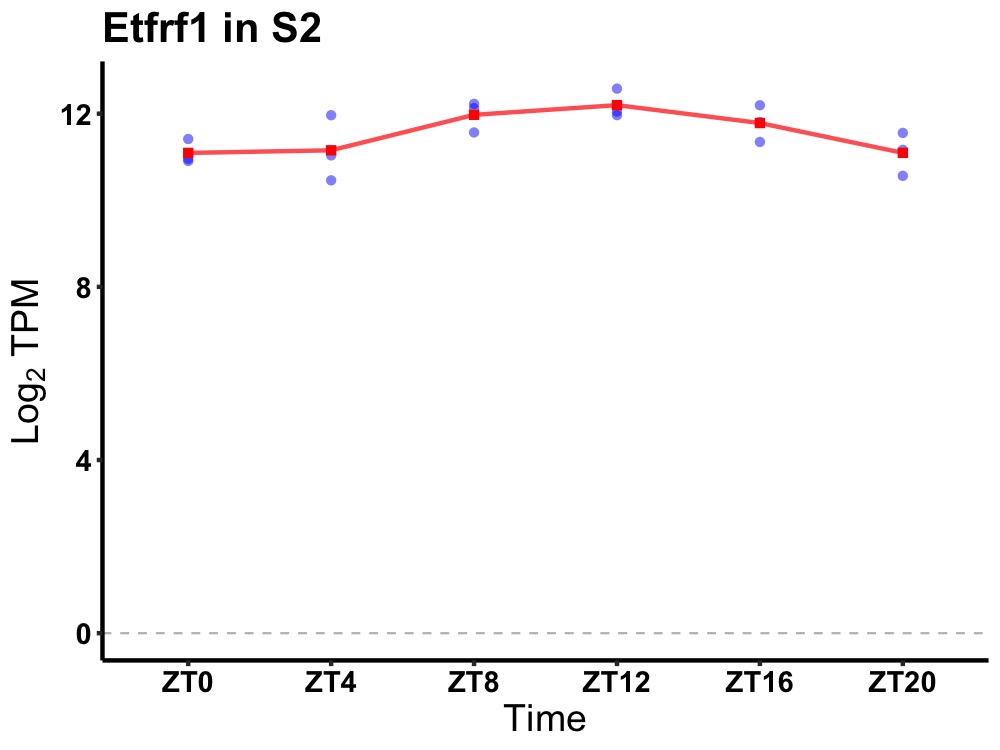 |
electron transfer flavoprotein regulatory factor 1 | 0.027 | 24 | 12 | 0.64 |
| ENSMUSG00000058622 | Cdc5l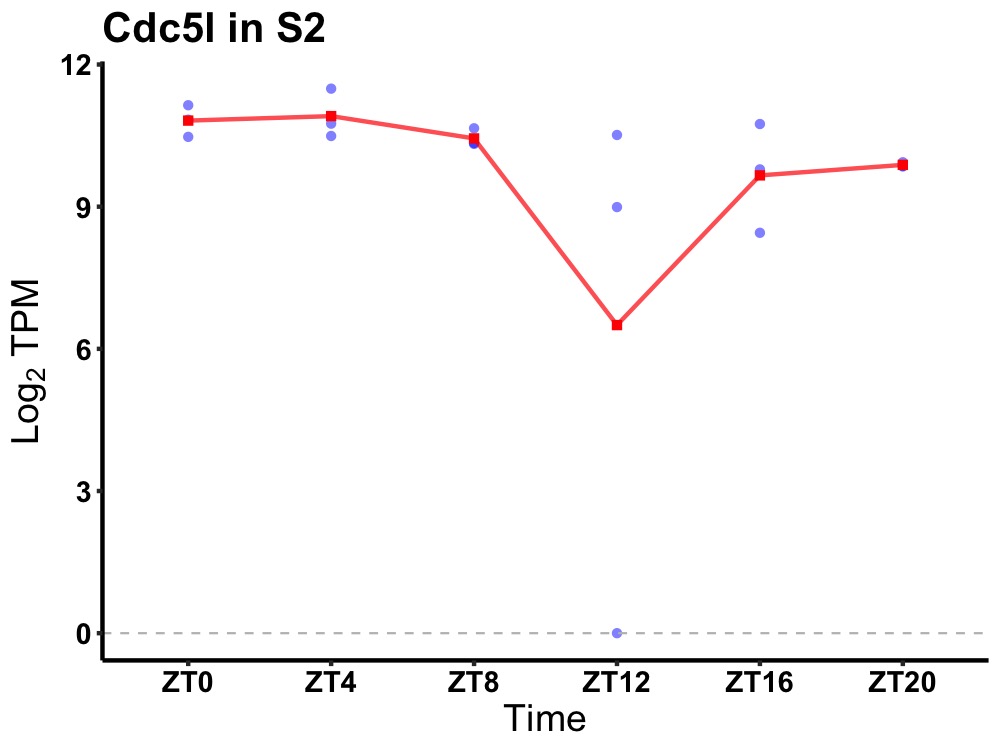 |
cell division cycle 5-like (S. pombe) | 0.027 | 24 | 4 | 0.64 |
| ENSMUSG00000117552 | Gm5686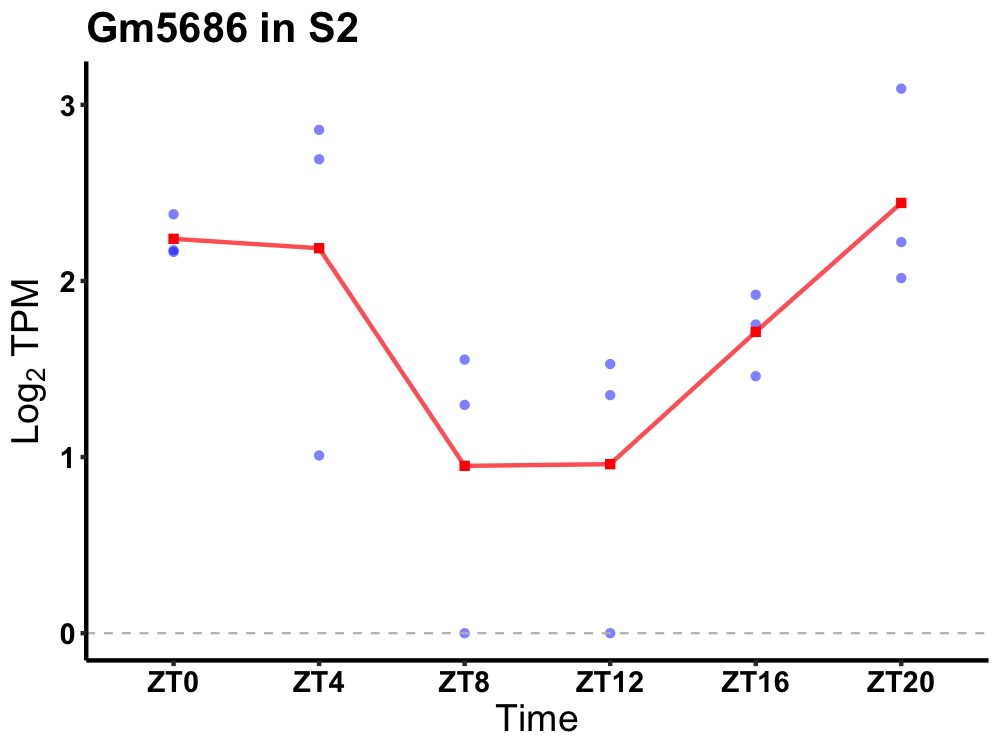 |
none | 0.031 | 20 | 2 | 0.64 |
| ENSMUSG00000083880 | Ildr2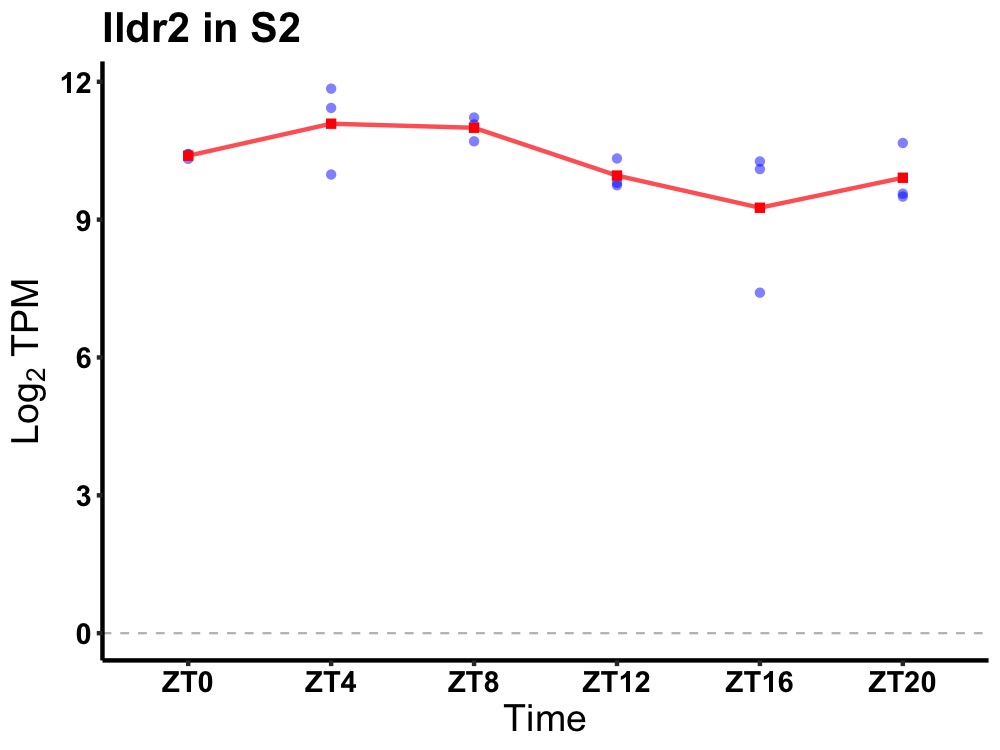 |
immunoglobulin-like domain containing receptor 2 | 0.014 | 24 | 6 | 0.63 |
| ENSMUSG00000083512 | Gm12749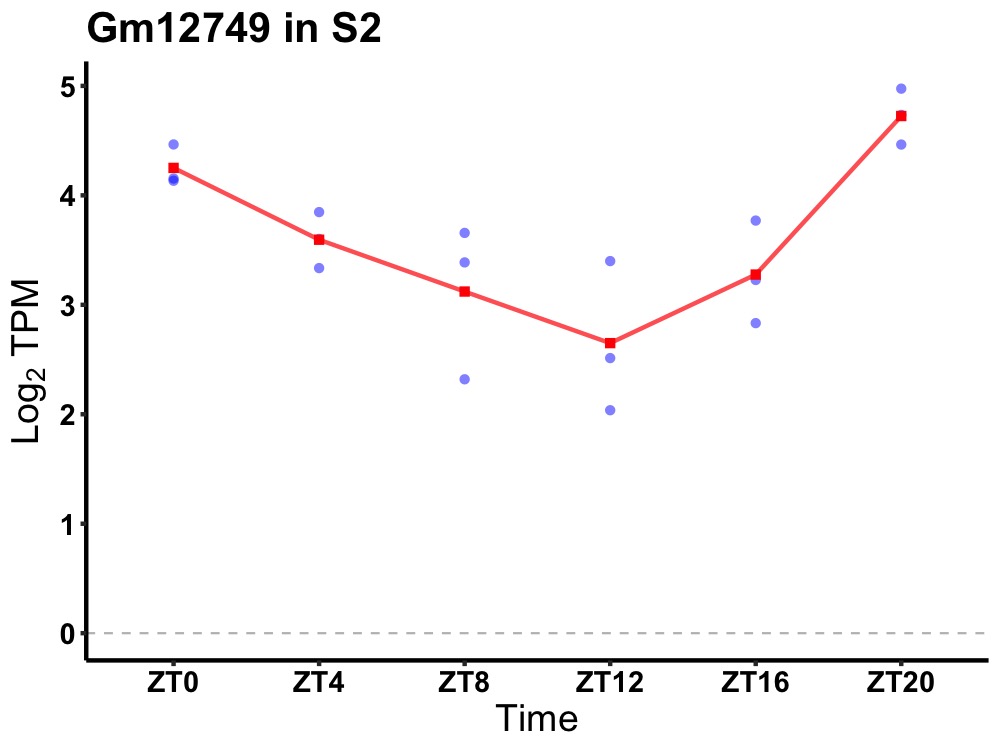 |
predicted gene 12749 | 0.000 | 20 | 2 | 0.63 |
| ENSMUSG00000026238 | Zdhhc12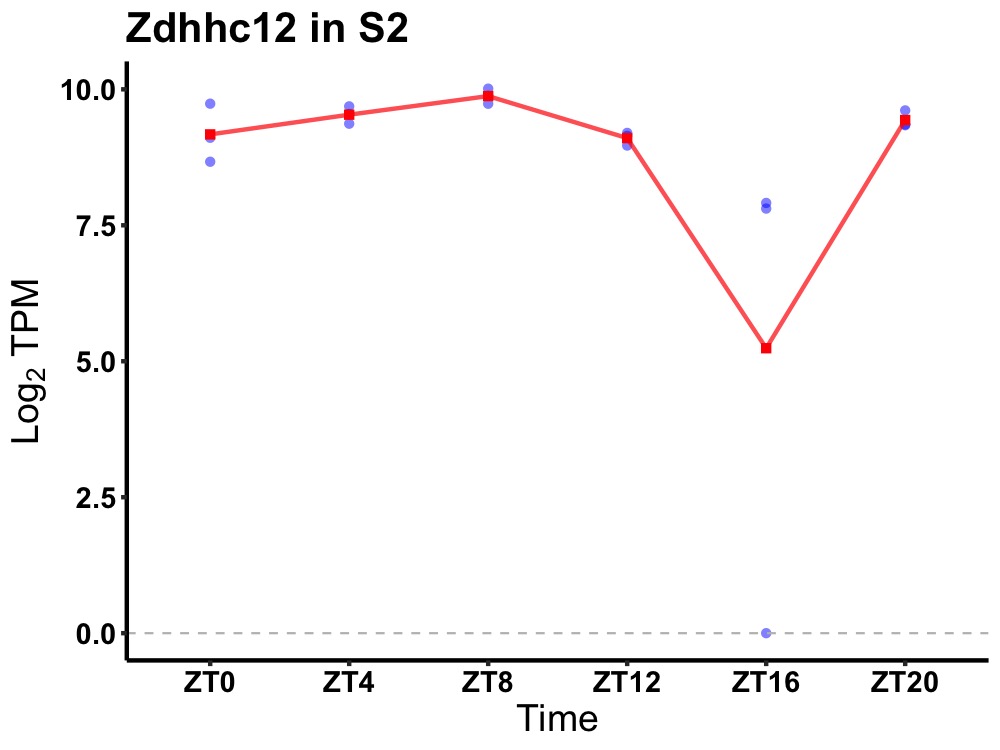 |
zinc finger, DHHC domain containing 12 | 0.005 | 20 | 8 | 0.63 |
| ENSMUSG00000031939 | Tmem219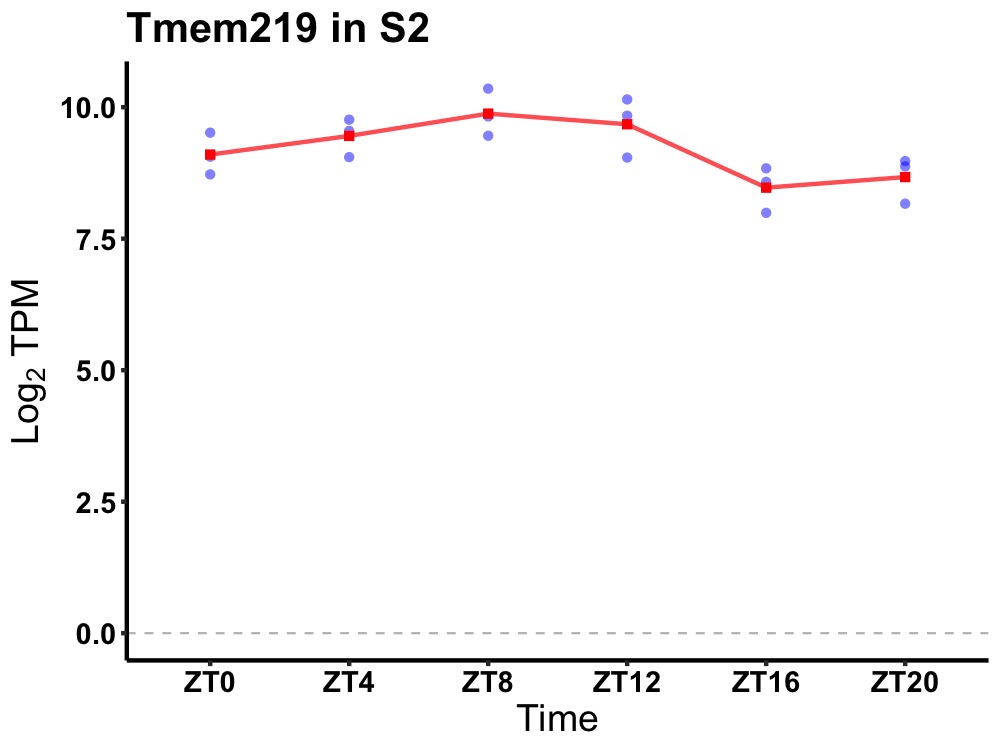 |
transmembrane protein 219 | 0.027 | 24 | 8 | 0.63 |
| ENSMUSG00000039917 | Micu3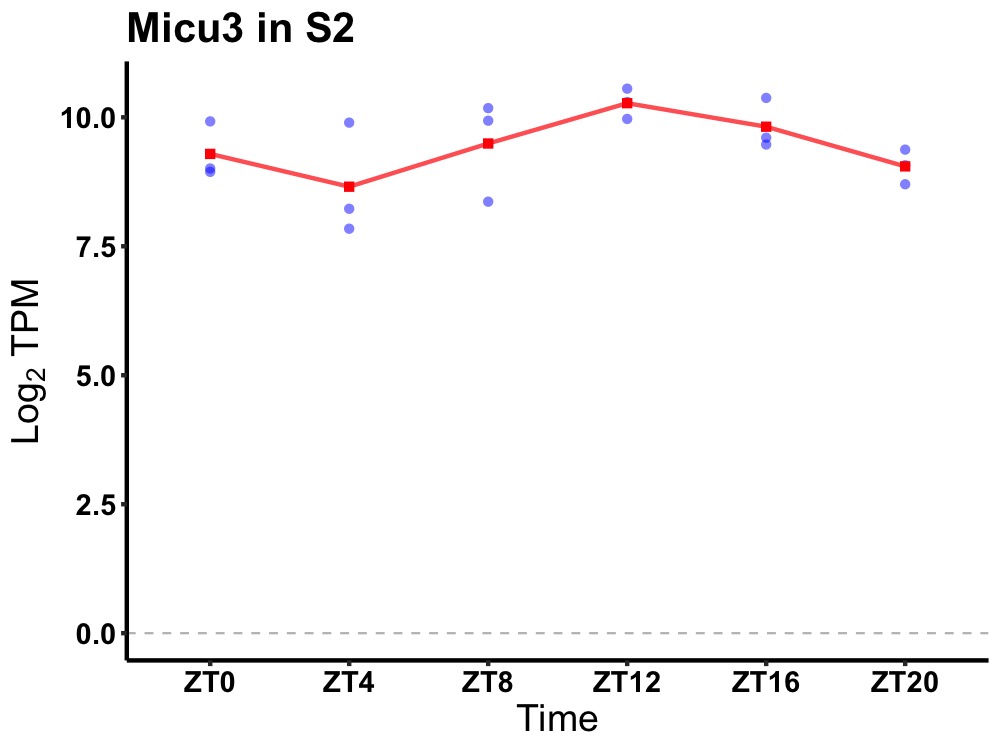 |
mitochondrial calcium uptake family, member 3 | 0.027 | 20 | 14 | 0.62 |
| ENSMUSG00000028266 | Pkig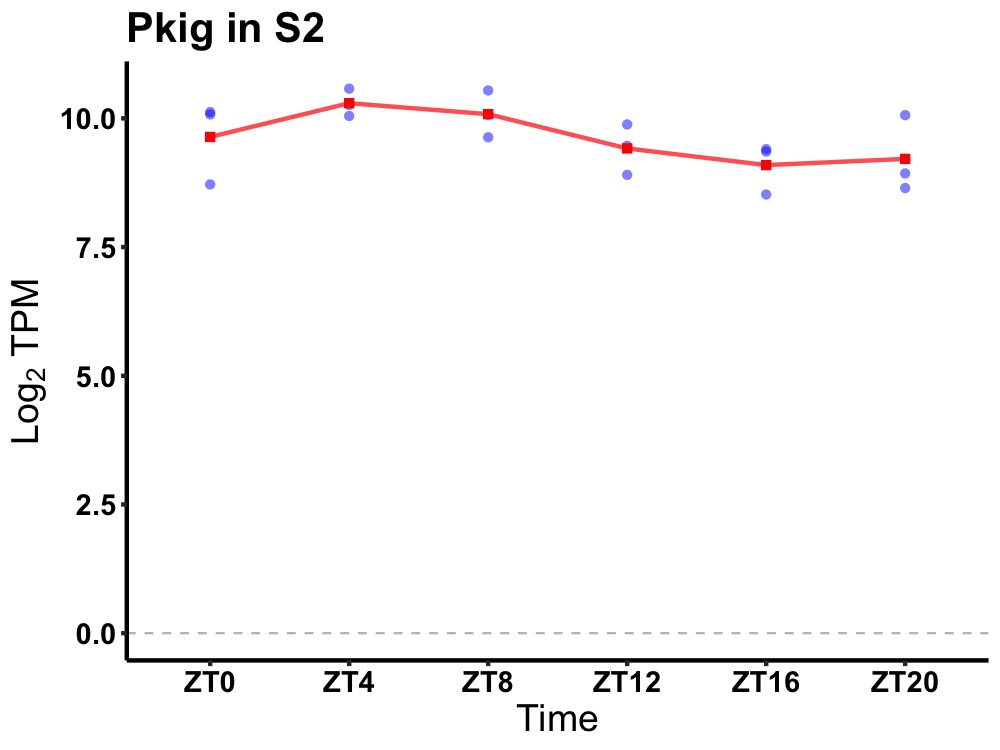 |
protein kinase inhibitor, gamma | 0.049 | 24 | 6 | 0.61 |
| ENSMUSG00000052397 | Ubiad1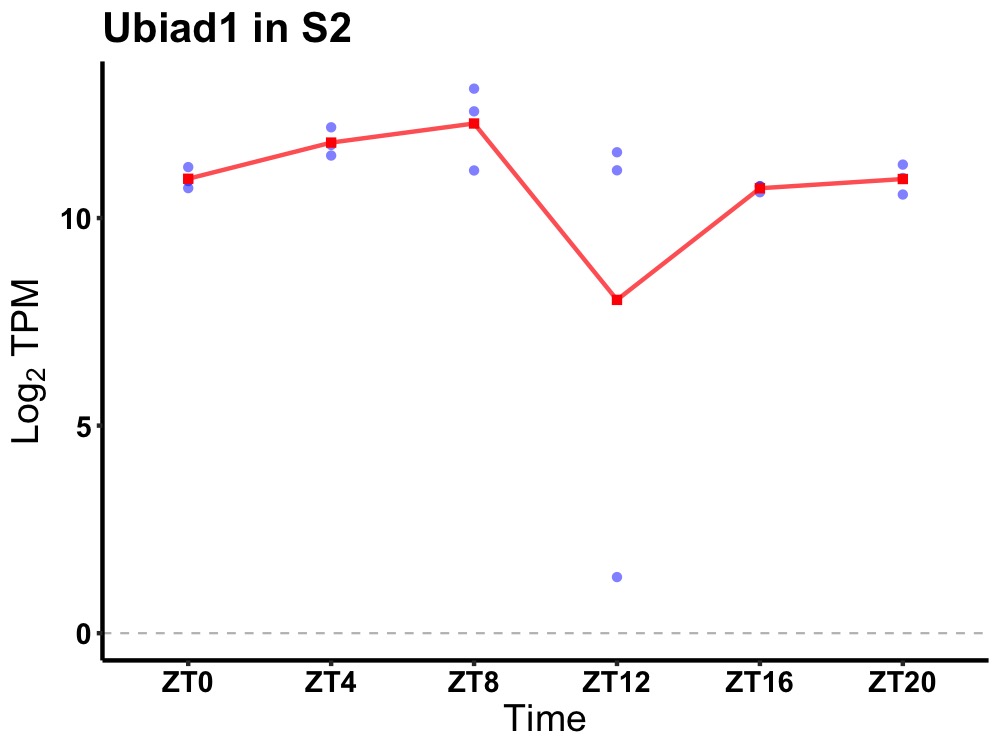 |
UbiA prenyltransferase domain containing 1 | 0.036 | 20 | 8 | 0.60 |
| ENSMUSG00000031077 | Mff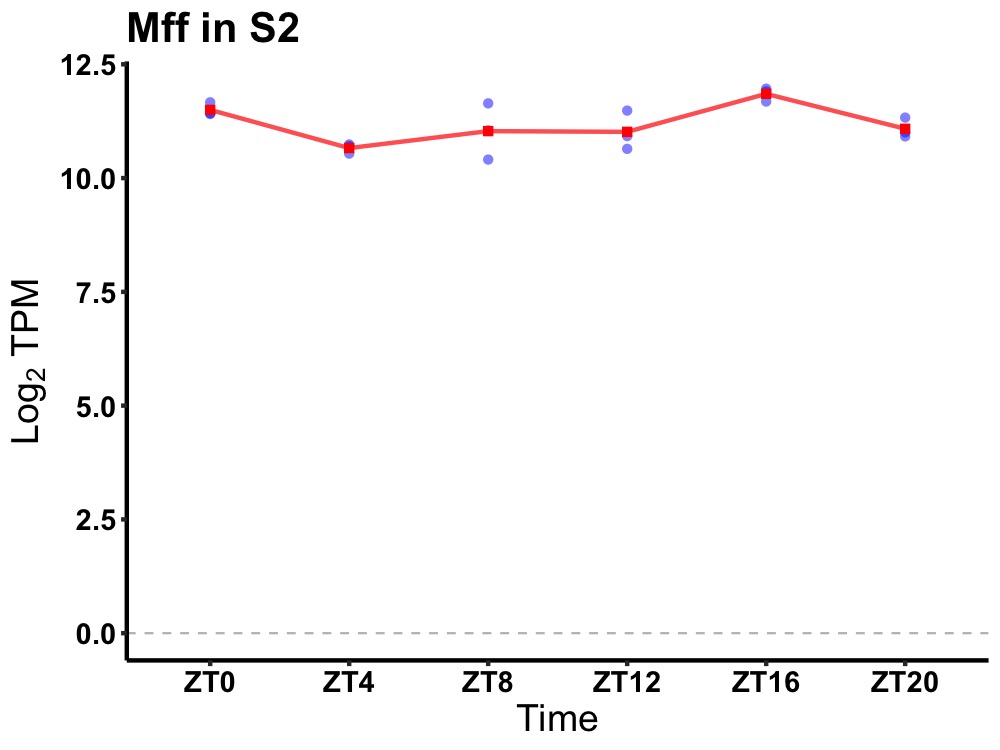 |
mitochondrial fission factor | 0.019 | 20 | 18 | 0.60 |
| ENSMUSG00000058488 | Cyhr1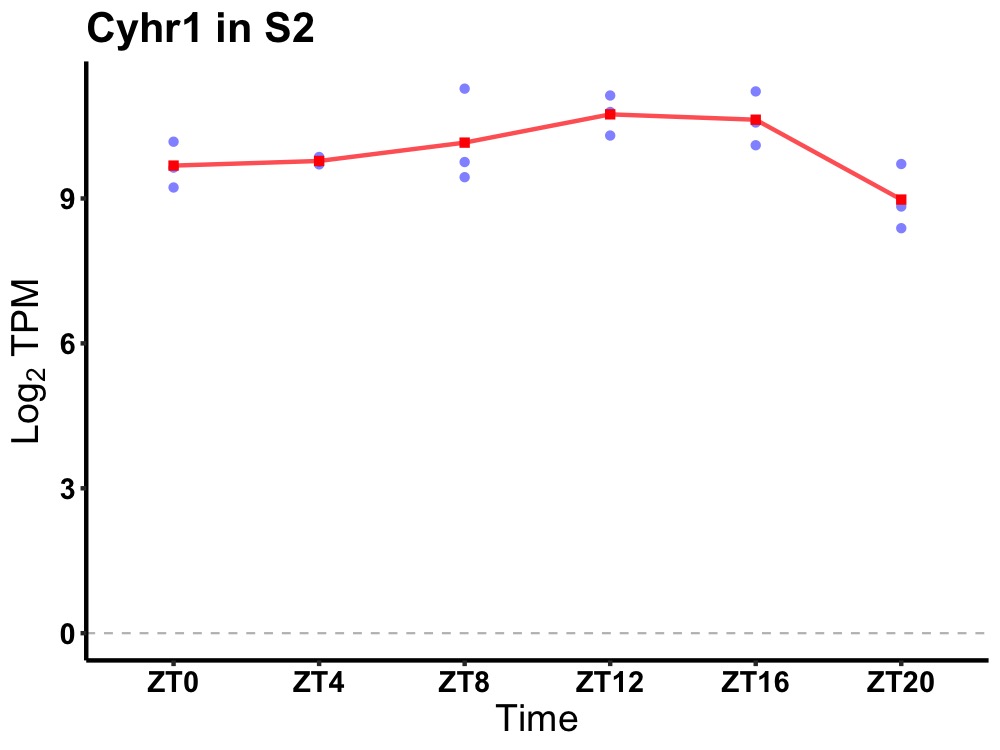 |
cysteine and histidine rich 1 | 0.049 | 20 | 12 | 0.59 |
| ENSMUSG00000100033 | Hikeshi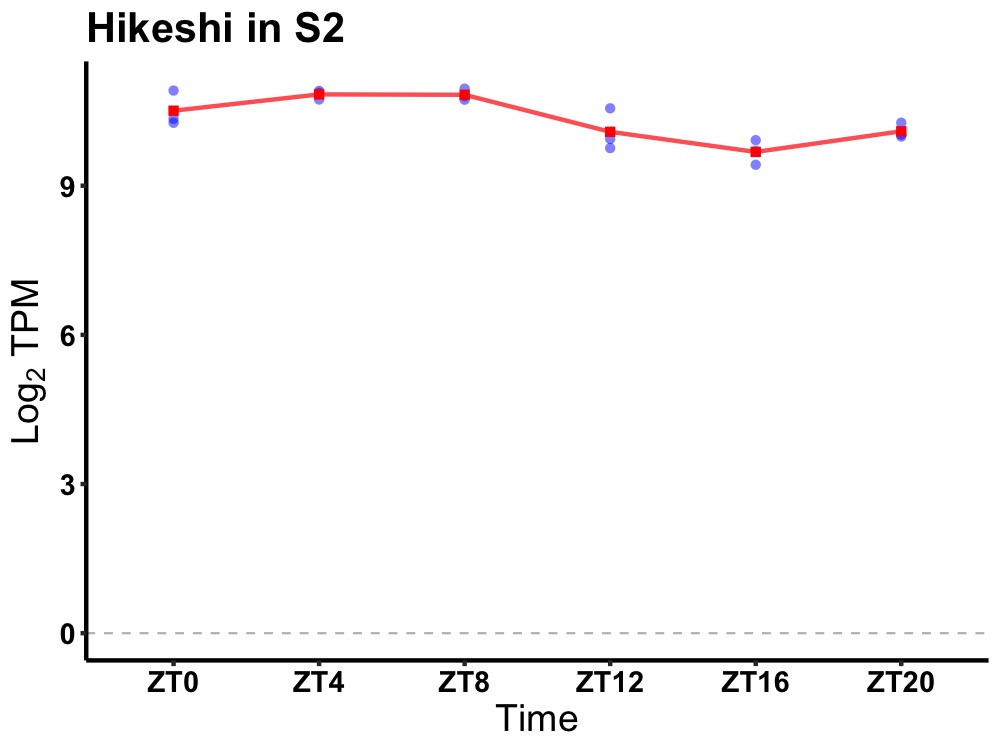 |
heat shock protein nuclear import factor | 0.001 | 24 | 6 | 0.59 |
| ENSMUSG00000056608 | Ethe1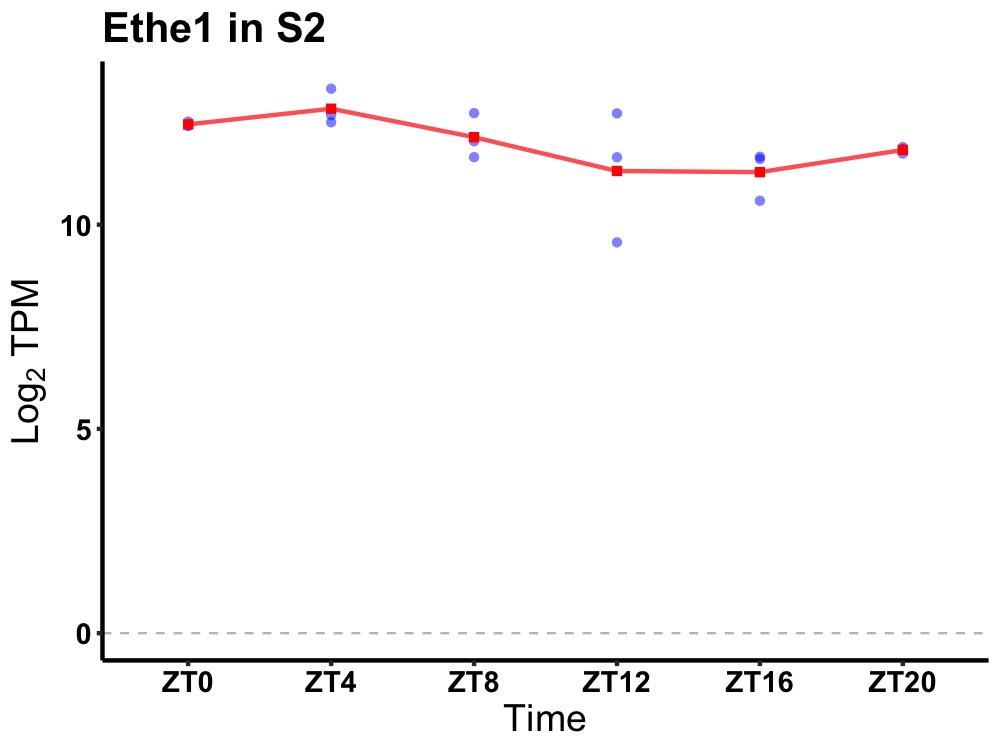 |
ethylmalonic encephalopathy 1 | 0.010 | 24 | 4 | 0.59 |
| ENSMUSG00000110331 | Nudc-ps1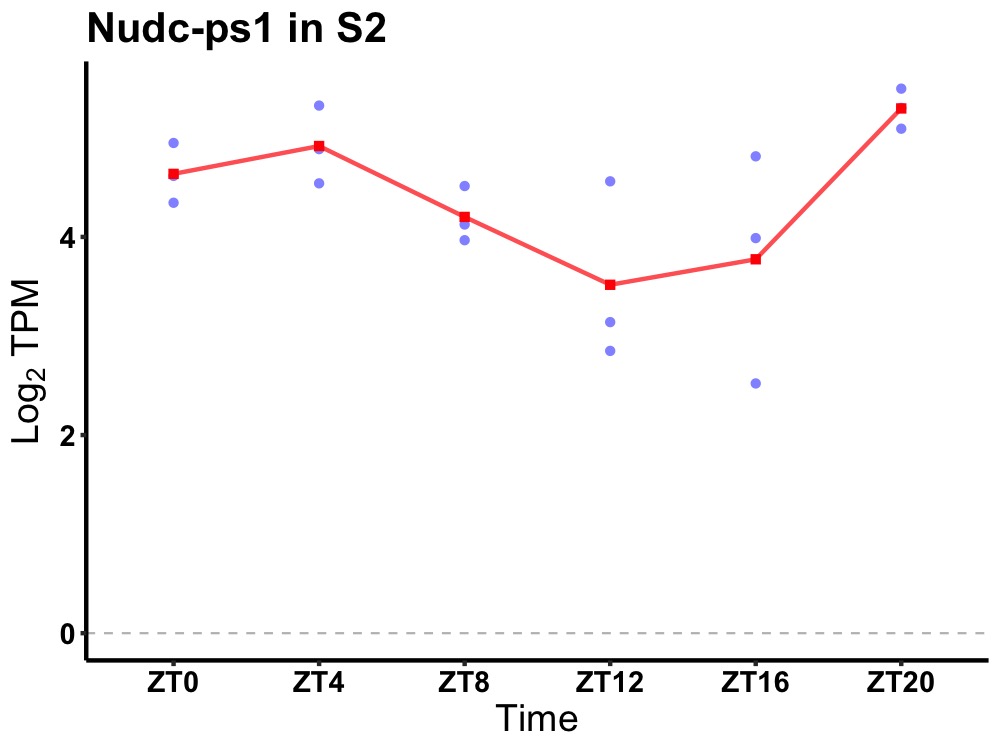 |
nuclear distribution gene C homolog (Aspergillus), pseudogene 1 | 0.019 | 20 | 2 | 0.59 |
| ENSMUSG00000024431 | Blvrb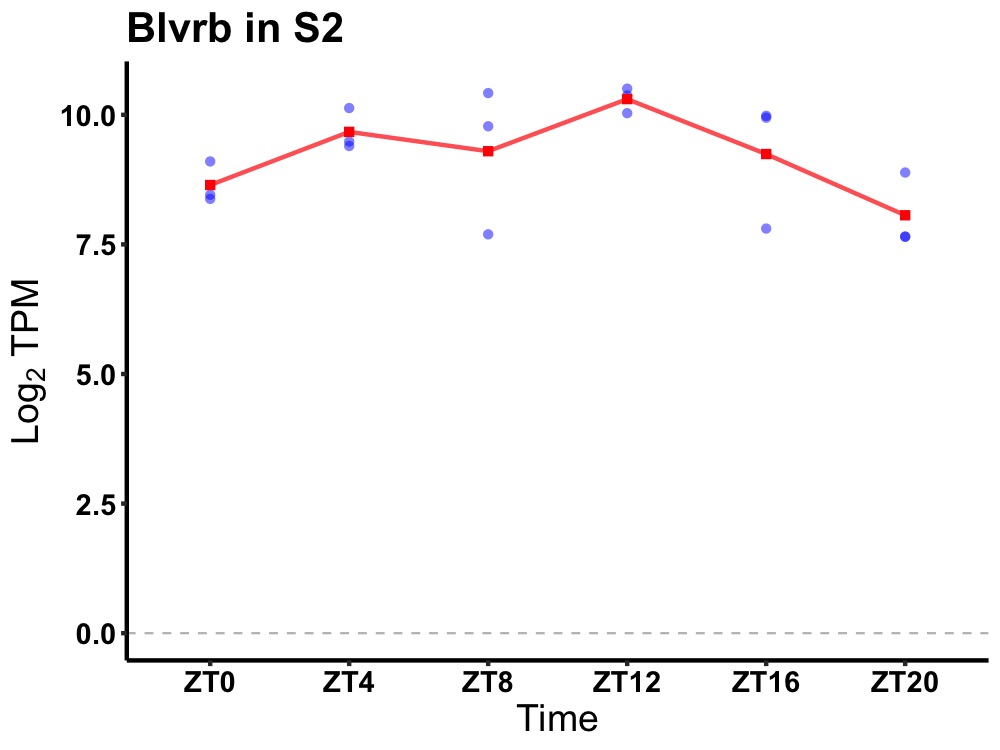 |
biliverdin reductase B (flavin reductase (NADPH)) | 0.027 | 20 | 12 | 0.58 |
| ENSMUSG00000082100 | Glns-ps1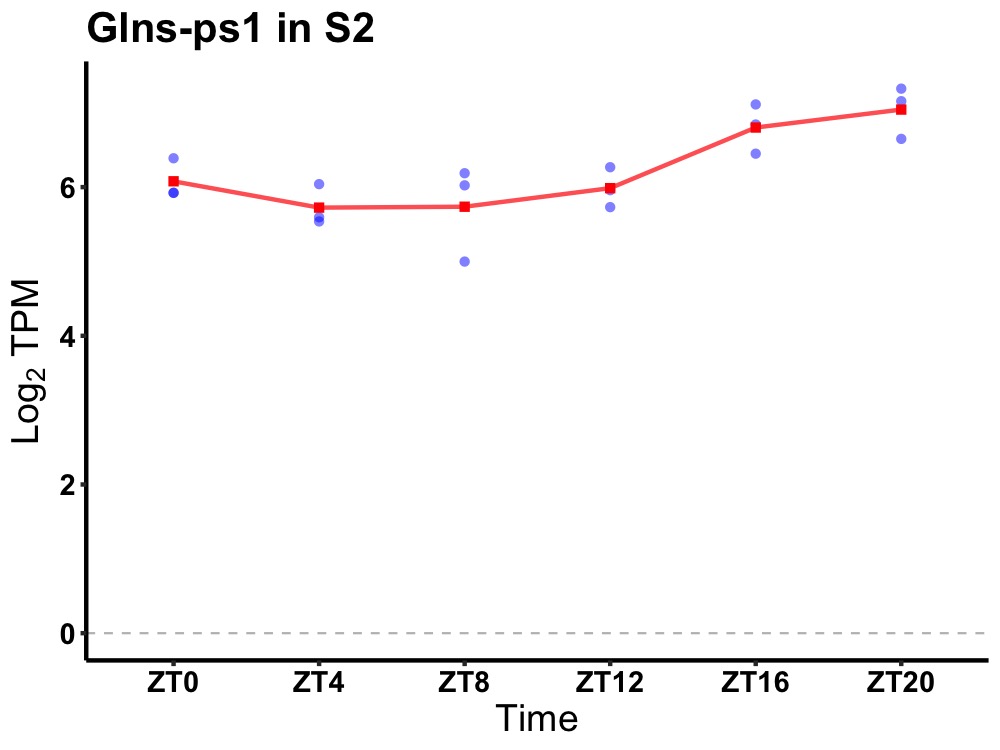 |
glutamine synthetase pseudogene 1 | 0.007 | 24 | 20 | 0.58 |
| ENSMUSG00000112341 | Rhov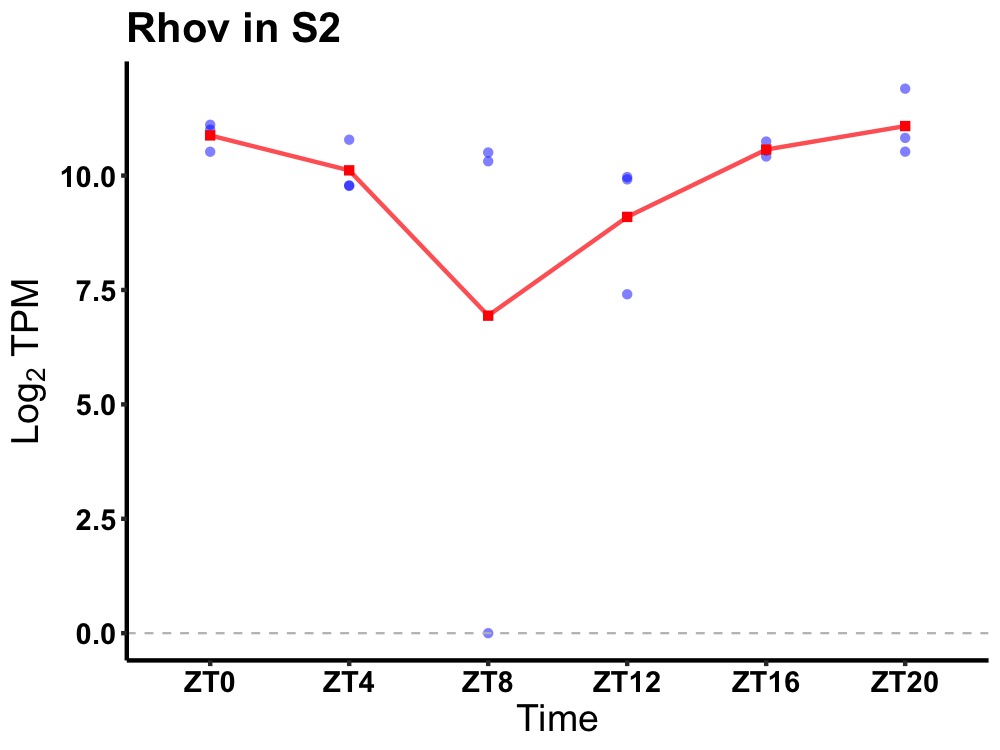 |
ras homolog family member V | 0.036 | 24 | 22 | 0.58 |
| ENSMUSG00000029610 | Gstt2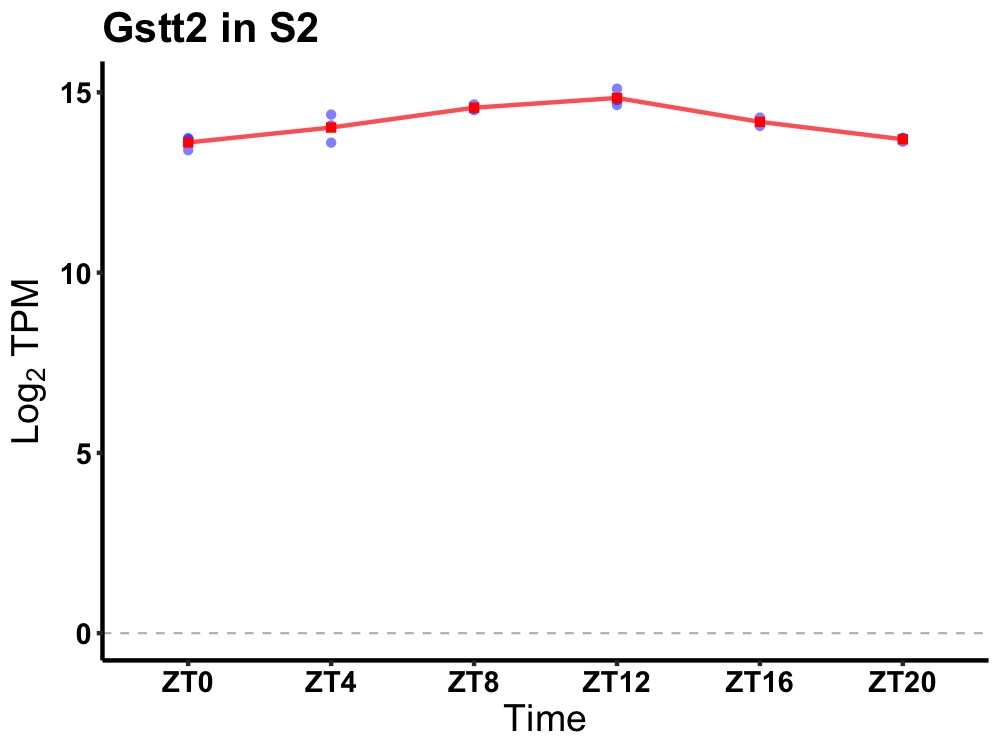 |
glutathione S-transferase, theta 2 | 0.000 | 24 | 12 | 0.58 |
| ENSMUSG00000090319 | Gm4462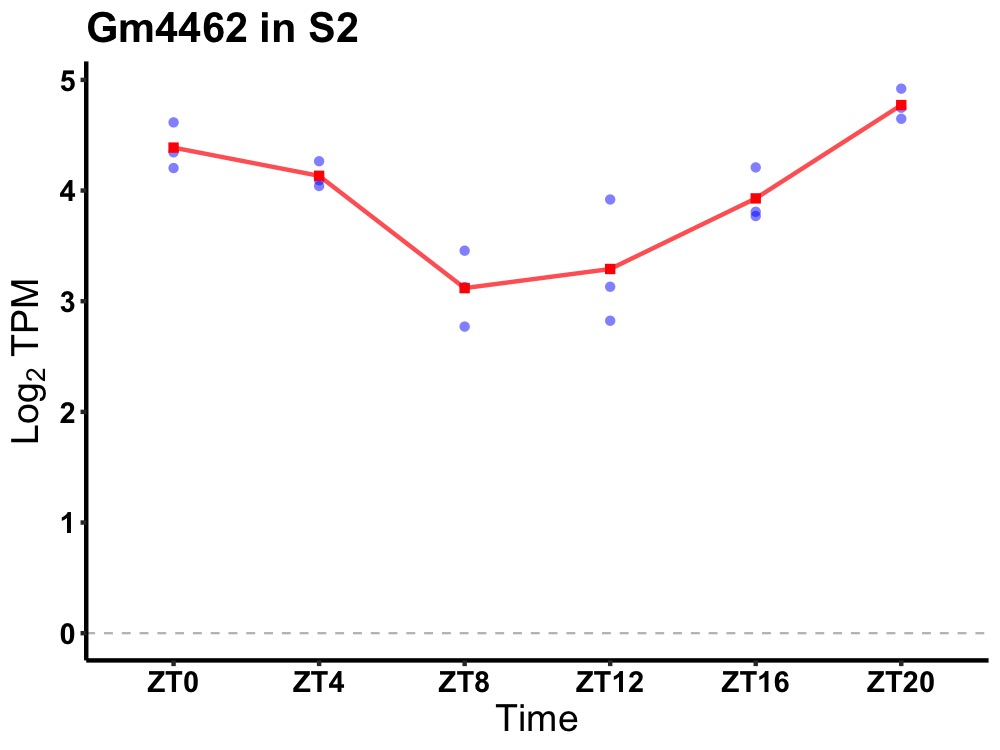 |
predicted gene 4462 | 0.000 | 20 | 2 | 0.57 |
| ENSMUSG00000057036 | Spsb4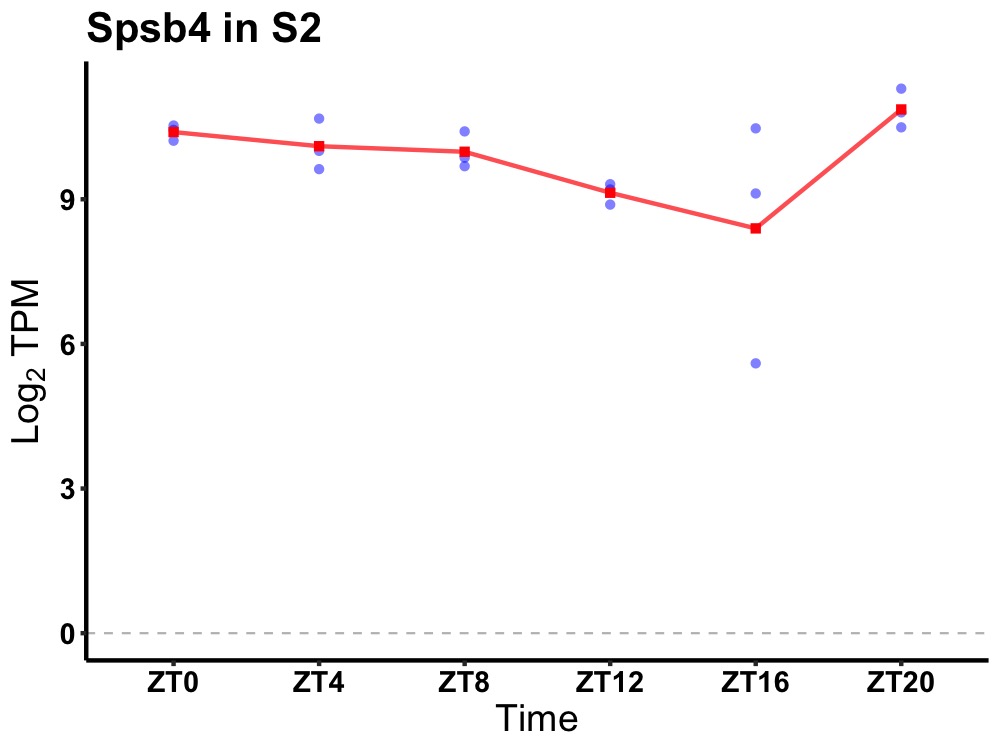 |
splA/ryanodine receptor domain and SOCS box containing 4 | 0.014 | 20 | 2 | 0.57 |
| ENSMUSG00000094886 | Gm10784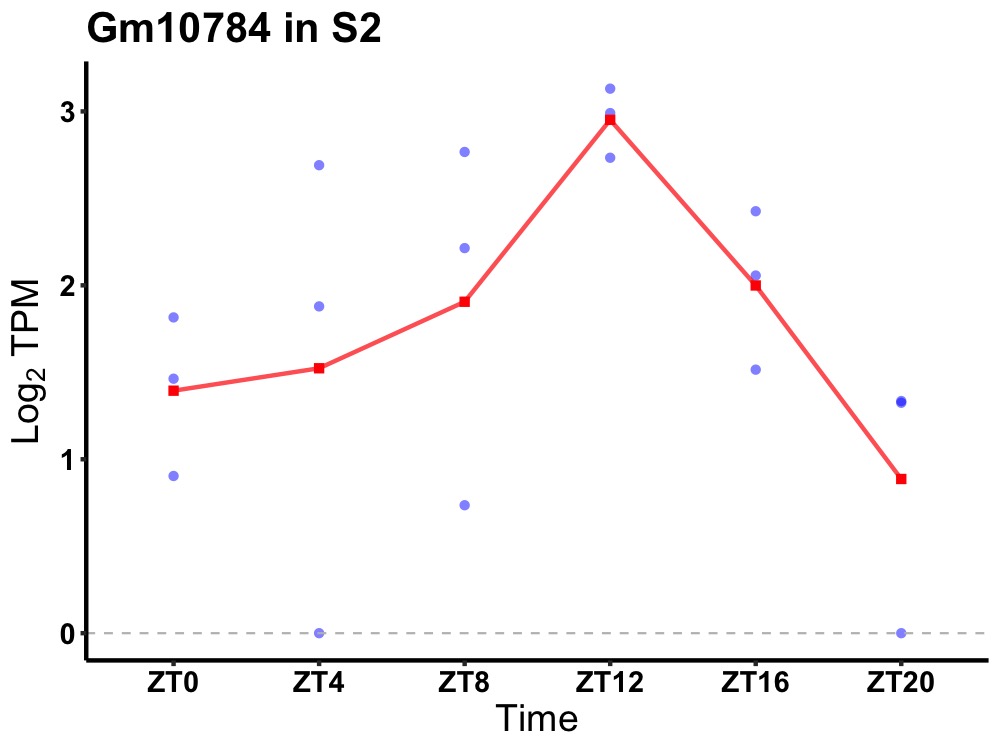 |
none | 0.016 | 20 | 12 | 0.57 |
| ENSMUSG00000007815 | Gm6210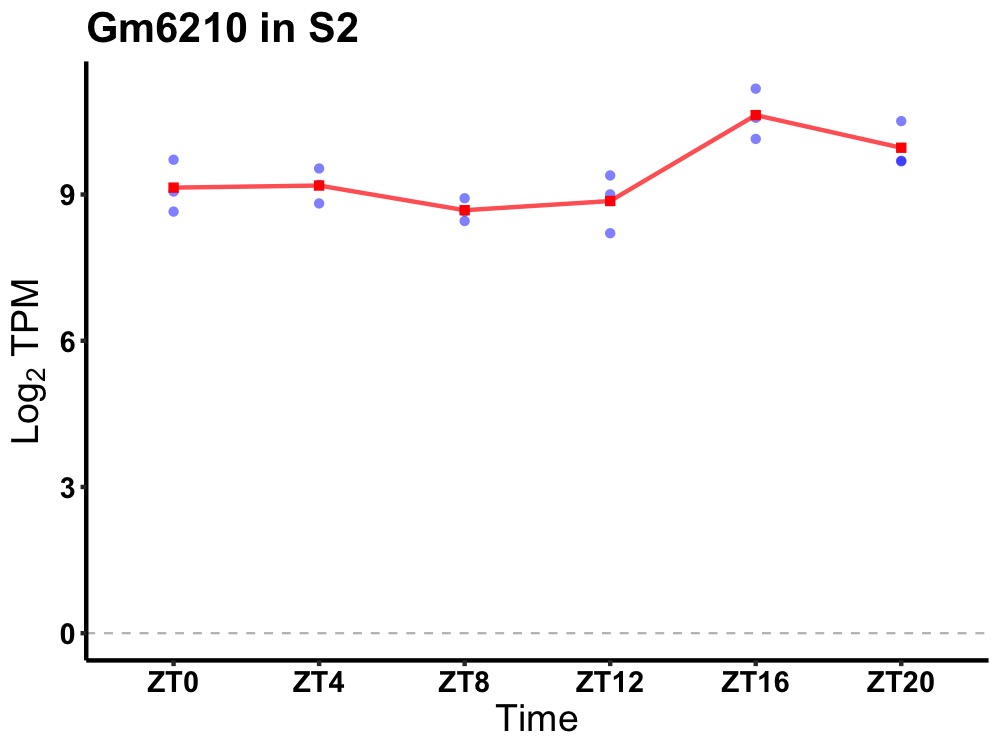 |
predicted gene 6210 | 0.036 | 20 | 18 | 0.56 |
| ENSMUSG00000085627 | Gm11222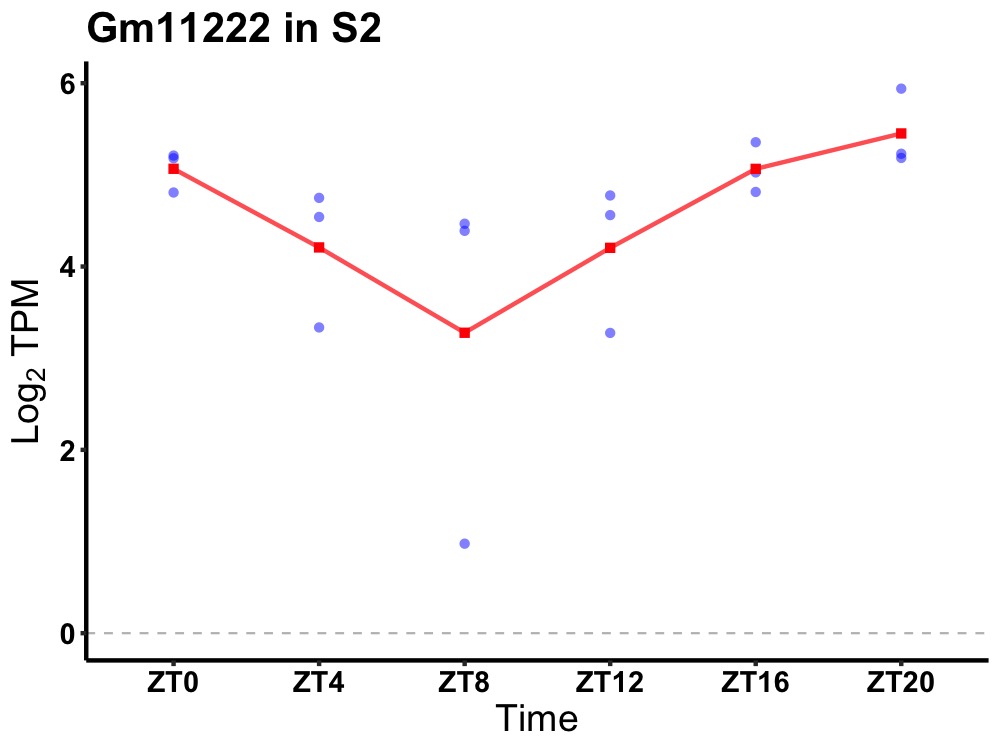 |
predicted gene 11222 | 0.000 | 24 | 20 | 0.56 |
| ENSMUSG00000115380 | Gm46453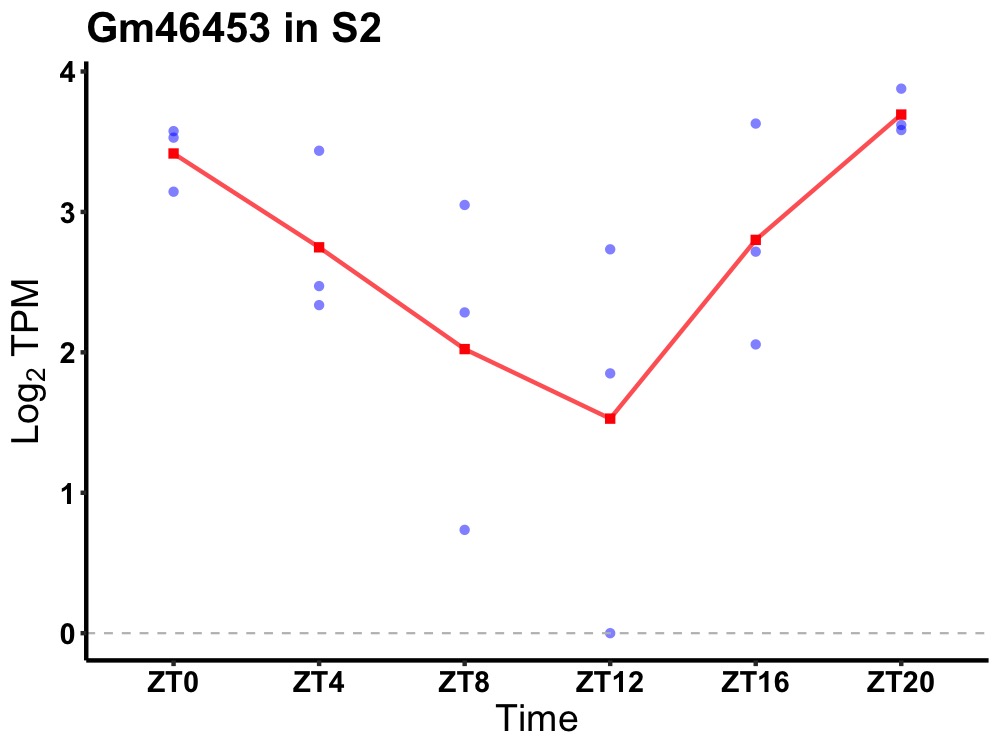 |
predicted gene, 46453 | 0.007 | 20 | 2 | 0.56 |
| ENSMUSG00000033624 | Ints14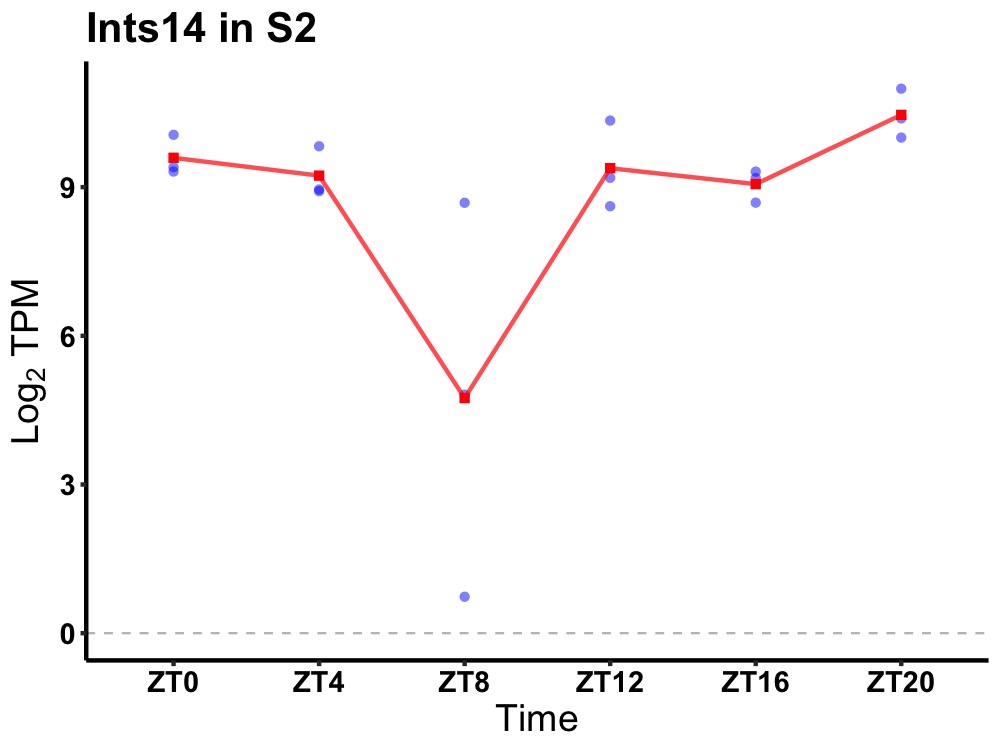 |
integrator complex subunit 14 | 0.007 | 24 | 22 | 0.56 |
| ENSMUSG00000046440 | Angptl4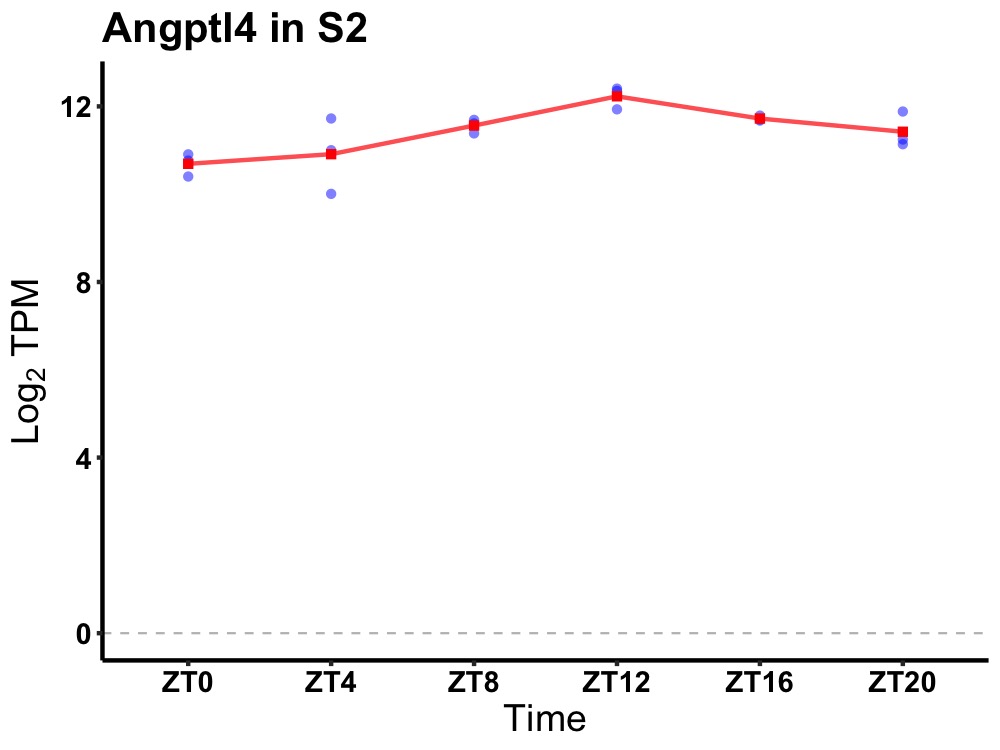 |
angiopoietin-like 4 | 0.001 | 24 | 14 | 0.55 |
| ENSMUSG00000081957 | Ak3l2-ps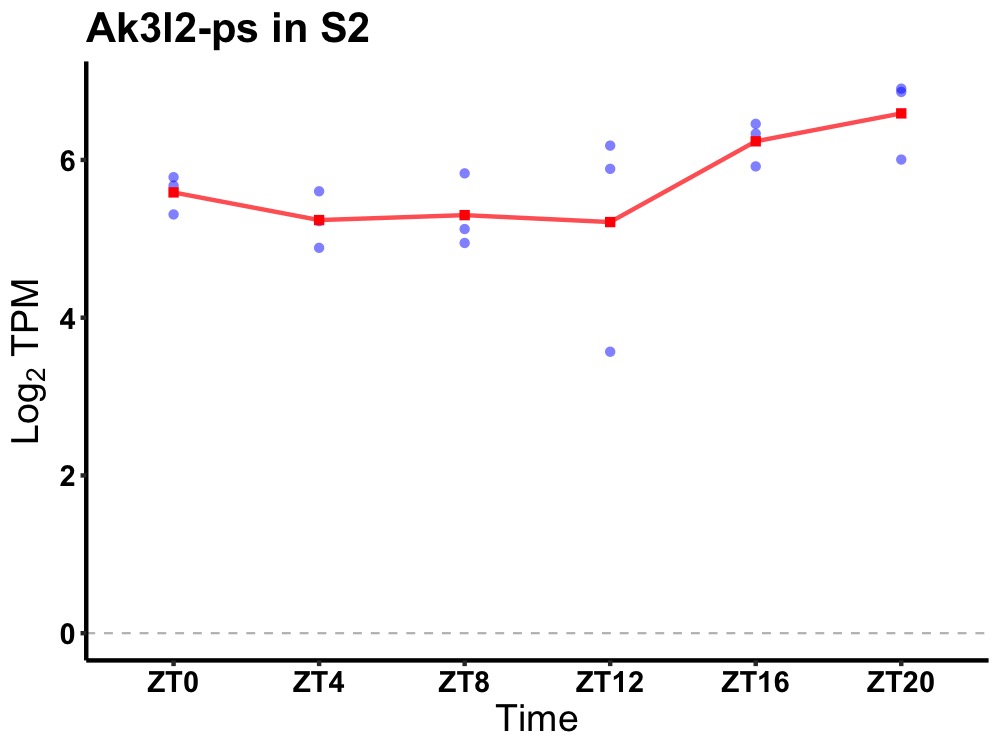 |
adenylate kinase 3-like 2, pseudogene | 0.014 | 24 | 20 | 0.55 |
| ENSMUSG00000080982 | Gm13549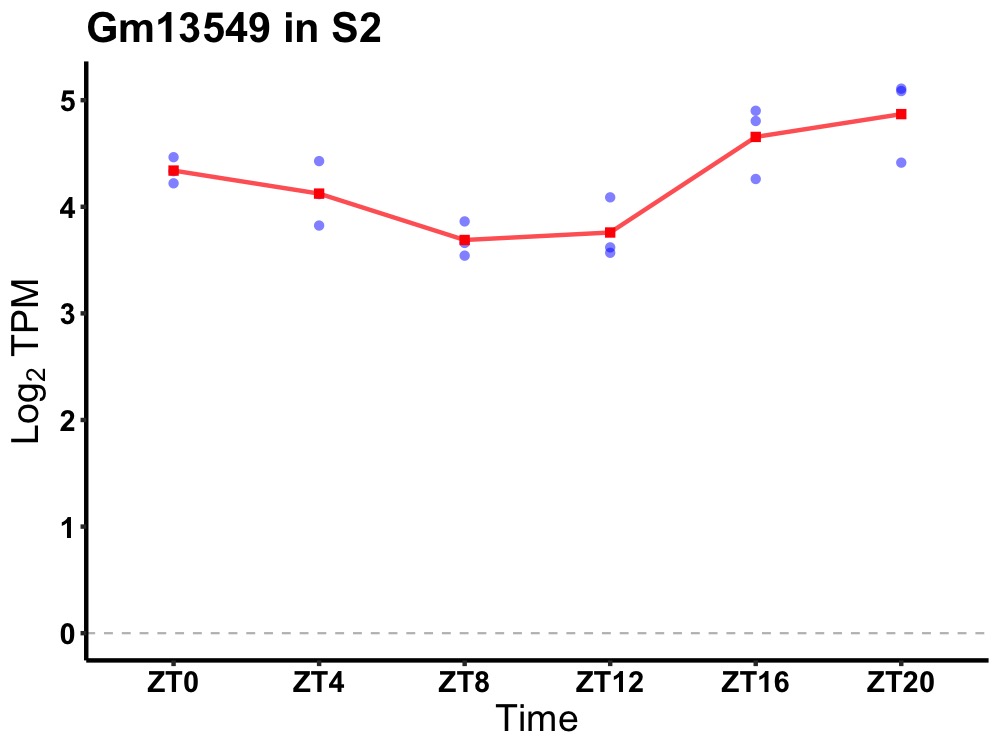 |
predicted gene 13549 | 0.002 | 24 | 22 | 0.55 |
| ENSMUSG00000098012 | Snapin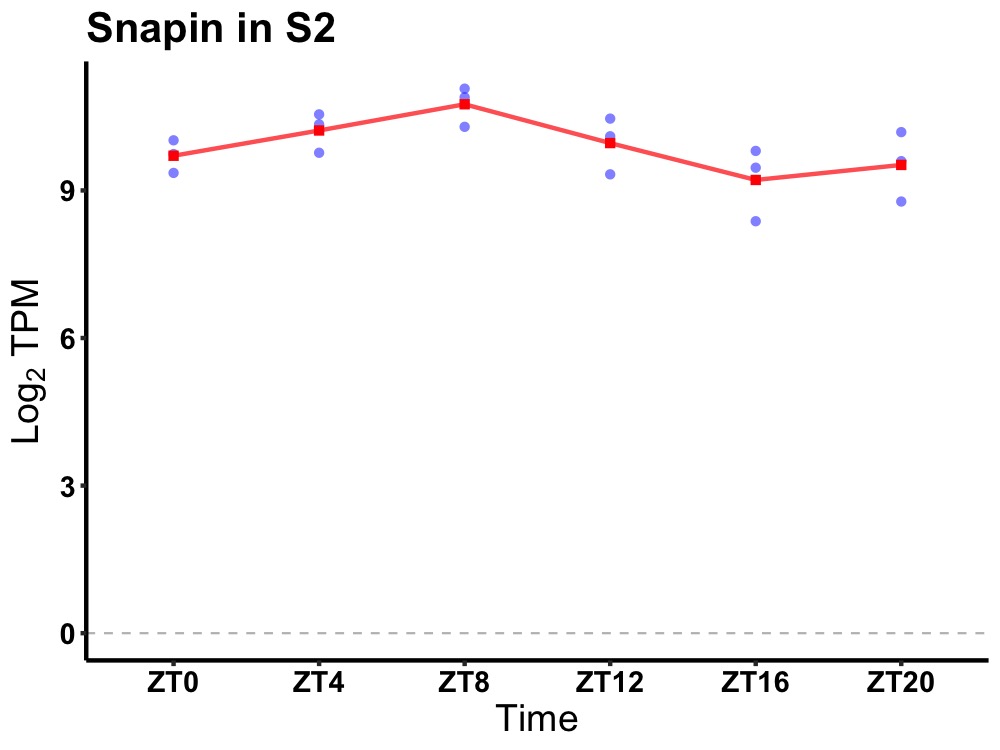 |
SNAP-associated protein | 0.036 | 20 | 8 | 0.54 |
| ENSMUSG00000081302 | Gm12020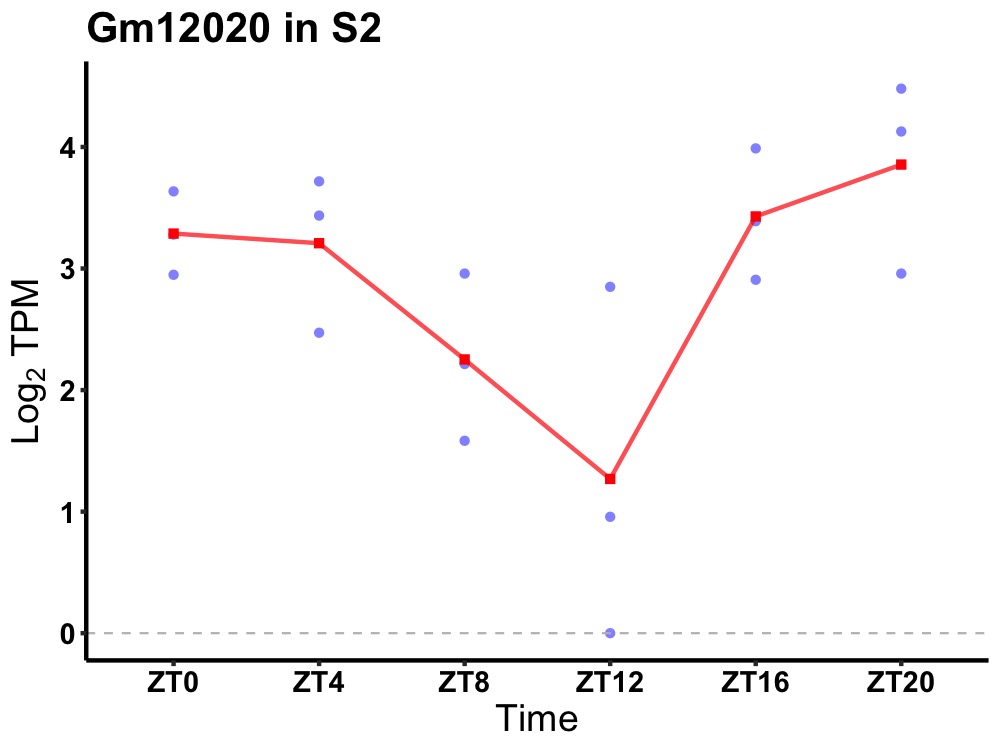 |
predicted gene 12020 | 0.036 | 20 | 2 | 0.54 |
| ENSMUSG00000113113 | Kars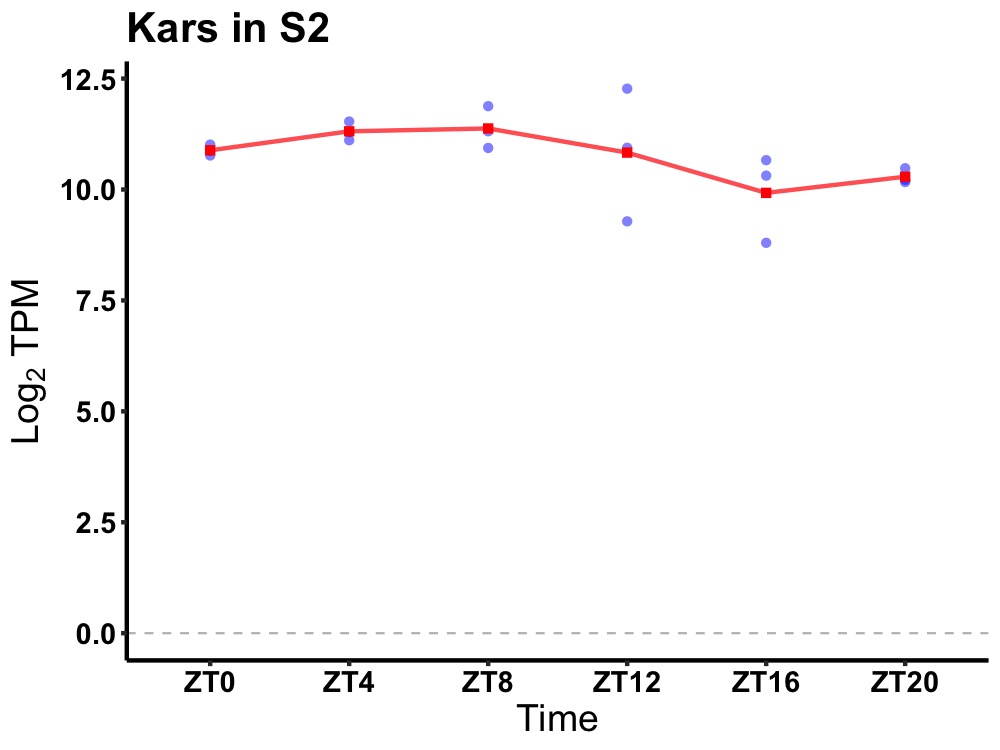 |
lysyl-tRNA synthetase | 0.014 | 24 | 8 | 0.54 |
| ENSMUSG00000090319 | Fcgr2b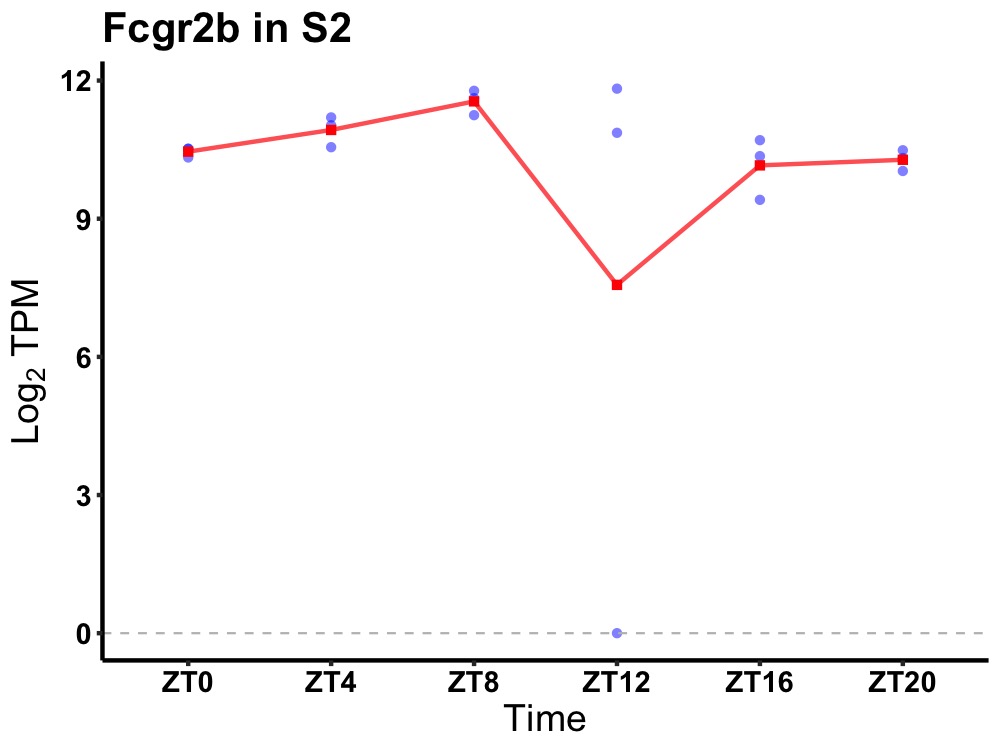 |
Fc receptor, IgG, low affinity IIb | 0.019 | 24 | 8 | 0.54 |
| ENSMUSG00000117515 | Actl6a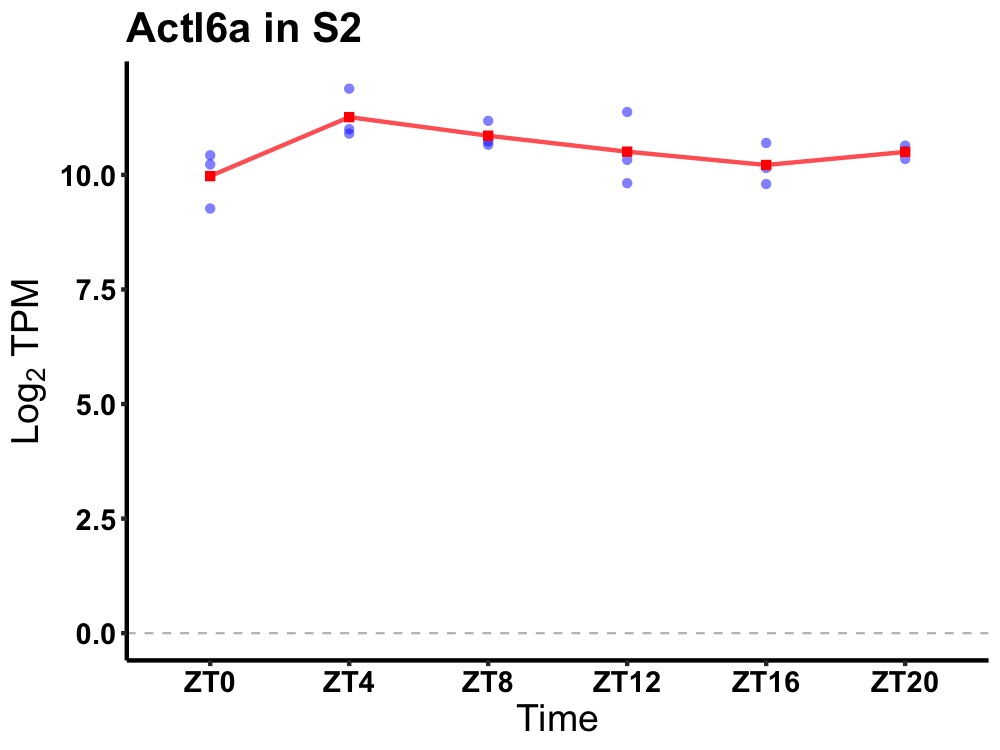 |
actin-like 6A | 0.049 | 20 | 6 | 0.54 |
| ENSMUSG00000063586 | Gm5513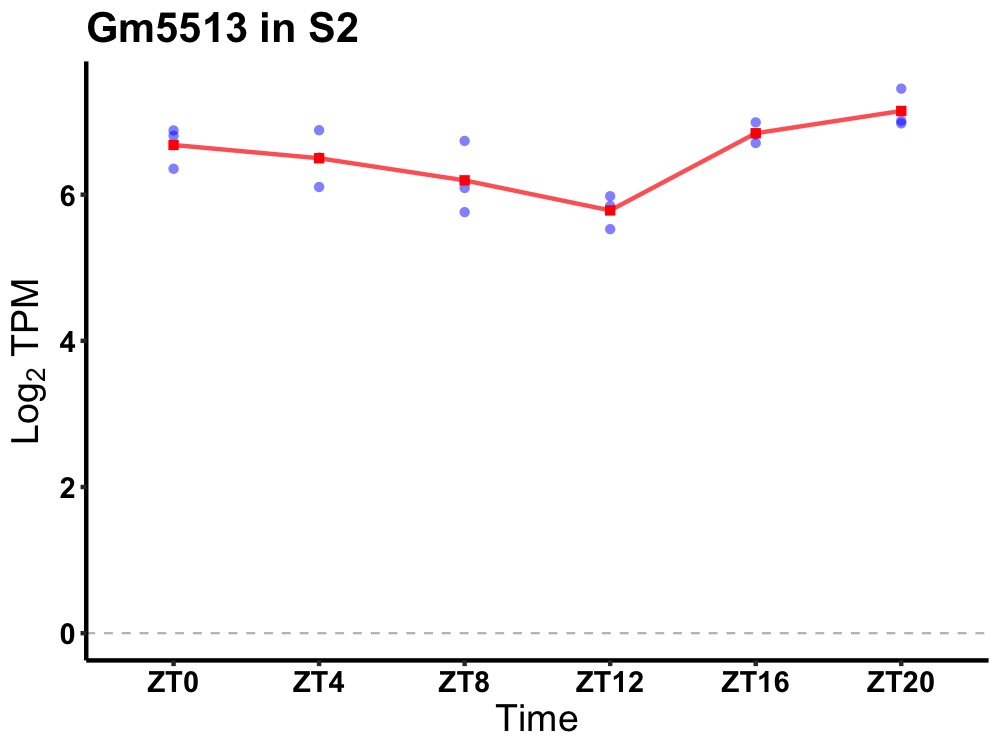 |
predicted pseudogene 5513 | 0.010 | 24 | 22 | 0.53 |
| ENSMUSG00000029535 | Snx24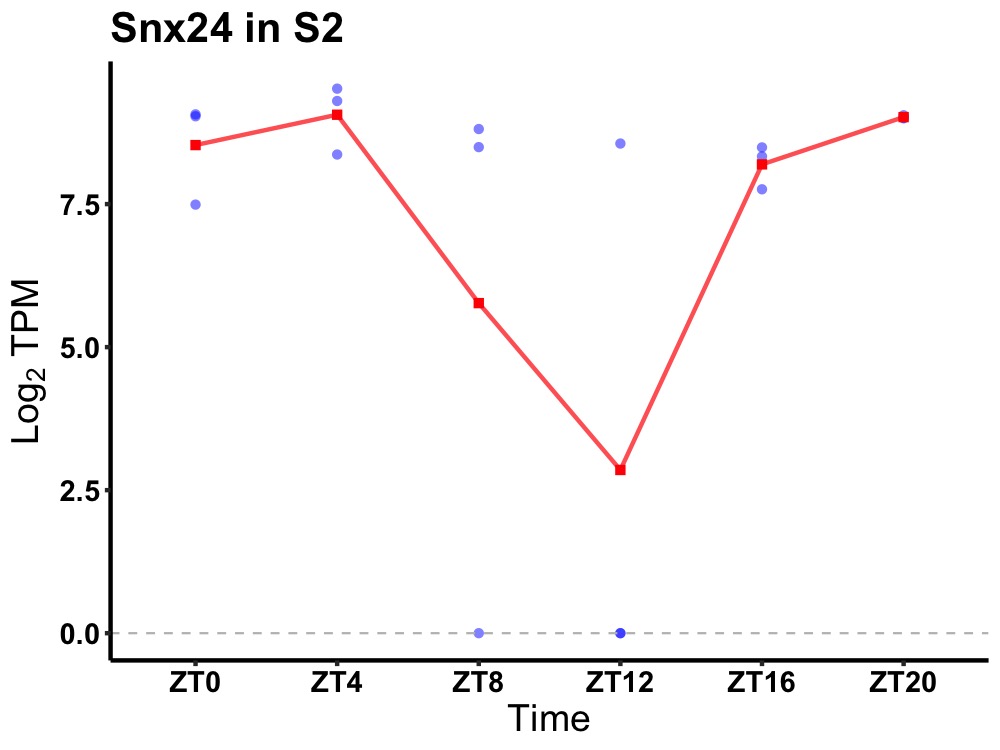 |
sorting nexin-24 | 0.049 | 20 | 4 | 0.53 |
| ENSMUSG00000039048 | Cpq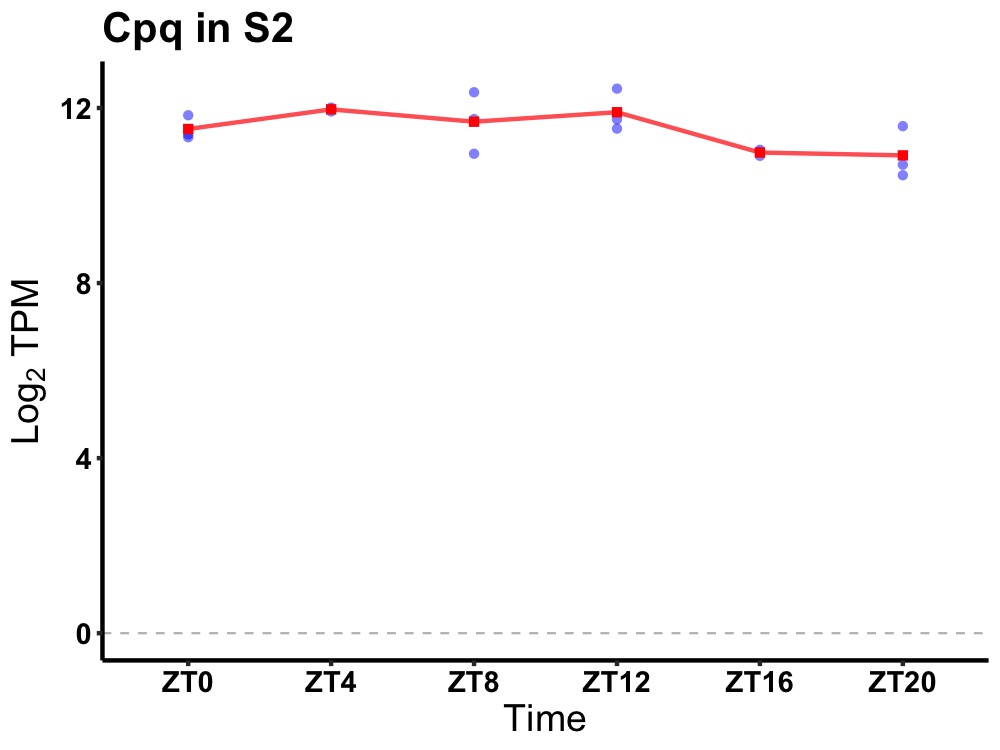 |
carboxypeptidase Q | 0.049 | 24 | 8 | 0.53 |
| ENSMUSG00000055762 | Chordc1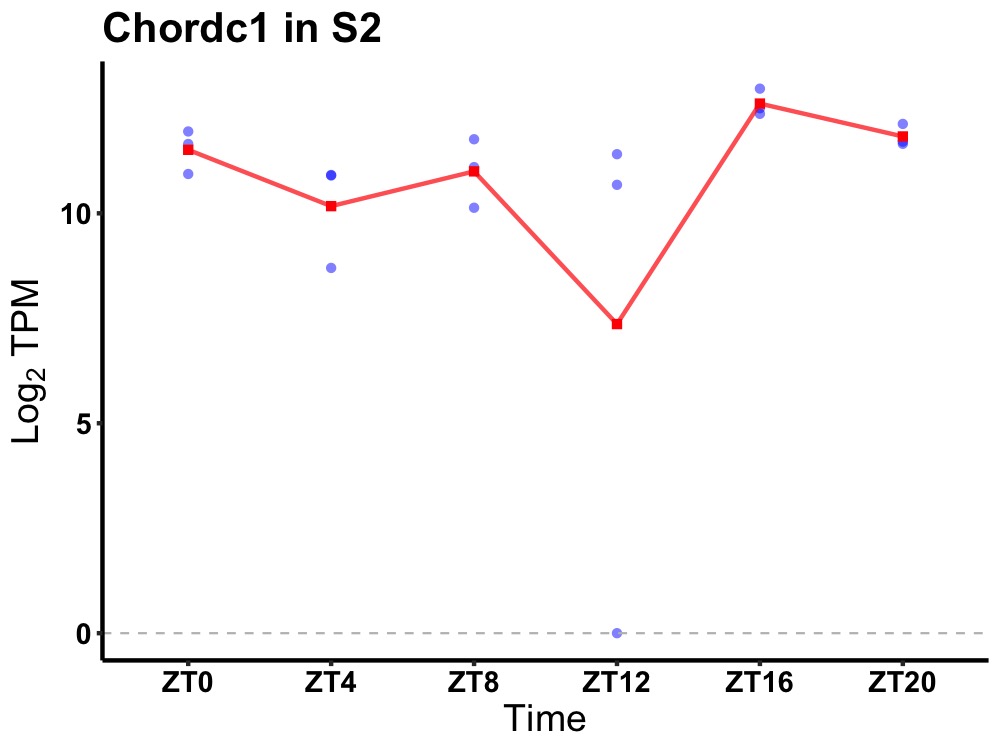 |
cysteine and histidine-rich domain (CHORD)-containing, zinc-binding protein 1 | 0.036 | 24 | 18 | 0.52 |
| ENSMUSG00000036932 | mt-Nd6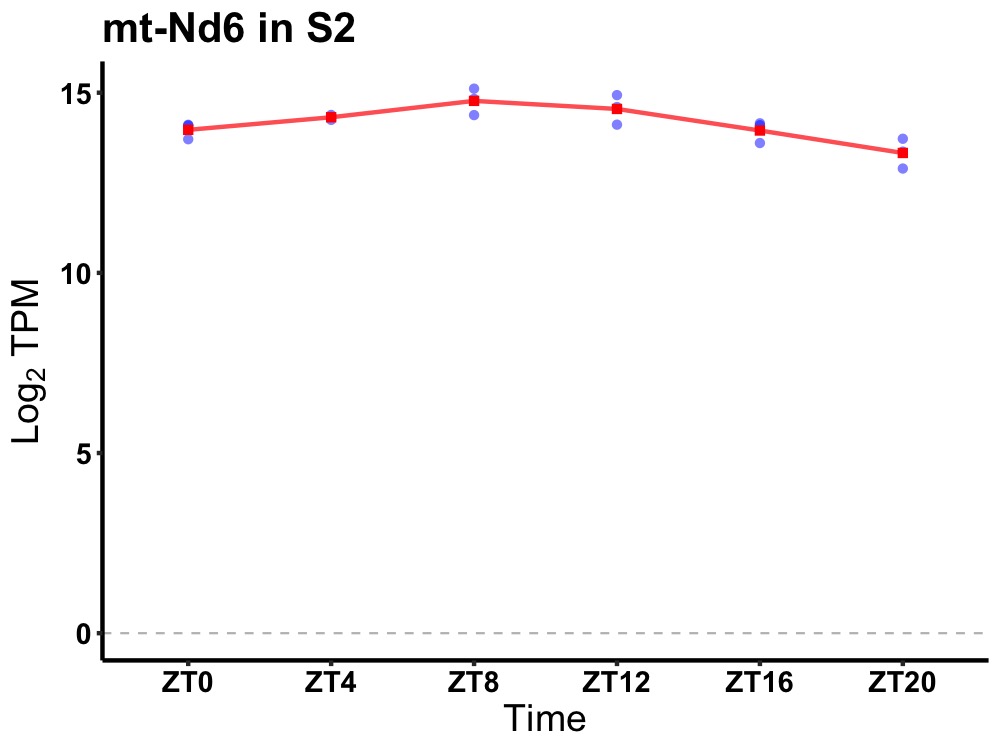 |
mitochondrially encoded NADH dehydrogenase 6 | 0.000 | 24 | 10 | 0.52 |
| ENSMUSG00000063543 | Mrpl49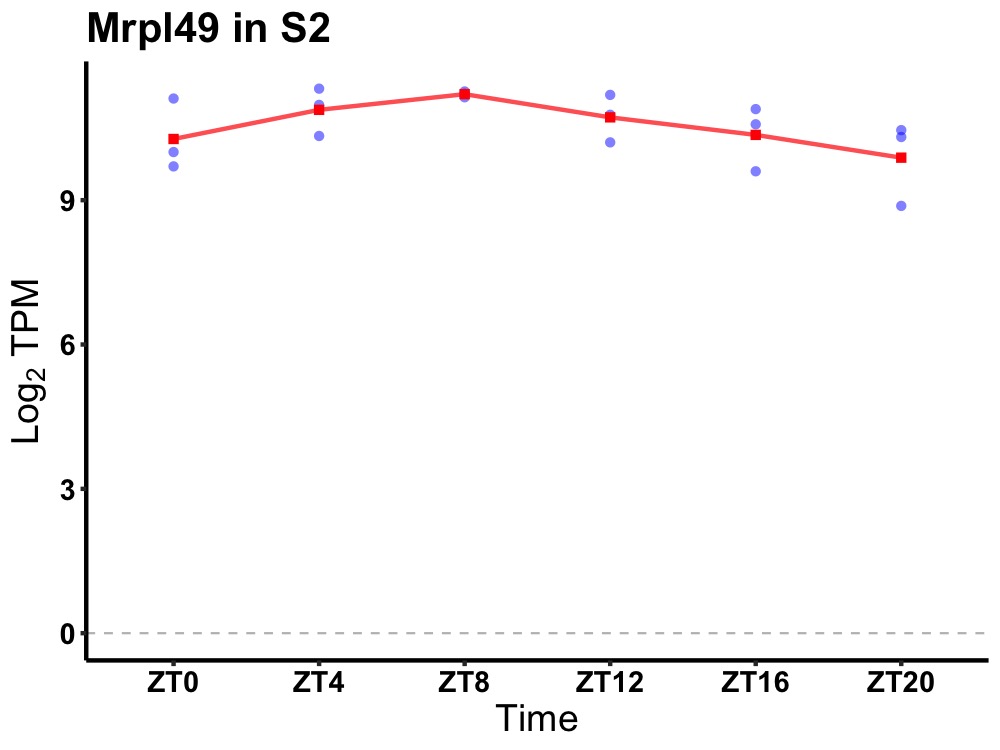 |
mitochondrial ribosomal protein L49 | 0.049 | 24 | 8 | 0.52 |
| ENSMUSG00000113617 | Gm8655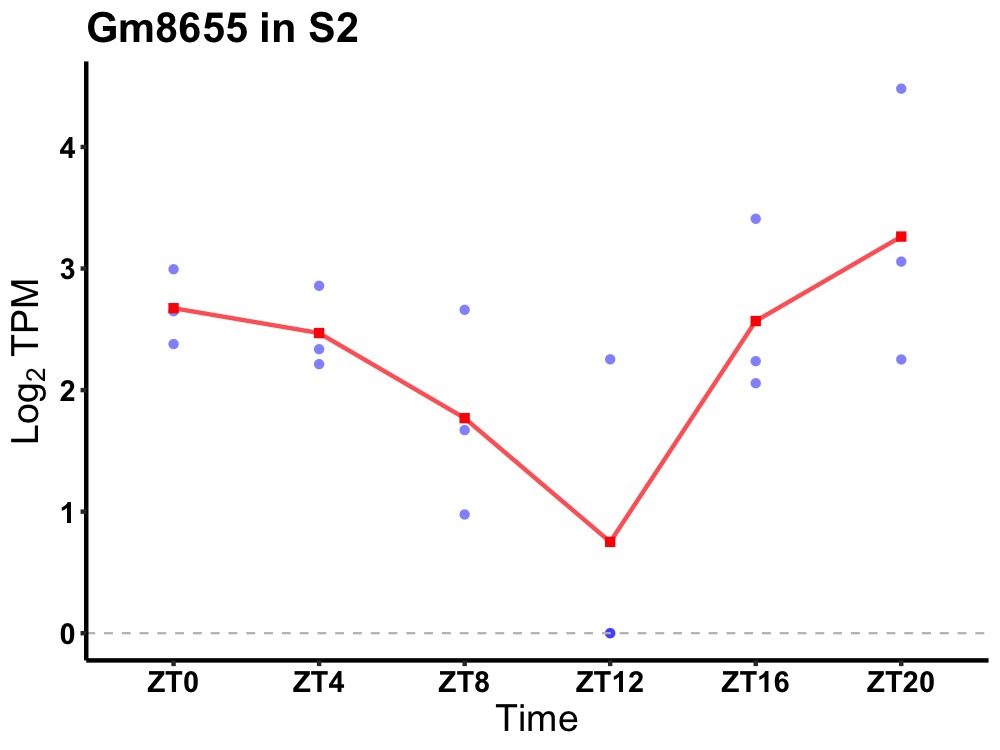 |
none | 0.049 | 20 | 2 | 0.52 |
| ENSMUSG00000117283 | Gm4471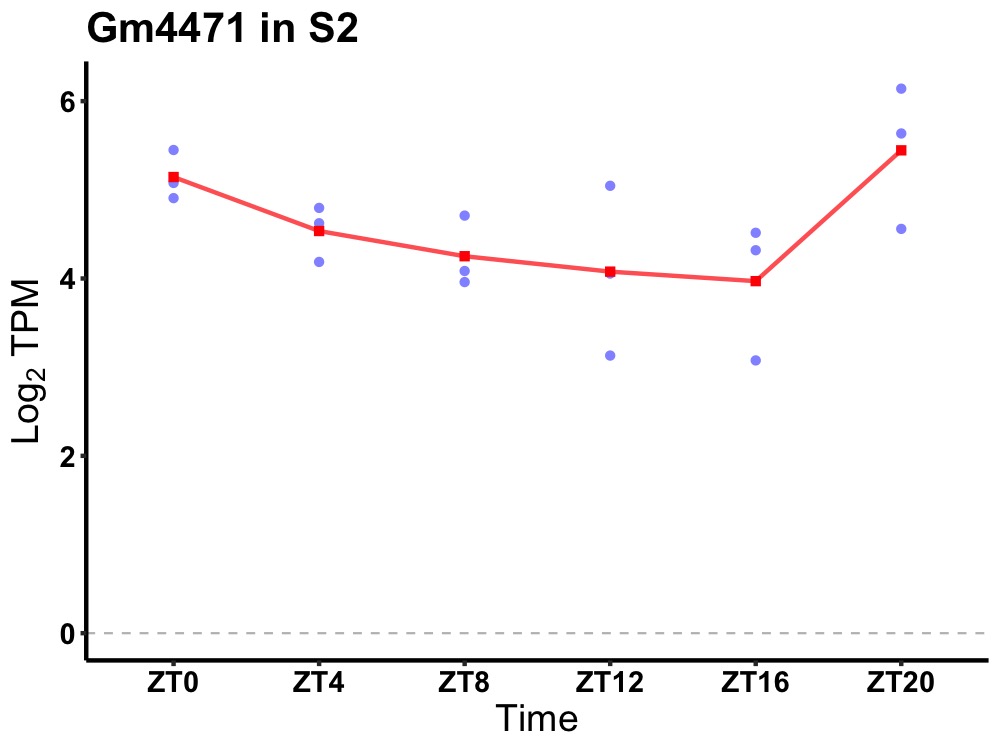 |
novel pseudogene | 0.049 | 20 | 2 | 0.52 |
| ENSMUSG00000055334 | Fam92a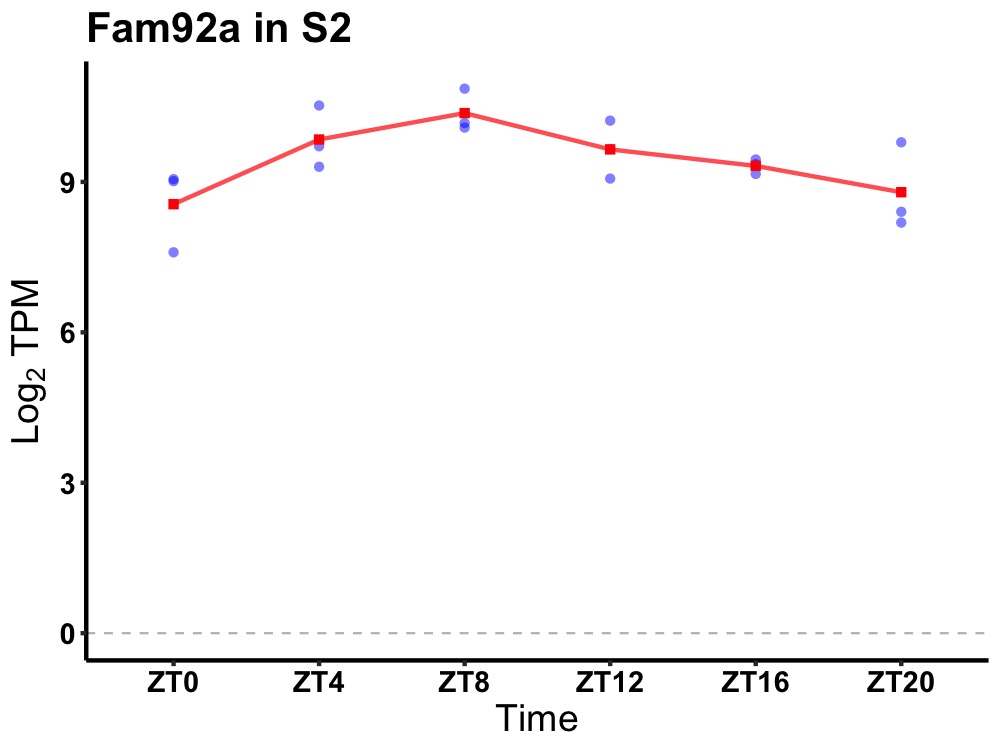 |
family with sequence similarity 92, member A | 0.019 | 20 | 10 | 0.52 |
| ENSMUSG00000038527 | Usp2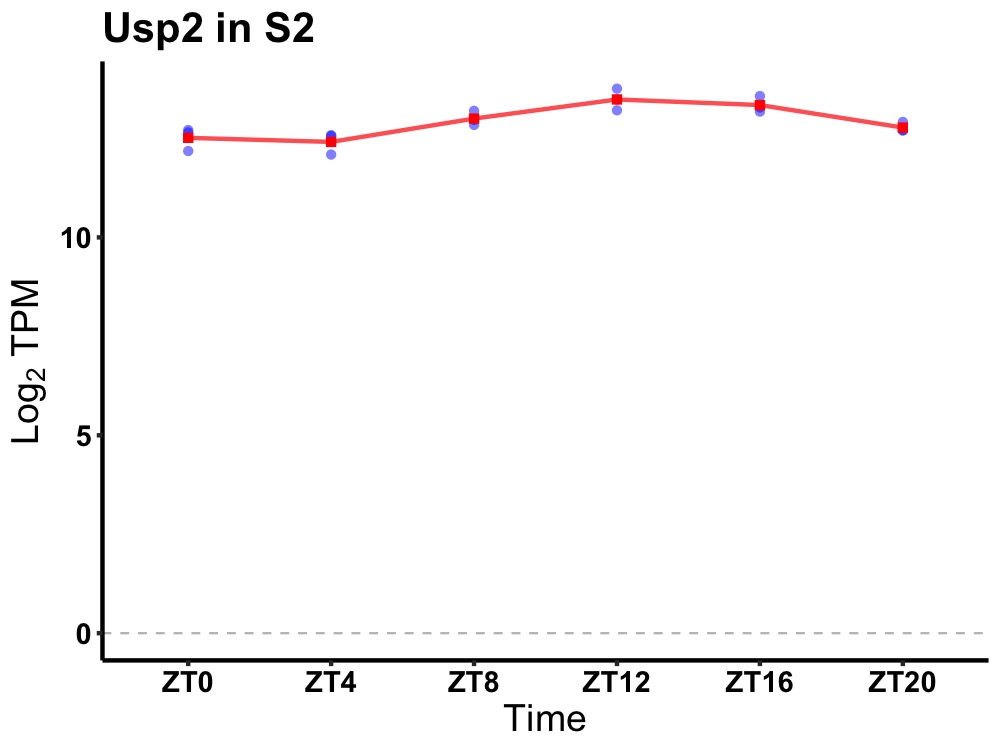 |
ubiquitin specific peptidase 2 | 0.000 | 20 | 14 | 0.51 |
| ENSMUSG00000104664 | Gm35570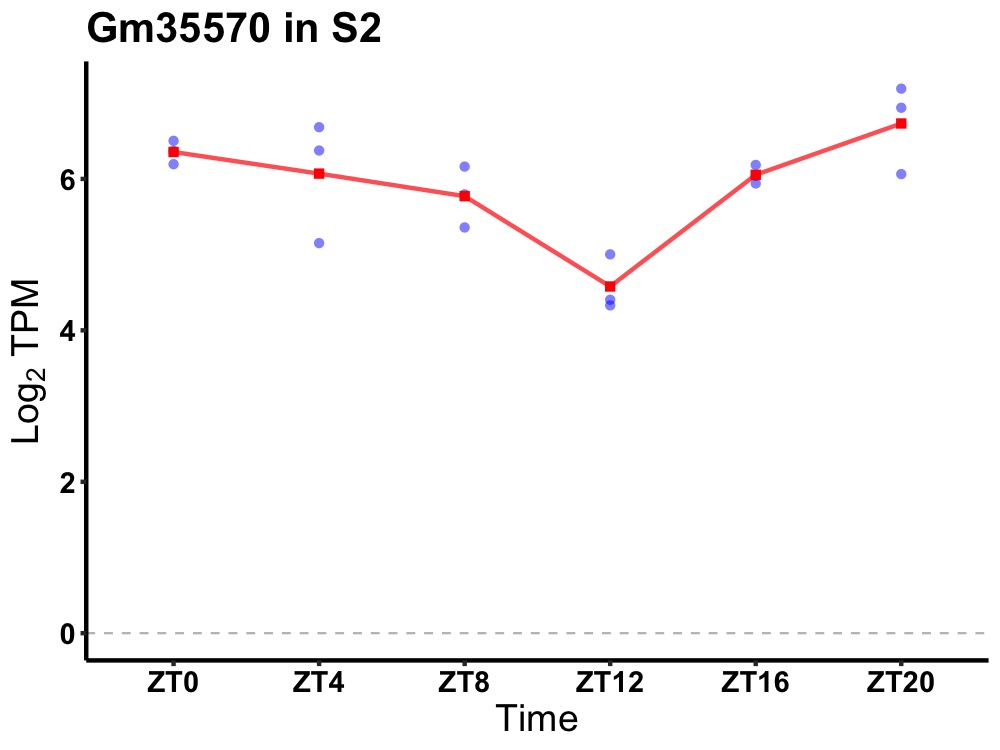 |
predicted gene, 35570 | 0.002 | 20 | 2 | 0.51 |
| ENSMUSG00000118345 | Pck1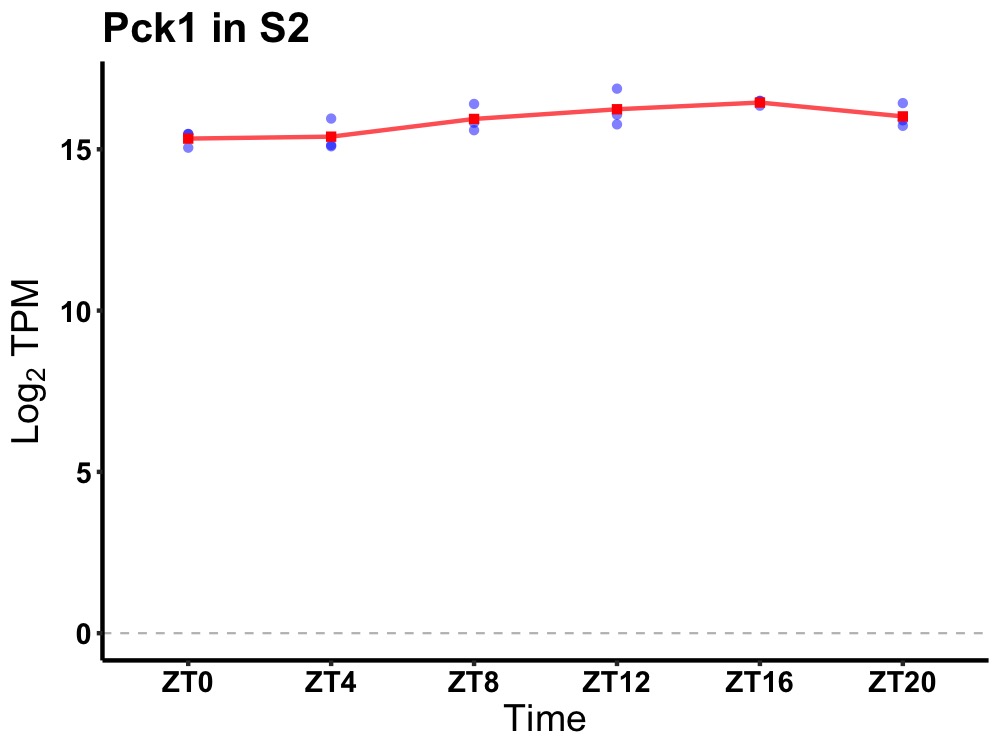 |
phosphoenolpyruvate carboxykinase 1, cytosolic | 0.010 | 24 | 16 | 0.51 |
| ENSMUSG00000019362 | Pdzd3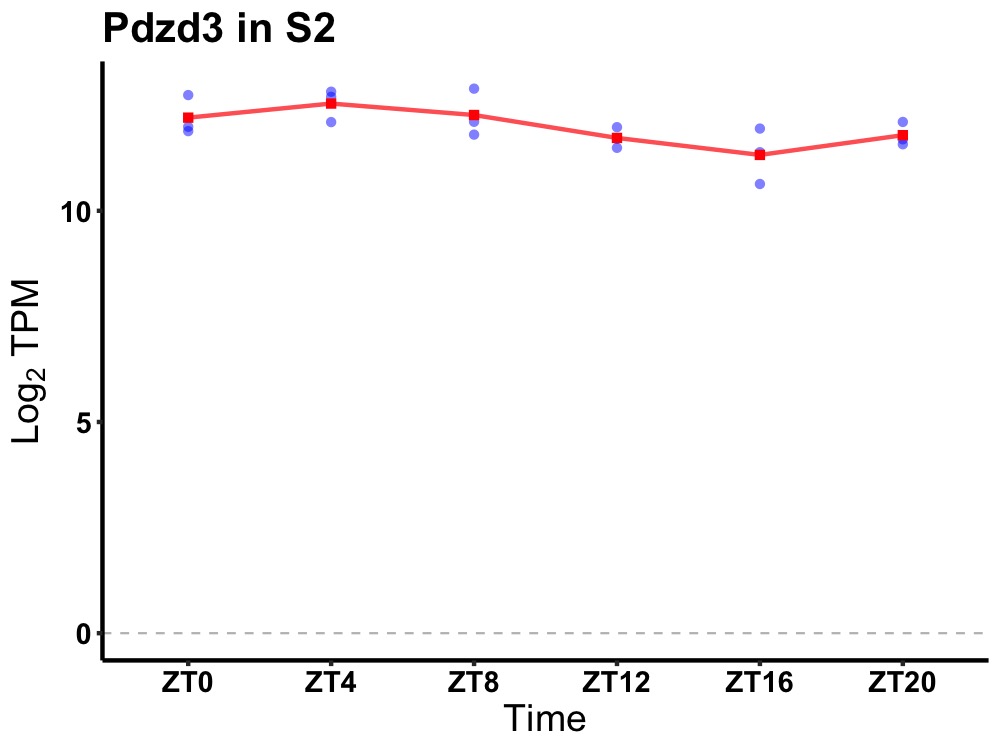 |
PDZ domain containing 3 | 0.027 | 24 | 6 | 0.50 |
| ENSMUSG00000003420 | St8sia1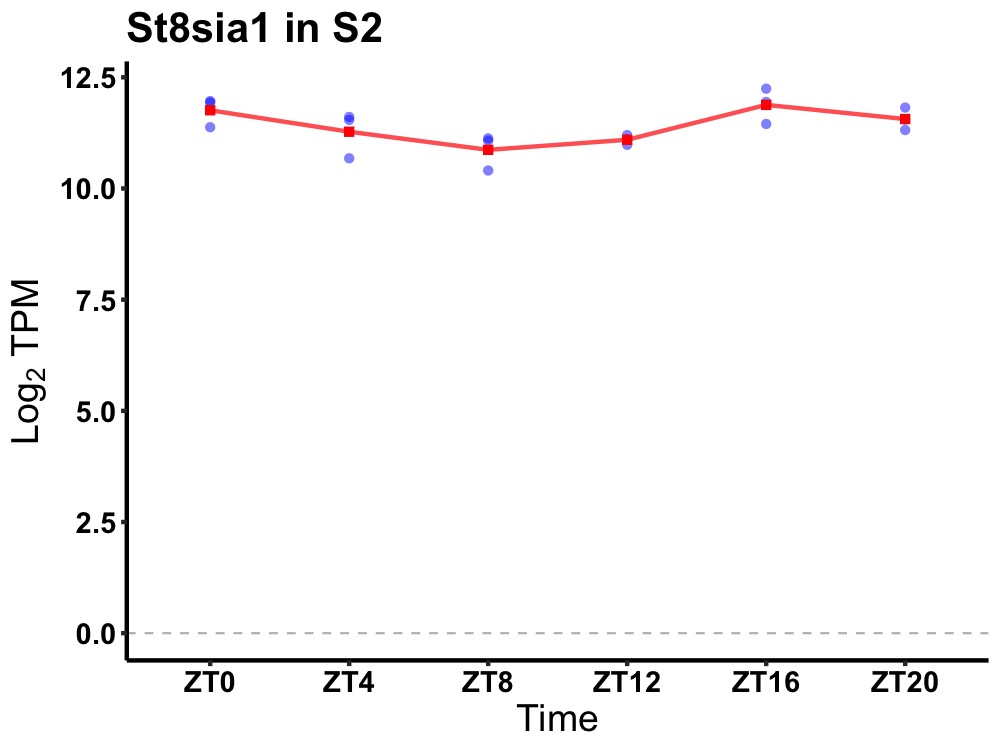 |
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 1 | 0.014 | 20 | 18 | 0.50 |
| ENSMUSG00000024158 | Gm5617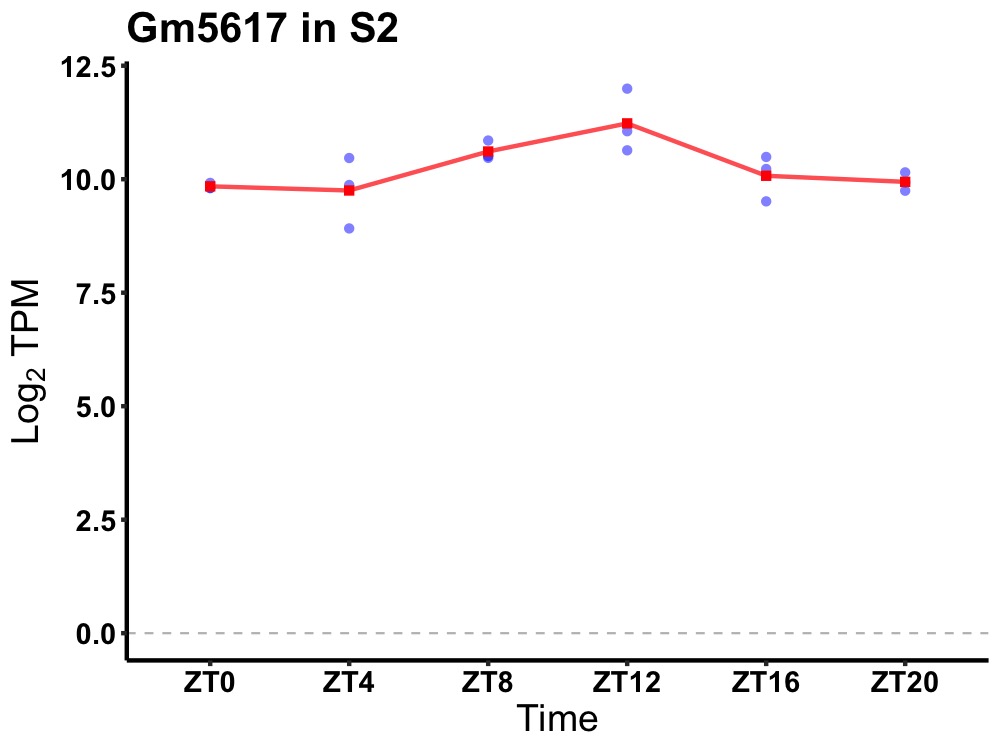 |
predicted gene 5617 | 0.007 | 24 | 12 | 0.50 |
| ENSMUSG00000032803 | Hnf1b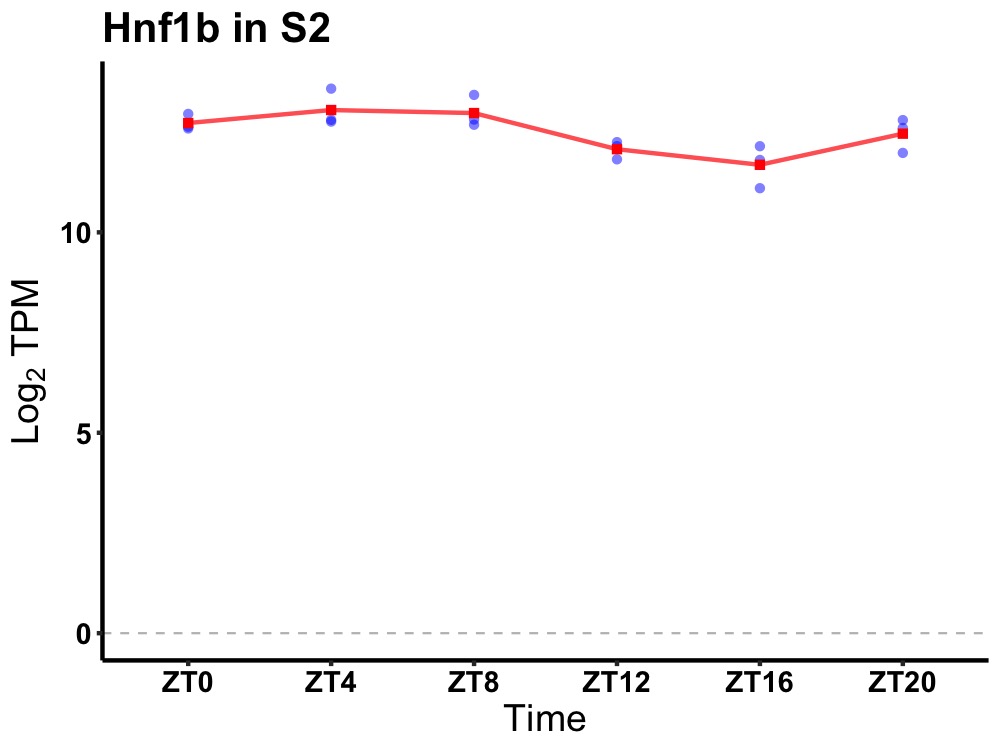 |
HNF1 homeobox B | 0.002 | 20 | 6 | 0.49 |
| ENSMUSG00000020570 | Emc9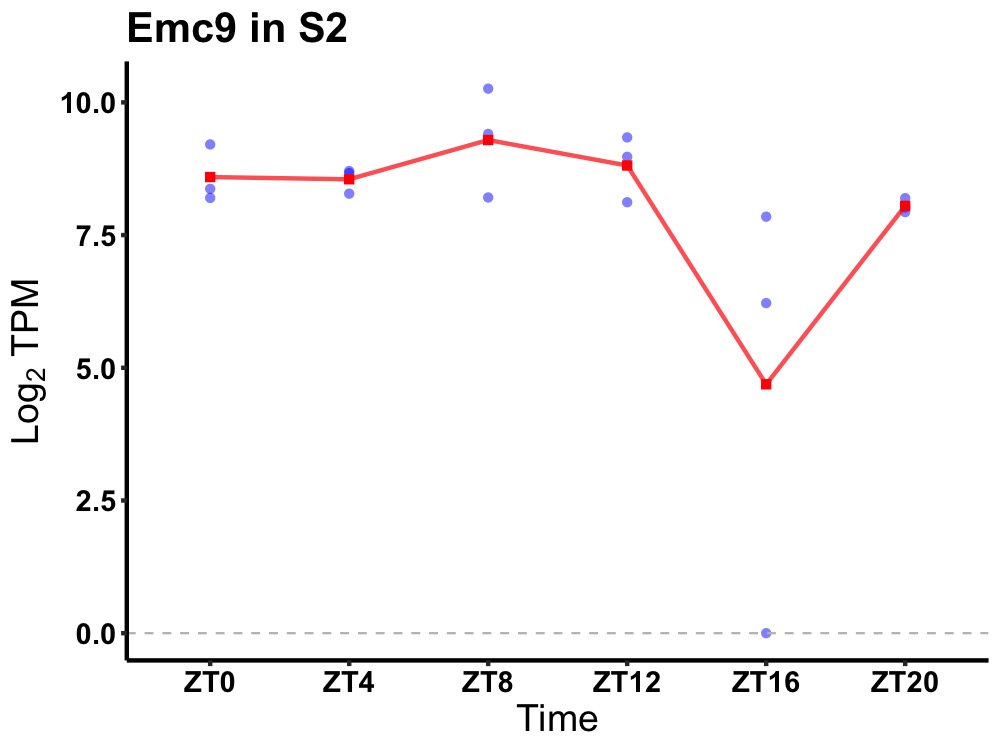 |
ER membrane protein complex subunit 9 | 0.019 | 24 | 6 | 0.49 |
| ENSMUSG00000082510 | Gm11793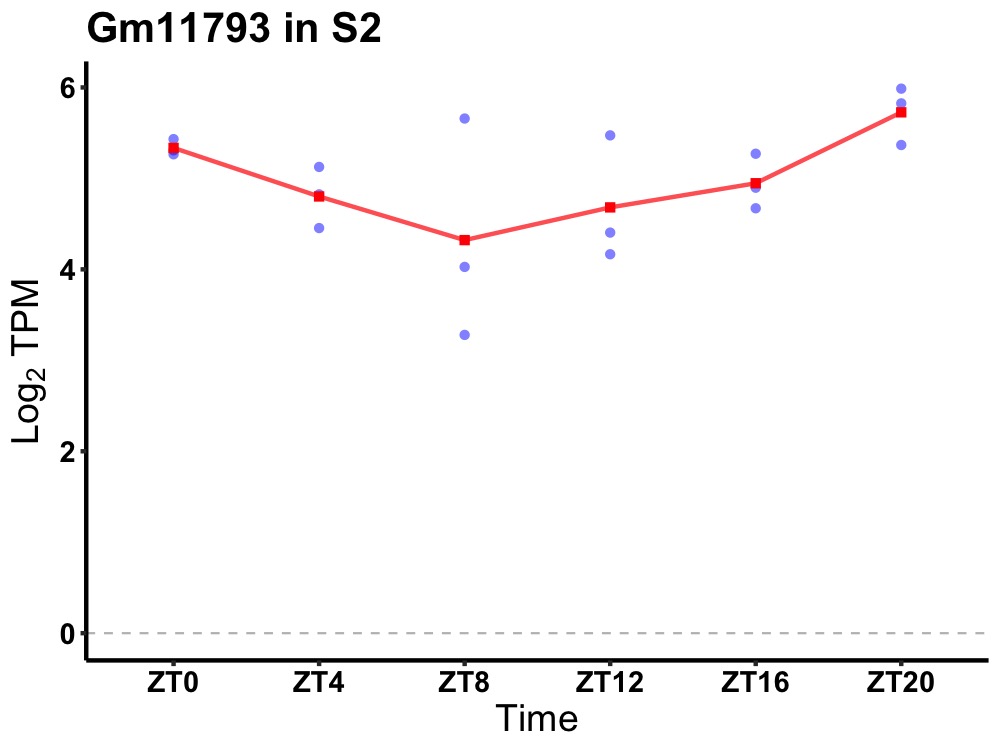 |
predicted gene 11793 | 0.036 | 24 | 22 | 0.49 |
| ENSMUSG00000085893 | Gm12091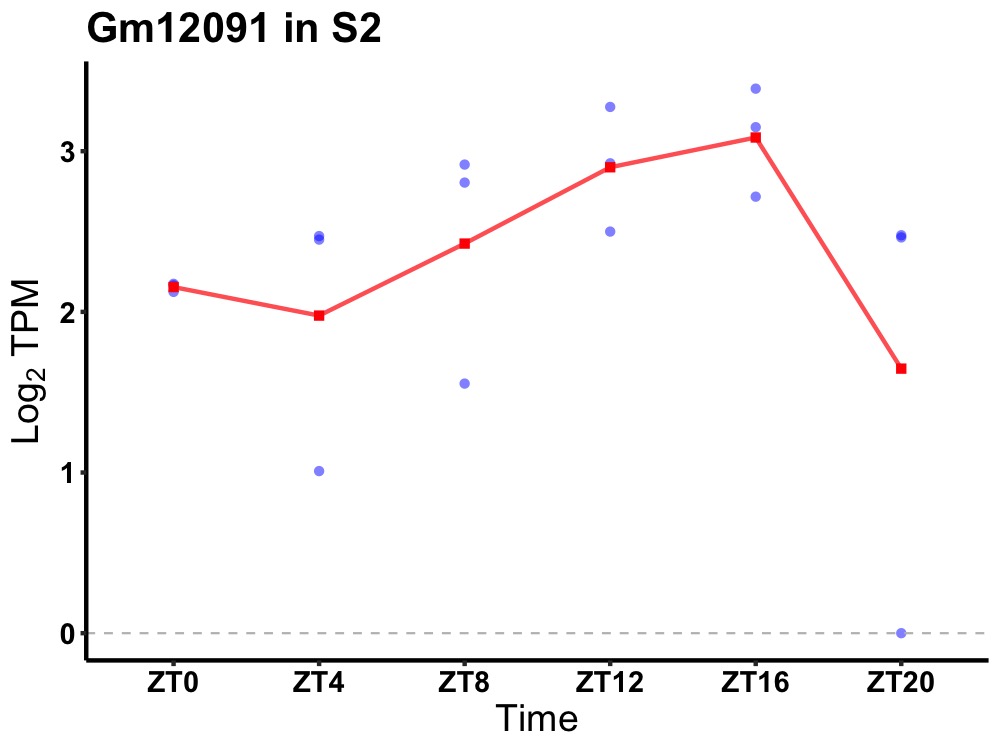 |
predicted gene 12091 | 0.027 | 24 | 14 | 0.49 |
| ENSMUSG00000104181 | Maf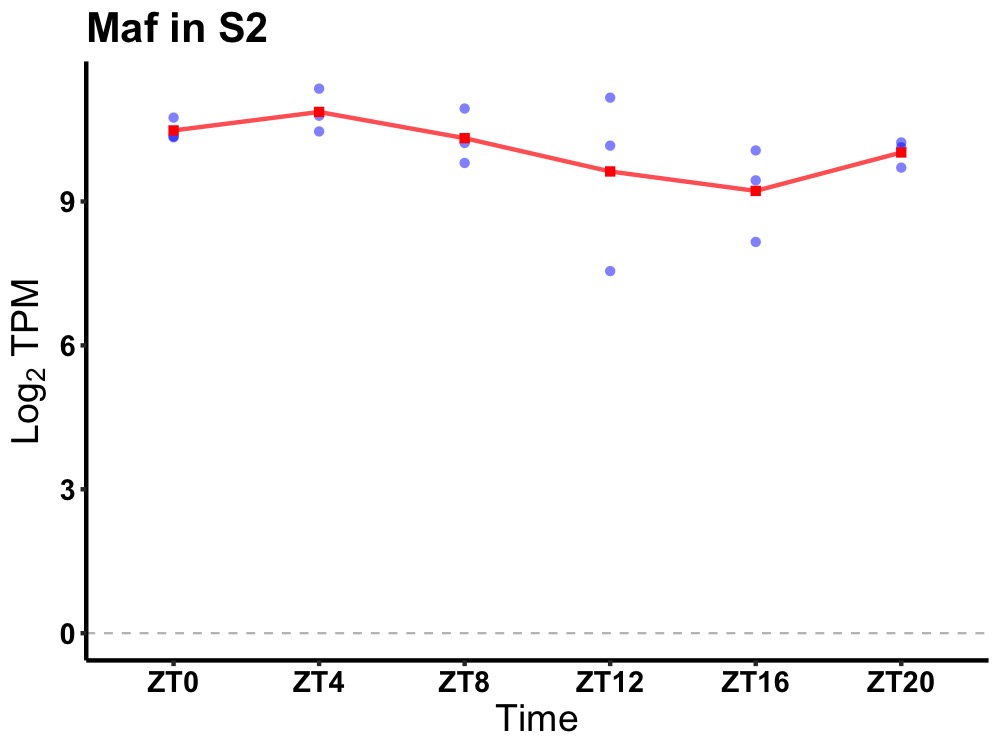 |
avian musculoaponeurotic fibrosarcoma oncogene homolog | 0.036 | 24 | 4 | 0.49 |
| ENSMUSG00000021850 | Pcyt2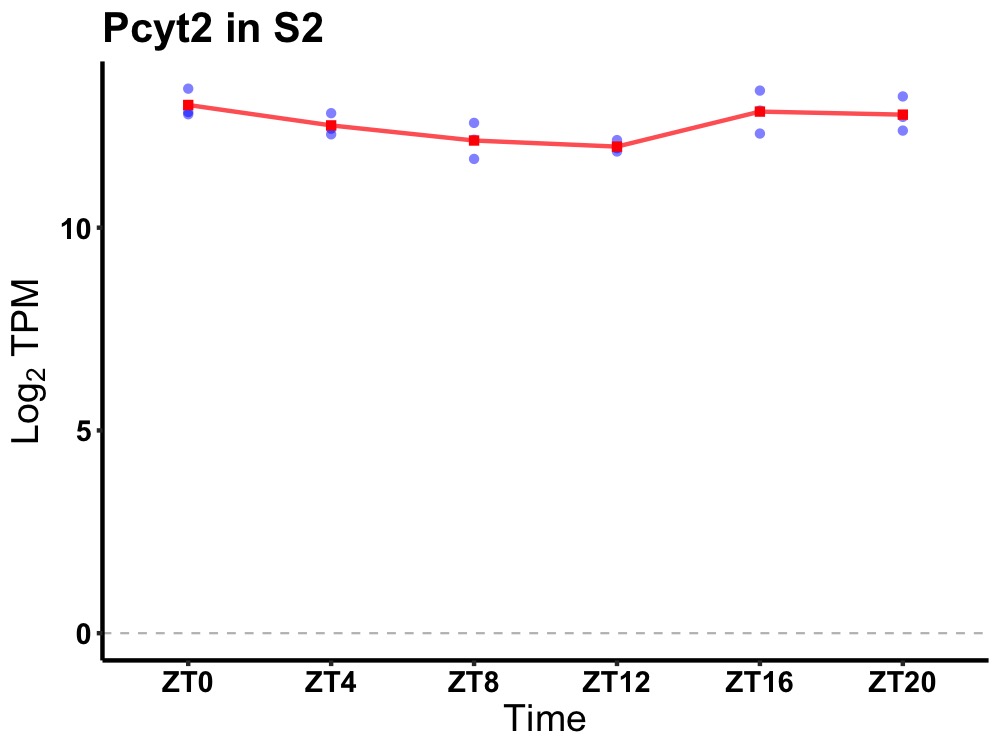 |
phosphate cytidylyltransferase 2, ethanolamine | 0.027 | 20 | 0 | 0.49 |
| ENSMUSG00000101523 | Gm10031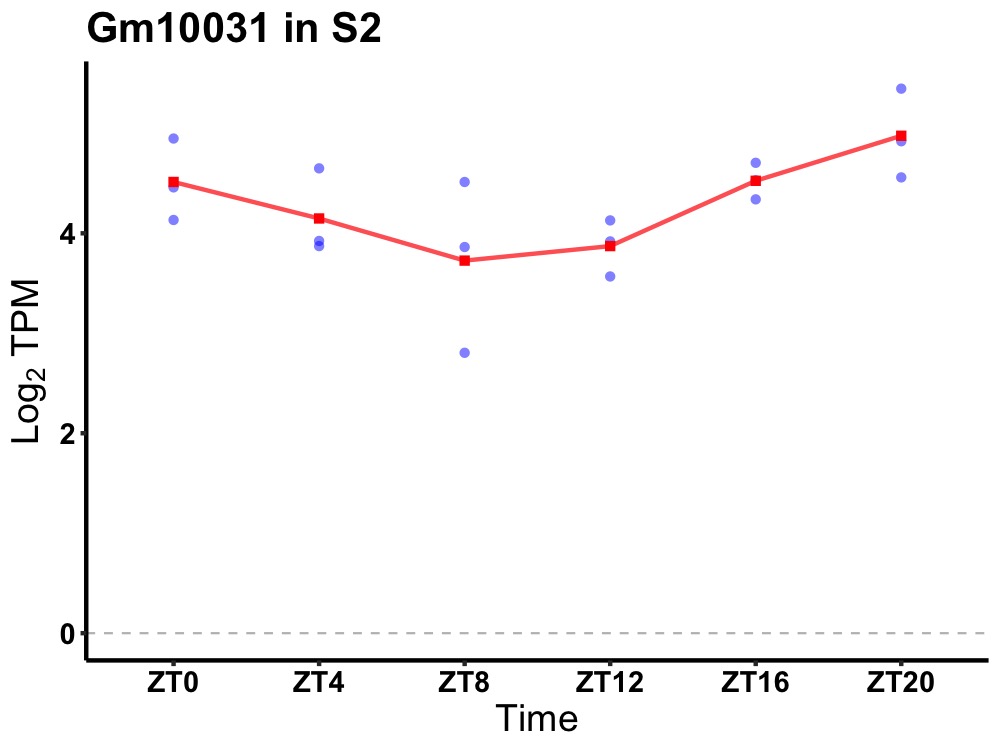 |
predicted pseudogene 10031 | 0.014 | 24 | 22 | 0.49 |
| ENSMUSG00000057666 | Stxbp3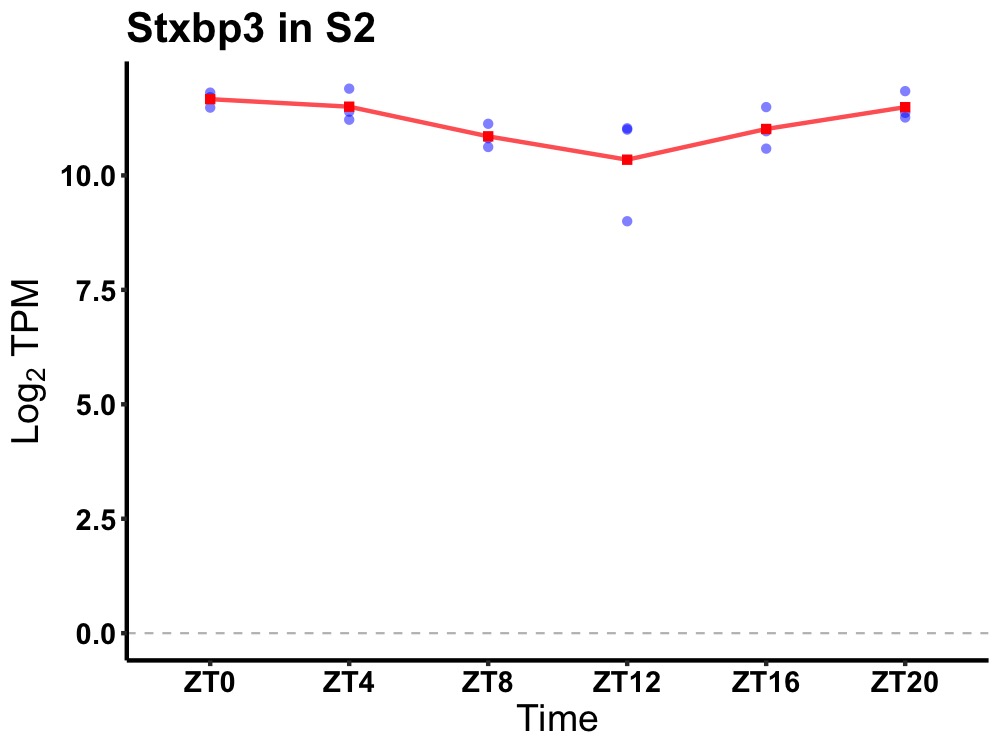 |
syntaxin-binding protein 3 | 0.036 | 24 | 0 | 0.48 |
| ENSMUSG00000005533 | Mrps31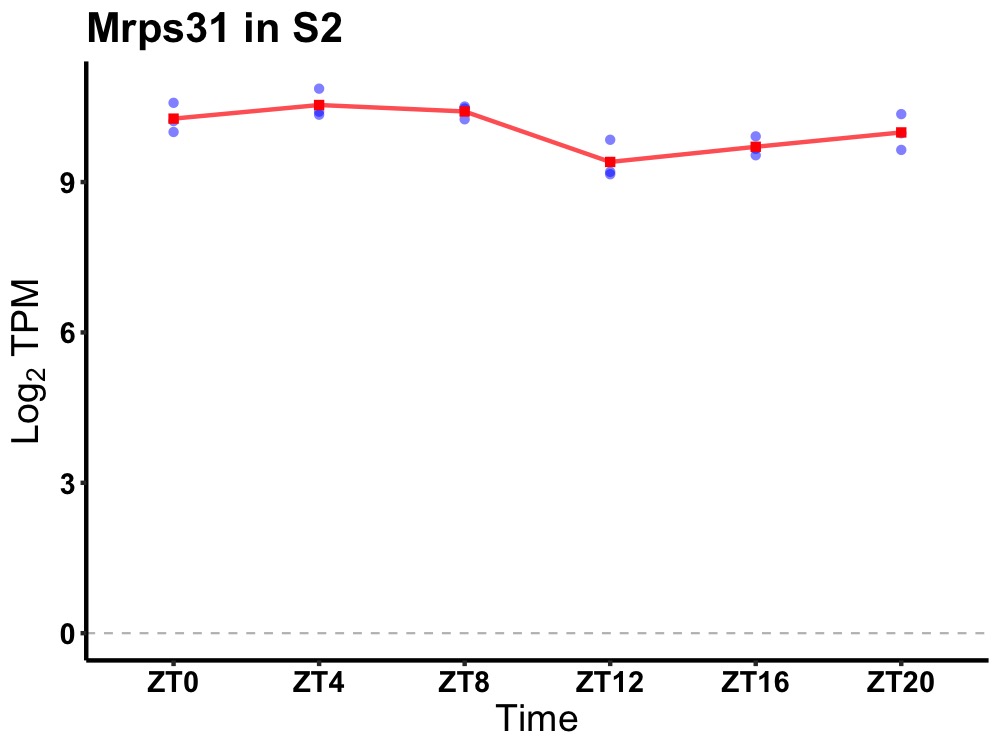 |
mitochondrial ribosomal protein S31 | 0.019 | 24 | 4 | 0.48 |
| ENSMUSG00000081451 | Gm12734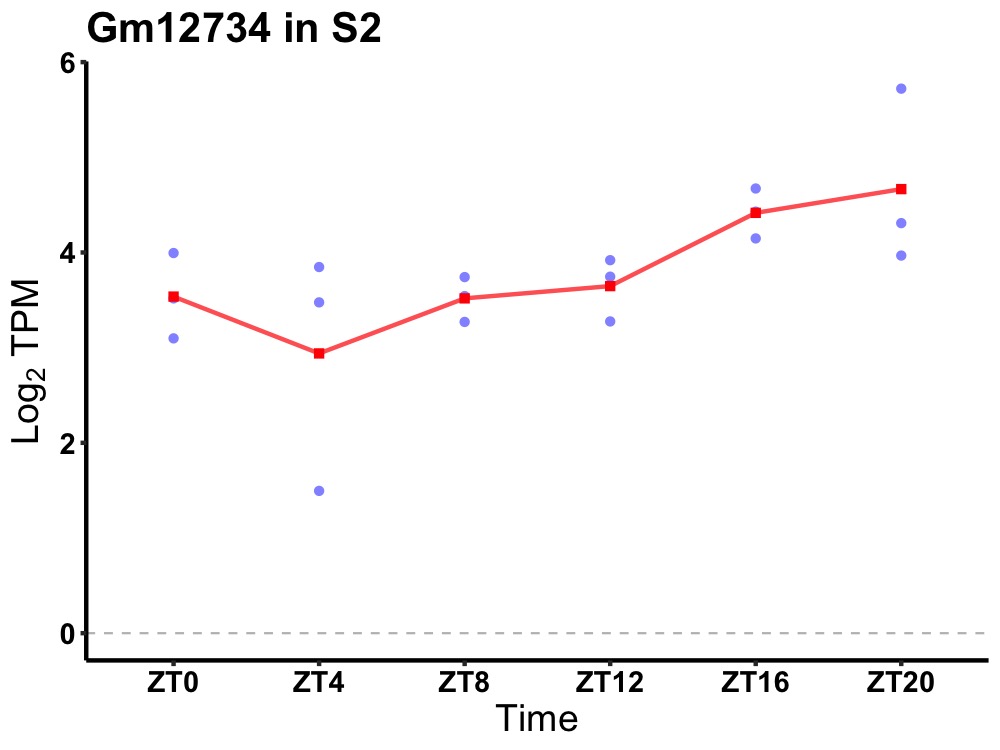 |
predicted gene 12734 | 0.019 | 24 | 18 | 0.48 |
| ENSMUSG00000084883 | Pdk2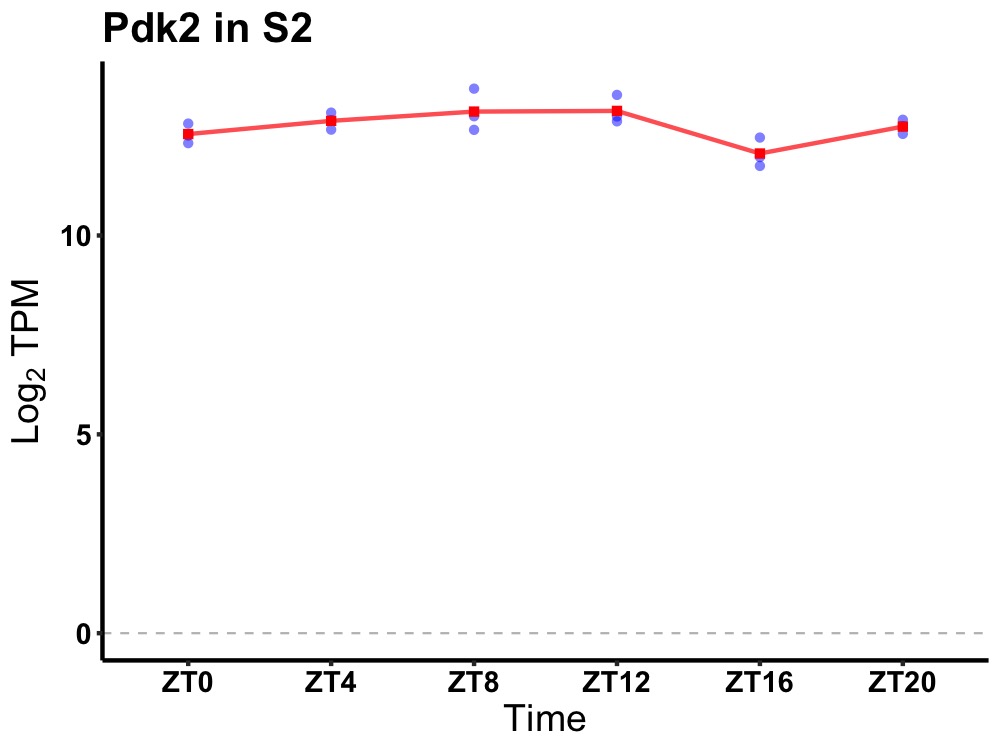 |
pyruvate dehydrogenase kinase, isoenzyme 2 | 0.014 | 20 | 8 | 0.47 |
| ENSMUSG00000031163 | Met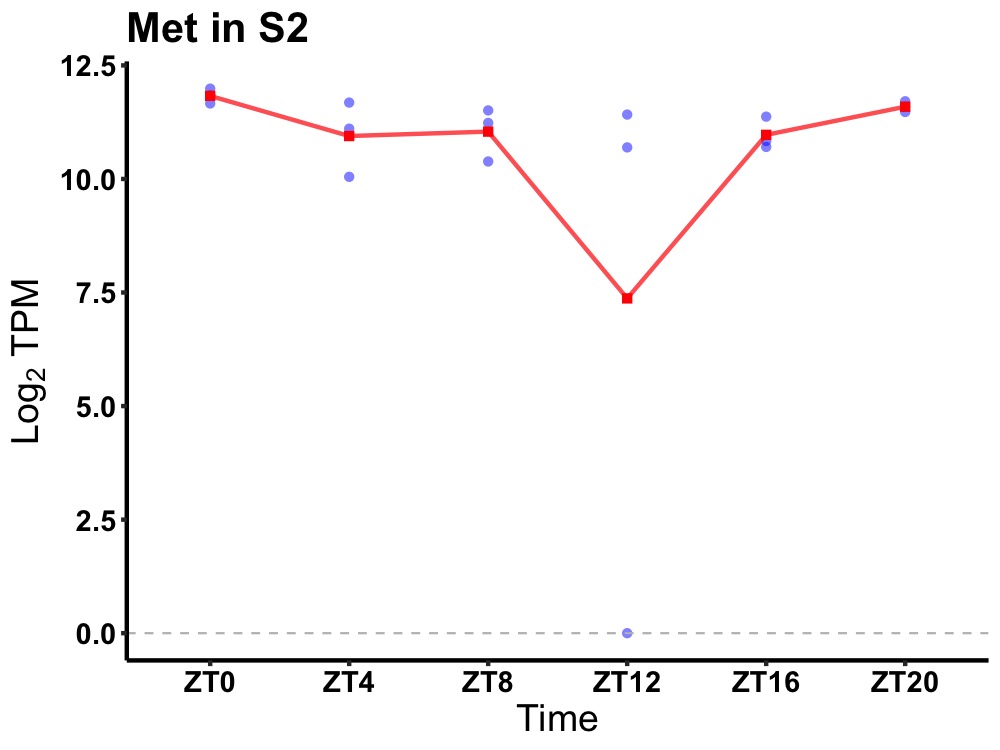 |
met proto-oncogene | 0.036 | 24 | 0 | 0.47 |
| ENSMUSG00000081600 | Mrpl46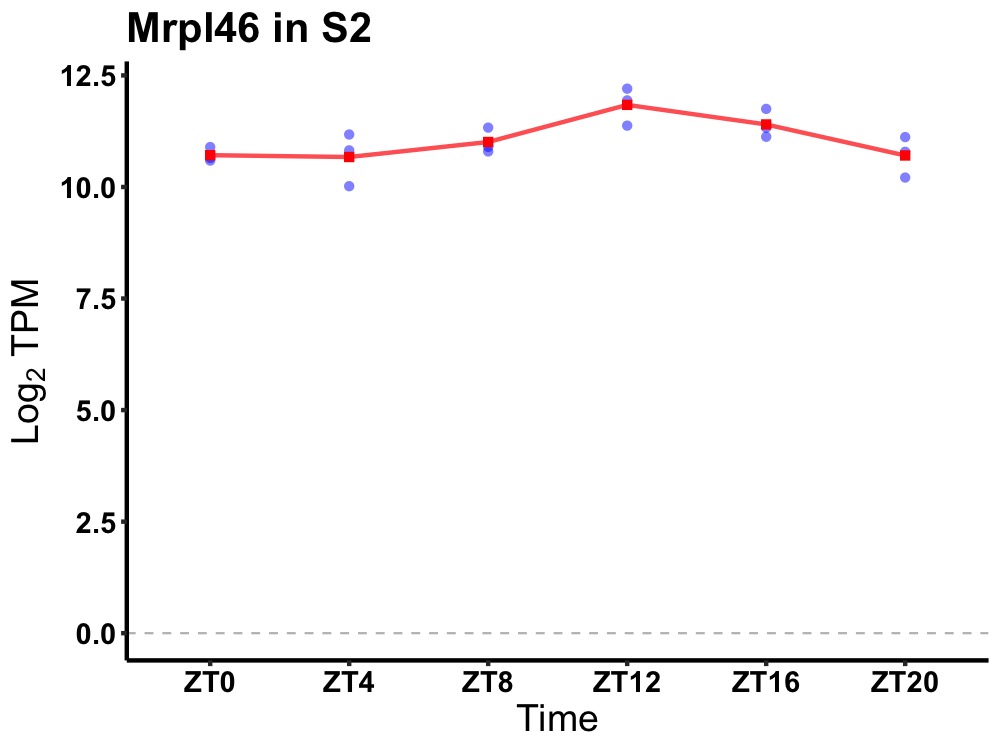 |
mitochondrial ribosomal protein L46 | 0.007 | 24 | 14 | 0.47 |
| ENSMUSG00000015749 | Herpud1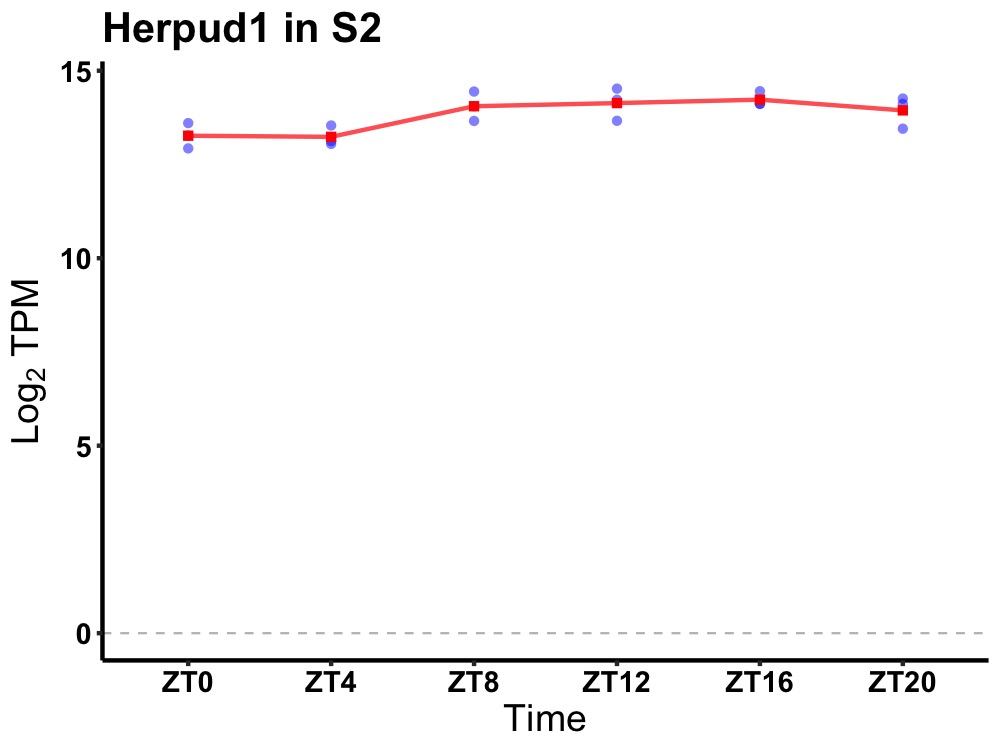 |
homocysteine-inducible, endoplasmic reticulum stress-inducible, ubiquitin-like domain member 1 | 0.019 | 24 | 14 | 0.47 |
| ENSMUSG00000026095 | Asnsd1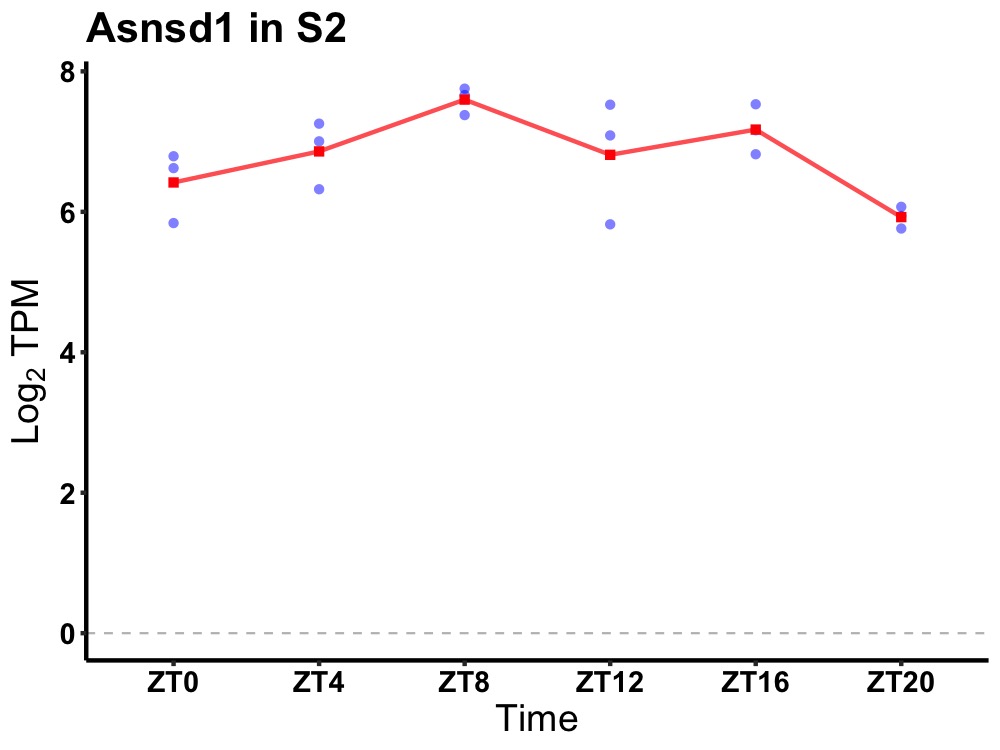 |
asparagine synthetase domain containing 1 | 0.027 | 24 | 10 | 0.47 |
| ENSMUSG00000091198 | Gm5218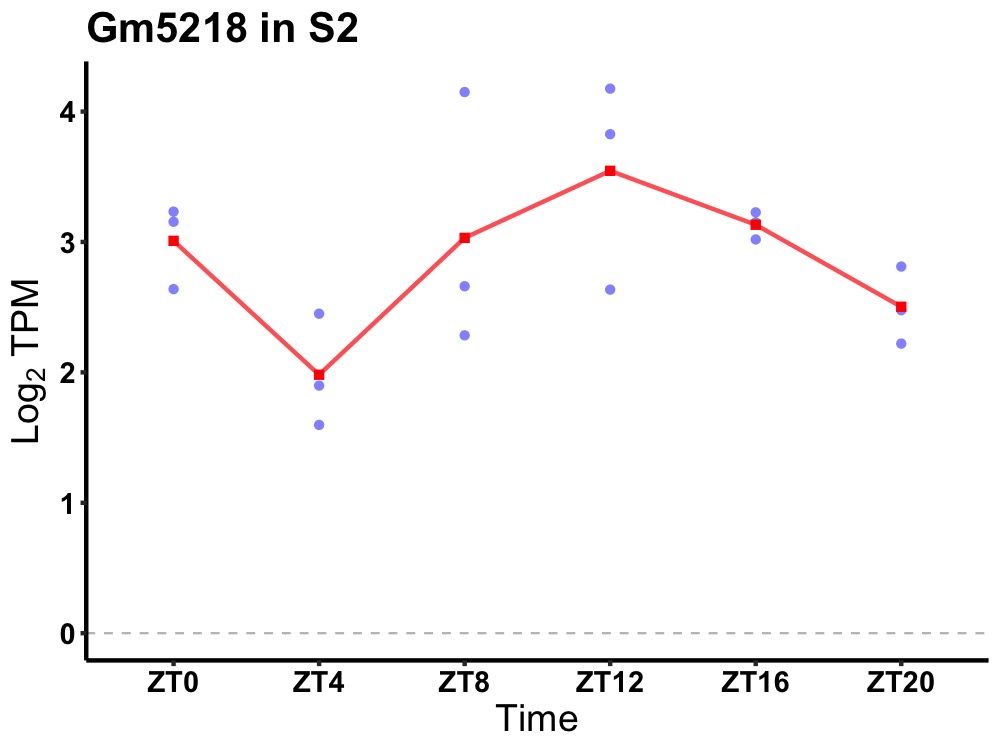 |
predicted gene 5218 | 0.049 | 20 | 14 | 0.47 |
| ENSMUSG00000074698 | CT010467.1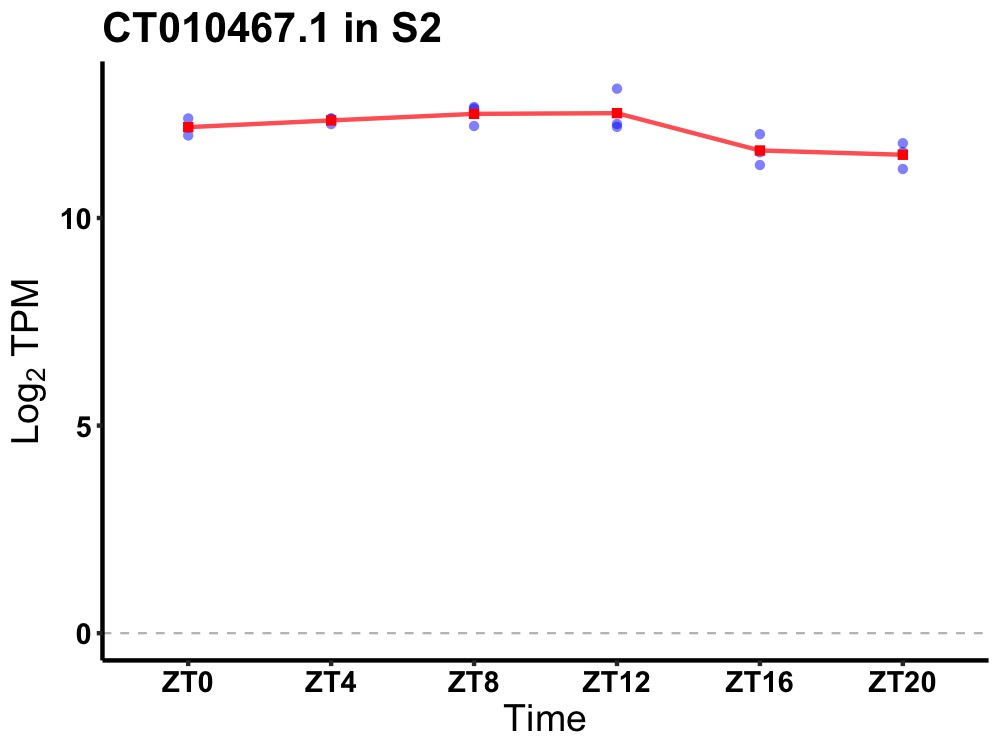 |
18s RNA, related sequence 5 | 0.003 | 24 | 8 | 0.46 |
| ENSMUSG00000069565 | 2610528J11Rik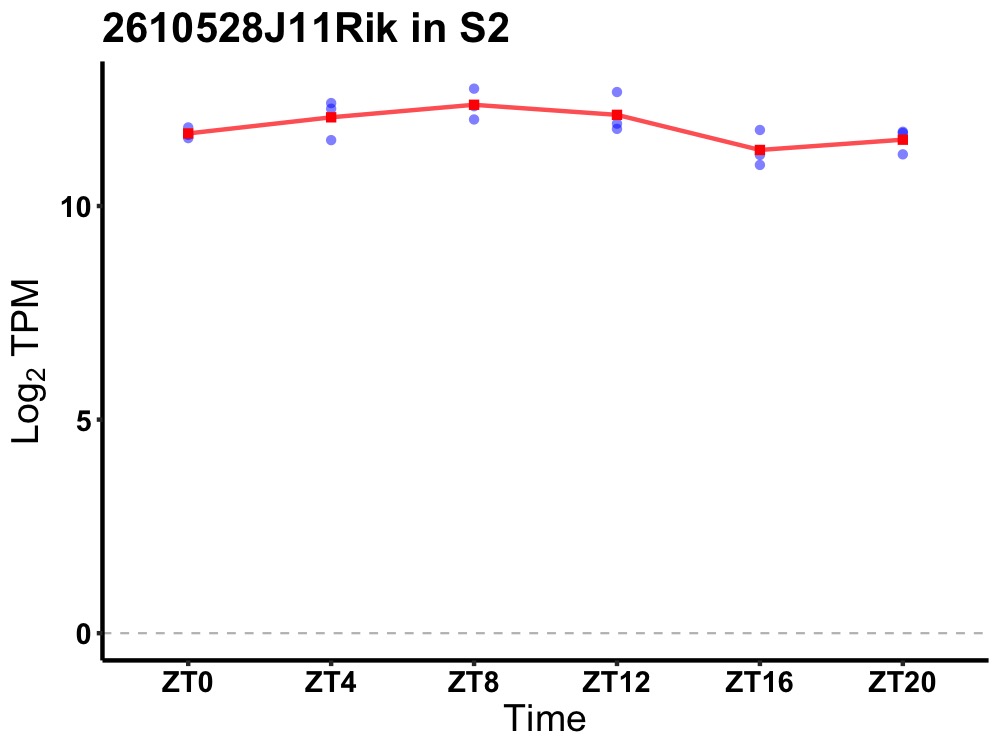 |
RIKEN cDNA 2610528J11 gene | 0.027 | 20 | 8 | 0.46 |
| ENSMUSG00000081251 | Gm15638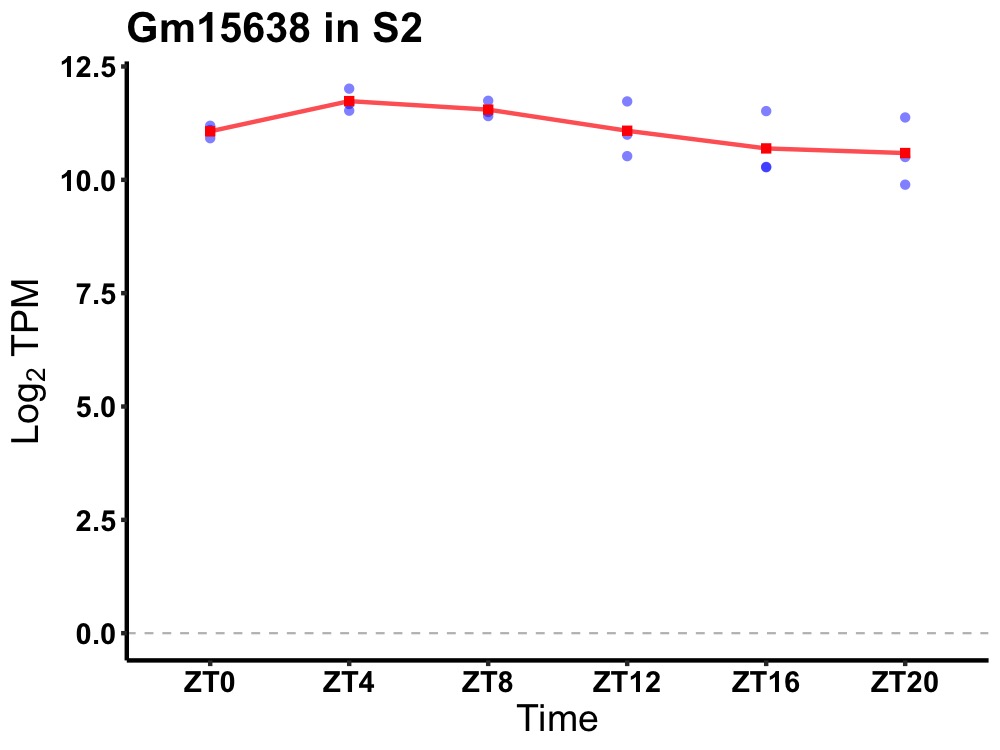 |
predicted gene 15638 | 0.027 | 24 | 6 | 0.46 |
| ENSMUSG00000110882 | Gm7642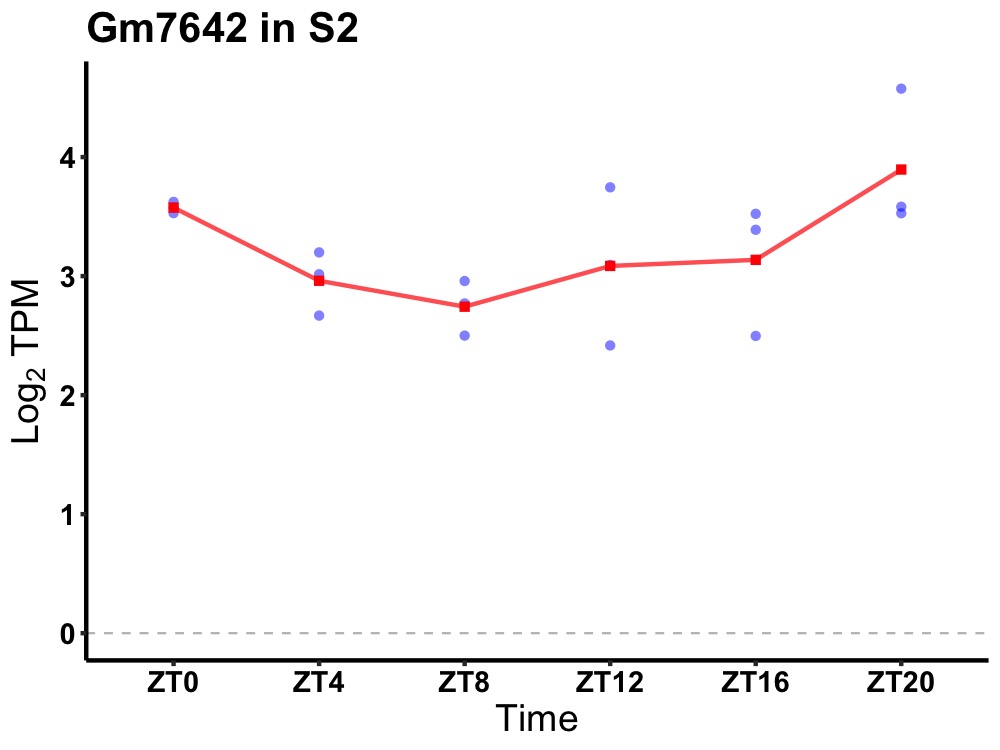 |
predicted gene 7642 | 0.049 | 24 | 22 | 0.45 |
| ENSMUSG00000074466 | Gm15417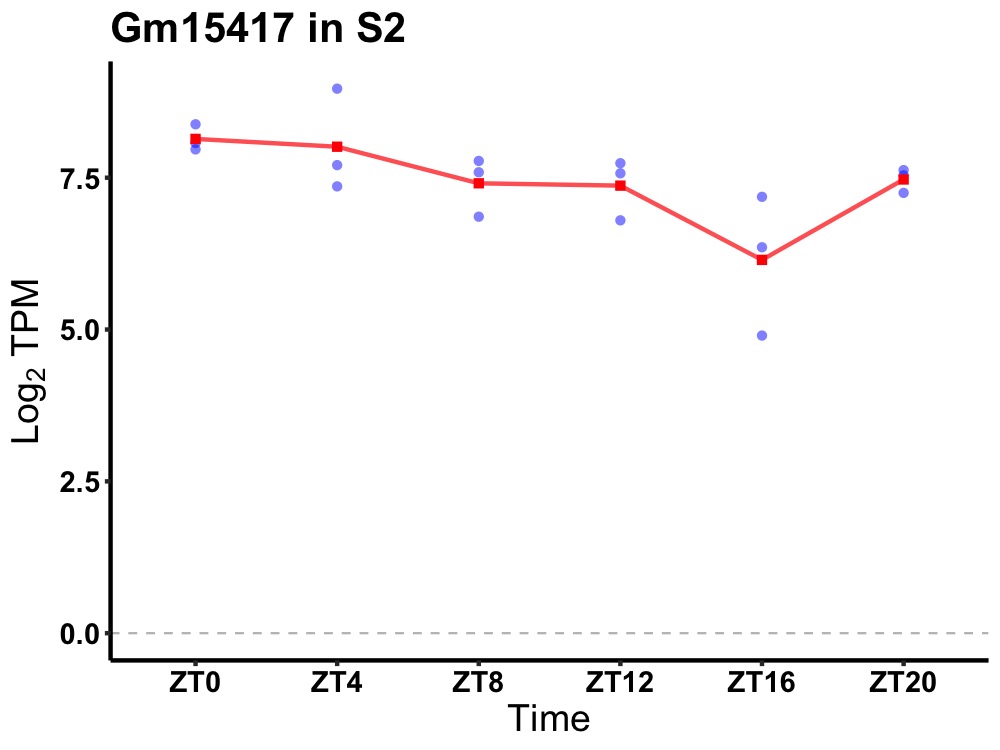 |
predicted gene 15417 | 0.036 | 24 | 4 | 0.45 |
| ENSMUSG00000083179 | Syf2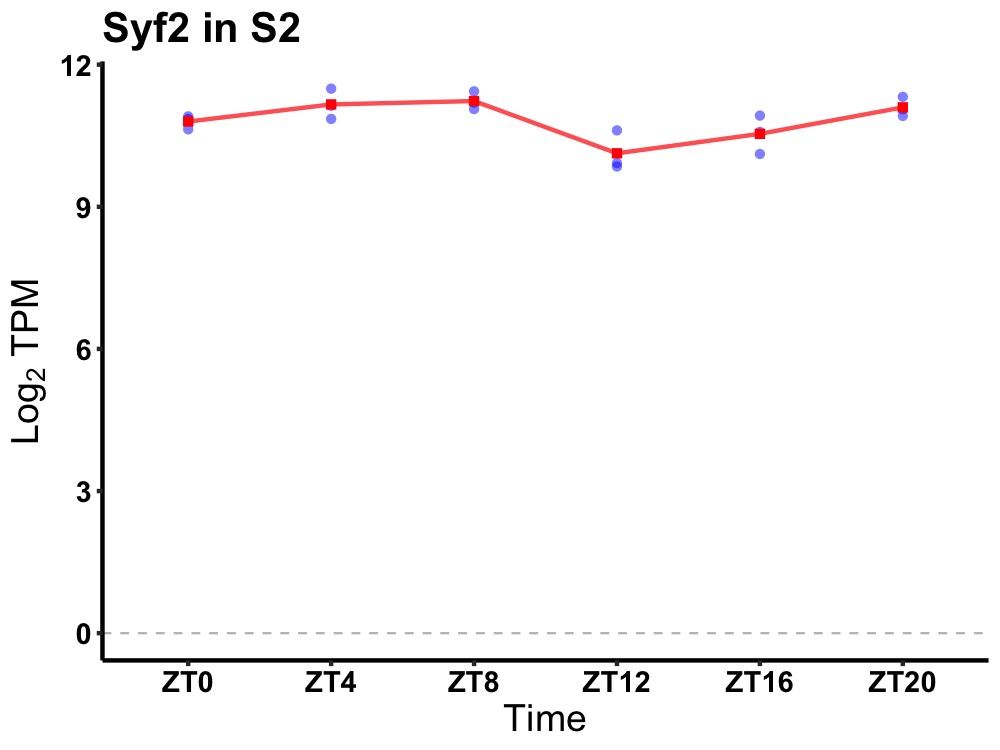 |
SYF2 homolog, RNA splicing factor (S. cerevisiae) | 0.014 | 20 | 6 | 0.45 |
| ENSMUSG00000082715 | Gm11633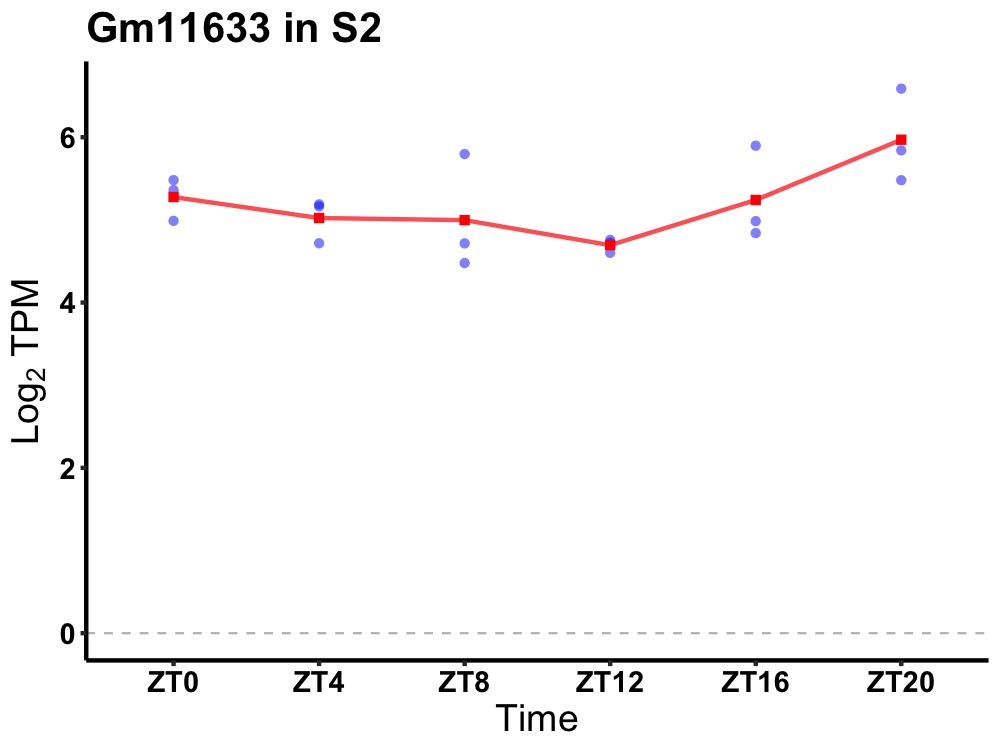 |
predicted gene 11633 | 0.027 | 24 | 22 | 0.45 |
| ENSMUSG00000080746 | Rpsa-ps12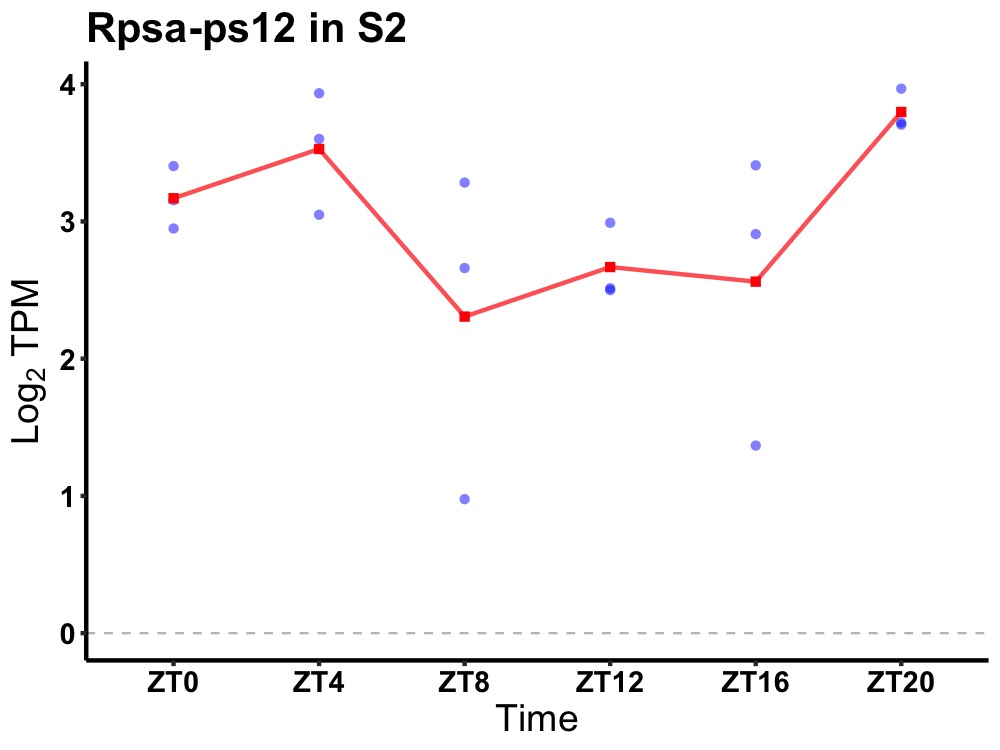 |
ribosomal protein SA, pseudogene 12 | 0.036 | 20 | 2 | 0.45 |
| ENSMUSG00000099997 | Tubb4b-ps2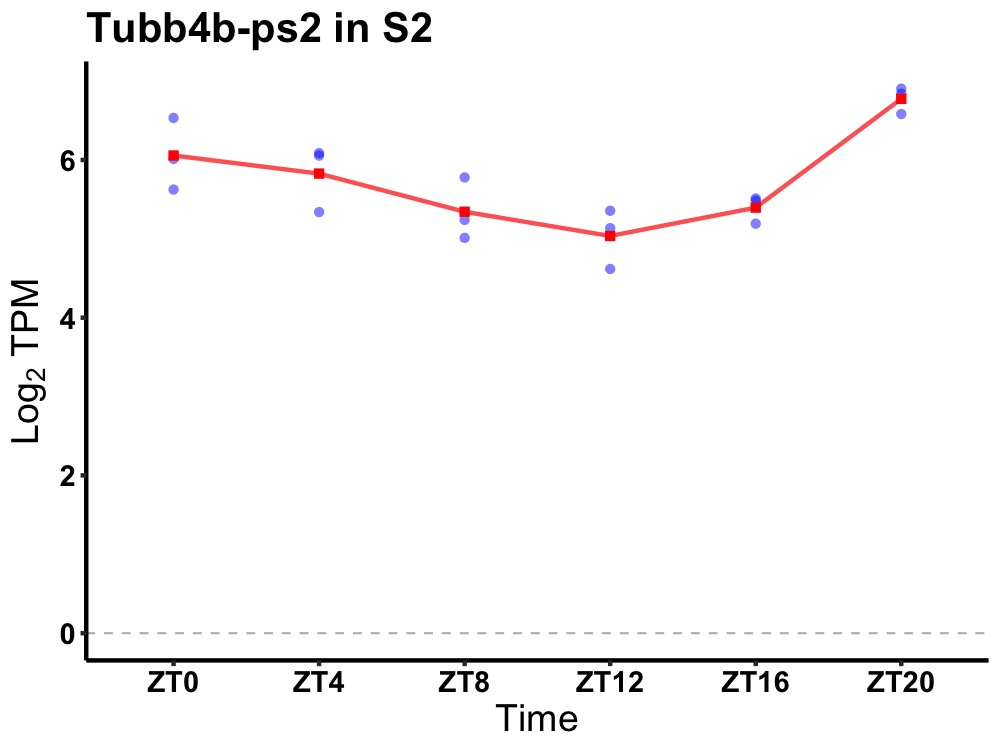 |
tubulin, beta 4B class IVB, pseudogene 2 | 0.001 | 20 | 2 | 0.45 |
| ENSMUSG00000082693 | Gm15190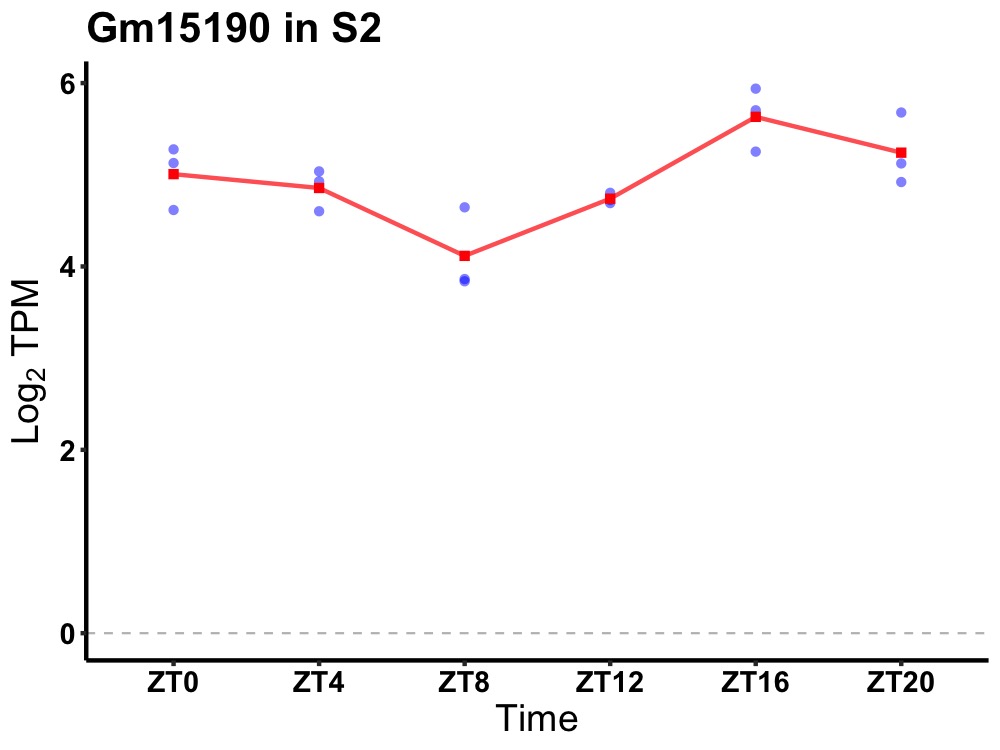 |
predicted gene 15190 | 0.005 | 20 | 18 | 0.45 |
| ENSMUSG00000117062 | Ldha-ps3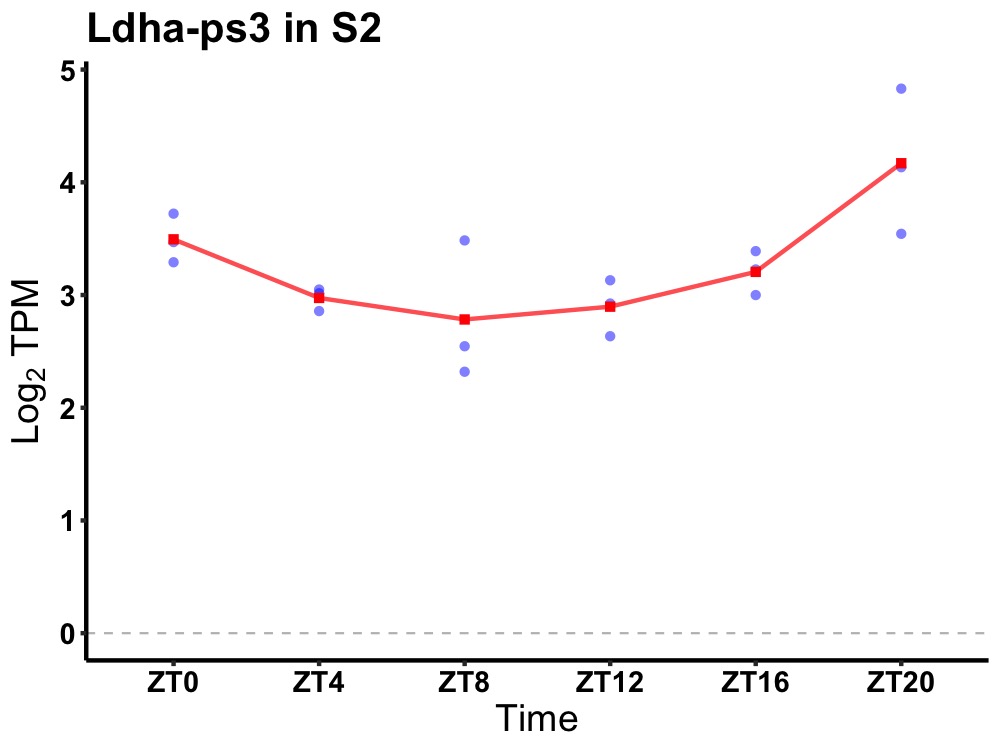 |
lactate dehydrogenase A (Ldha) pseudogene | 0.002 | 24 | 22 | 0.44 |
| ENSMUSG00000058355 | mt-Nd5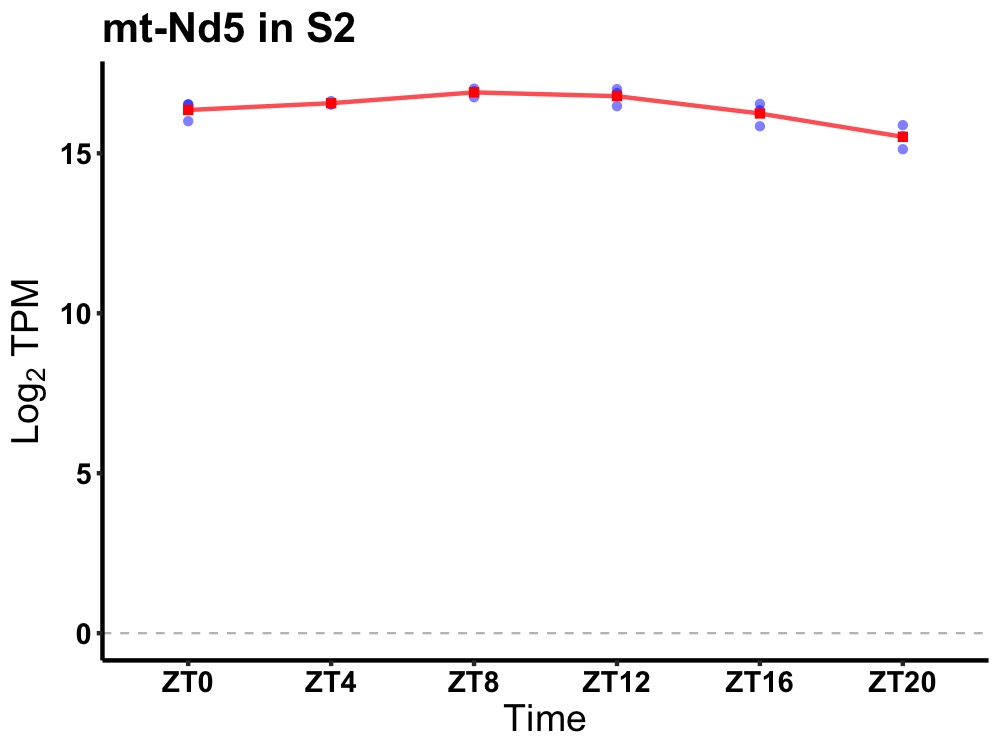 |
mitochondrially encoded NADH dehydrogenase 5 | 0.001 | 24 | 10 | 0.44 |
| ENSMUSG00000021824 | Hsp90b1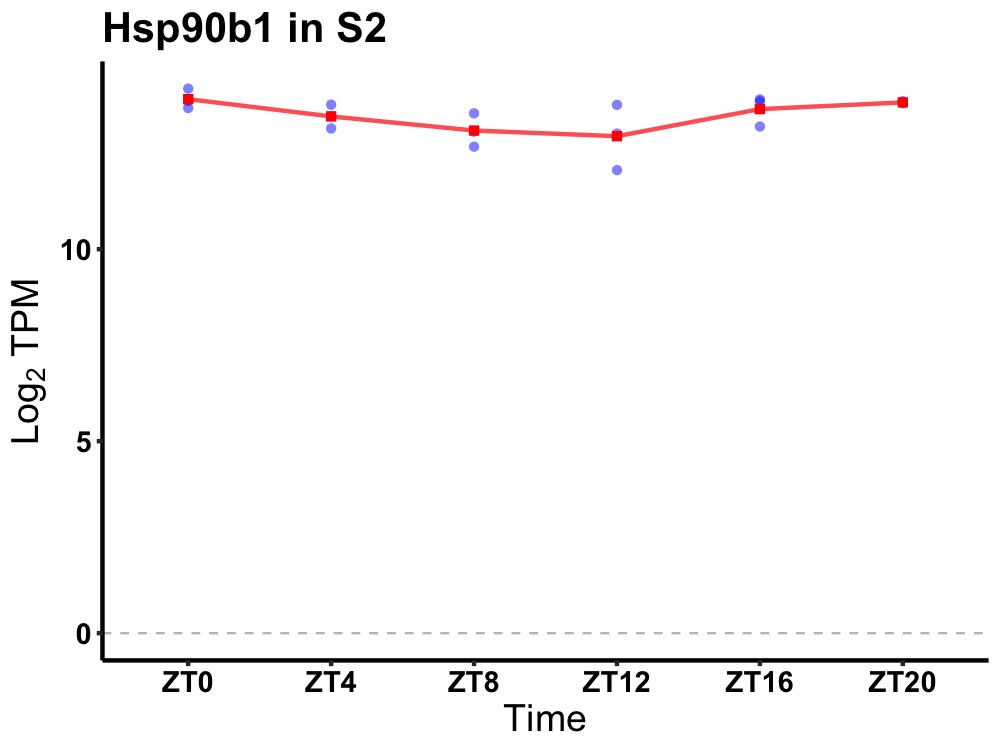 |
heat shock protein 90, beta (Grp94), member 1 | 0.019 | 20 | 0 | 0.43 |
| ENSMUSG00000096979 | Gm26880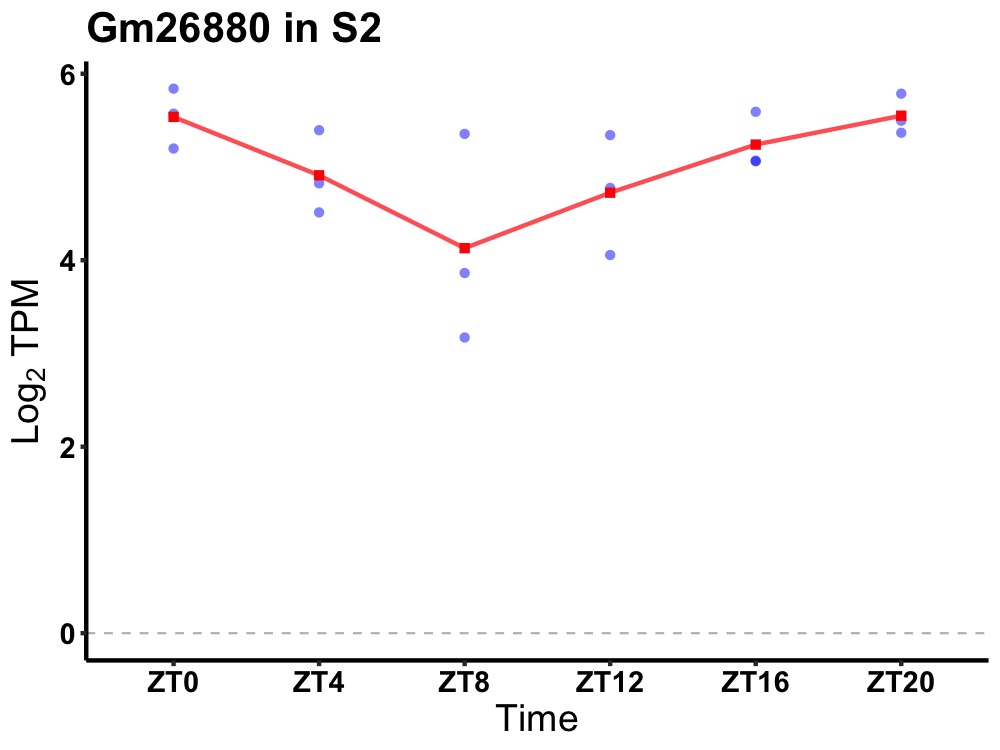 |
predicted gene, 26880 | 0.019 | 20 | 0 | 0.43 |
| ENSMUSG00000081953 | Gm12435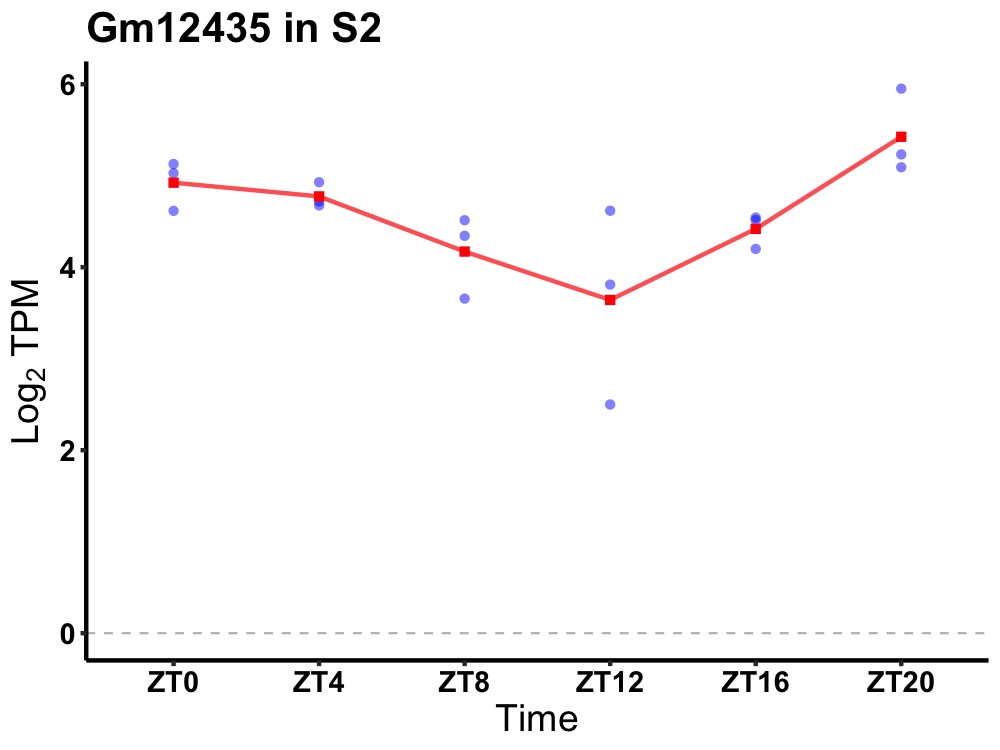 |
predicted gene 12435 | 0.000 | 20 | 2 | 0.43 |
| ENSMUSG00000117642 | Amd-ps7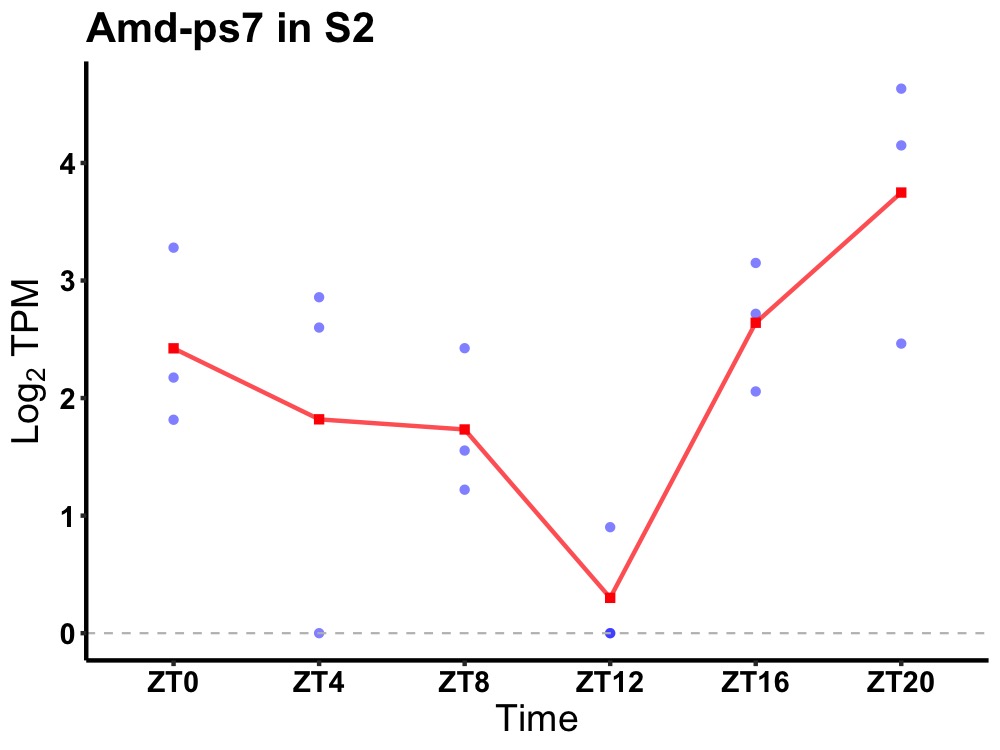 |
none | 0.036 | 20 | 2 | 0.43 |
| ENSMUSG00000090197 | Dnaja1-ps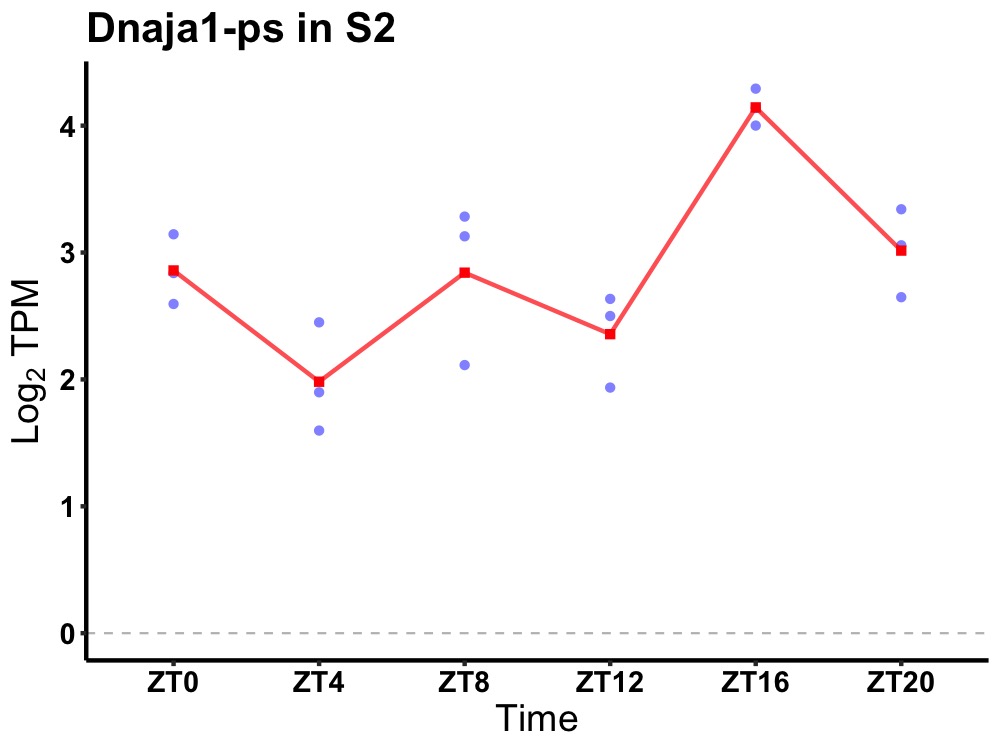 |
DnaJ heat shock protein family (Hsp40) member A1, pseudogene | 0.049 | 24 | 18 | 0.43 |
| ENSMUSG00000025779 | Ly96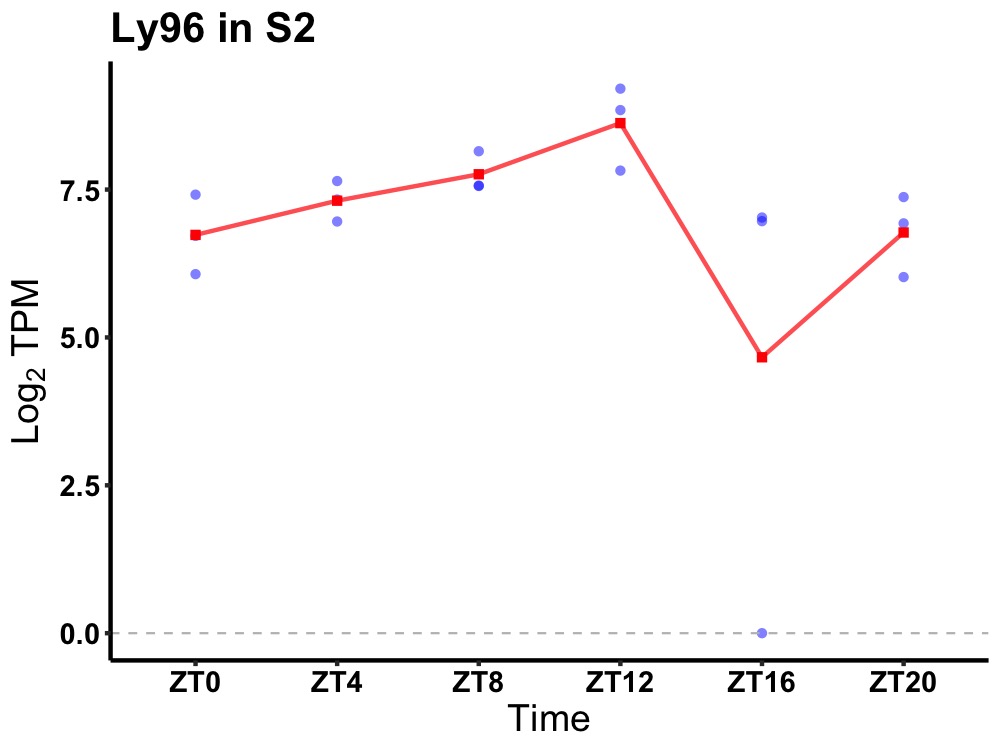 |
lymphocyte antigen 96 | 0.036 | 20 | 12 | 0.43 |
| ENSMUSG00000019278 | Rpl8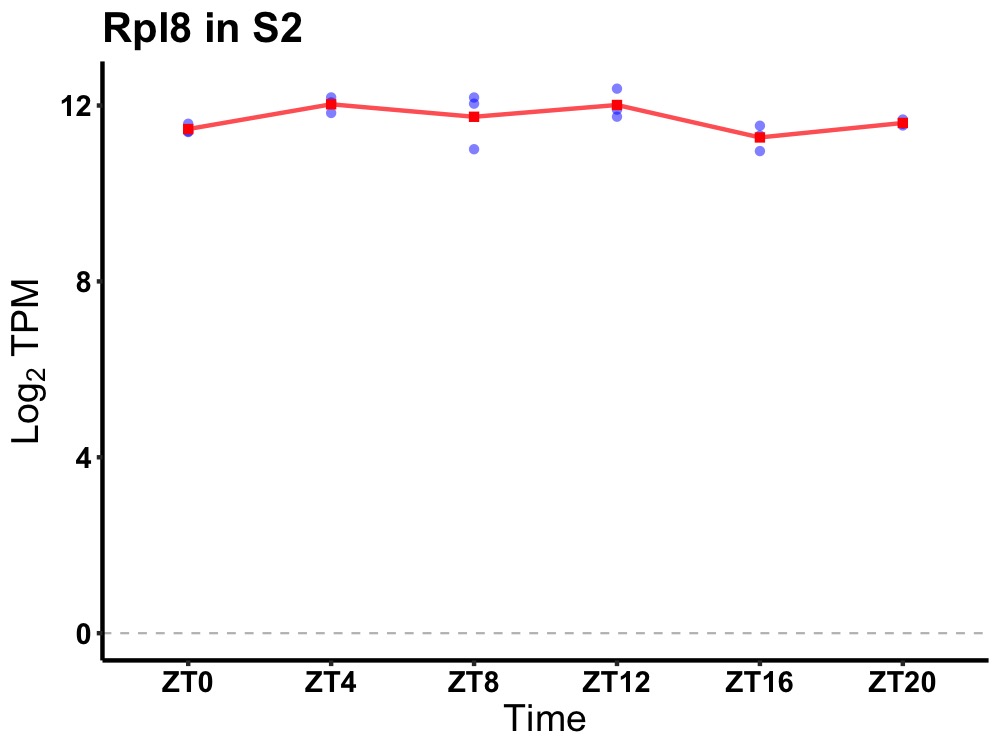 |
ribosomal protein L8 | 0.027 | 20 | 8 | 0.42 |
| ENSMUSG00000081329 | Mvd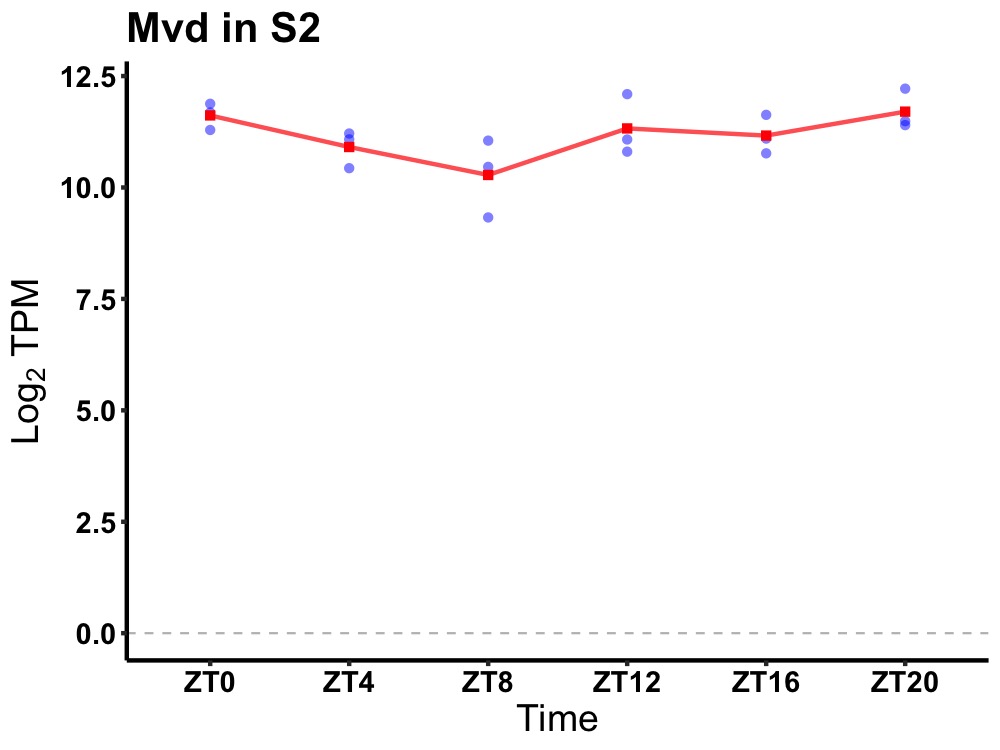 |
mevalonate (diphospho) decarboxylase | 0.049 | 24 | 22 | 0.42 |
| ENSMUSG00000109768 | Atp2c1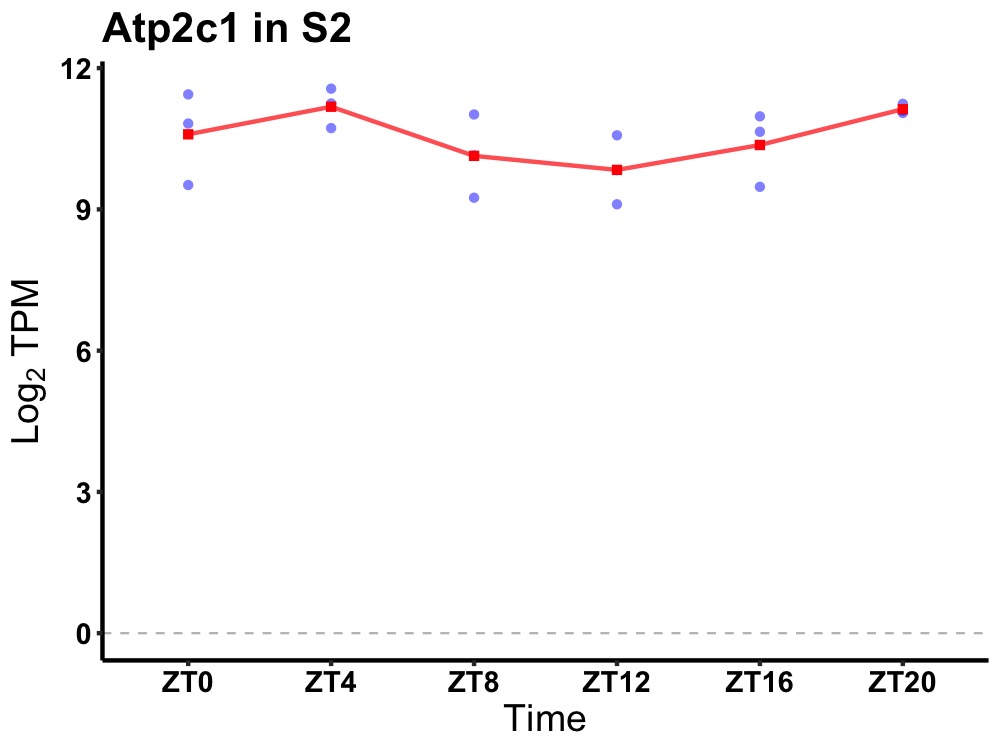 |
ATPase, Ca++-sequestering | 0.049 | 20 | 4 | 0.42 |
| ENSMUSG00000022790 | Hmox2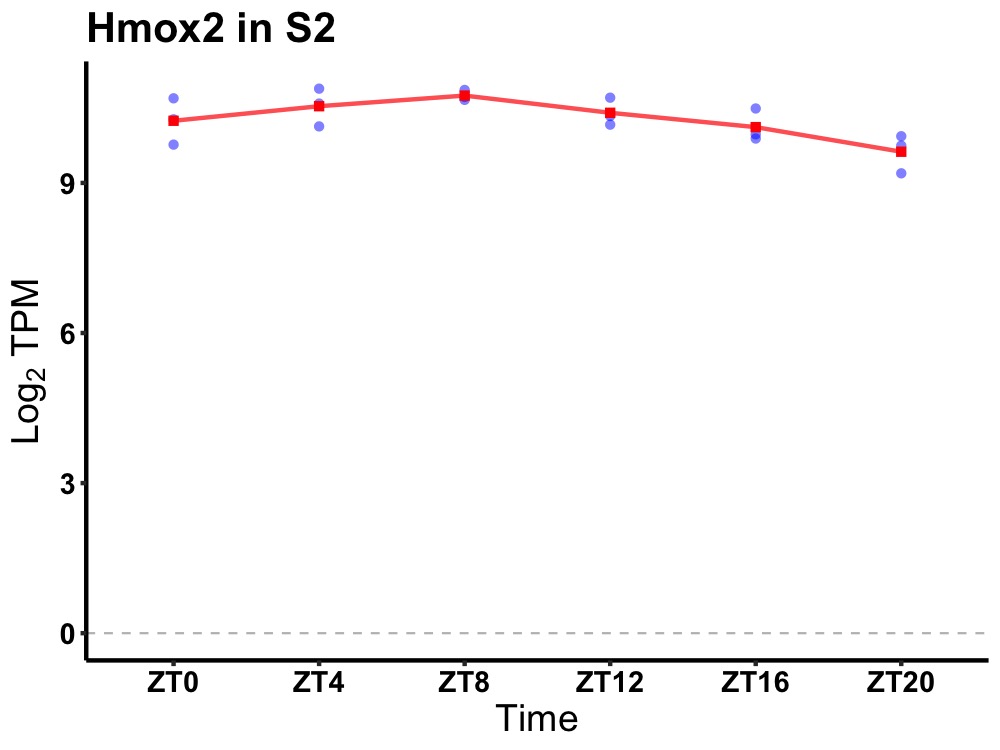 |
heme oxygenase 2 | 0.010 | 24 | 8 | 0.42 |
| ENSMUSG00000038914 | Tubb4b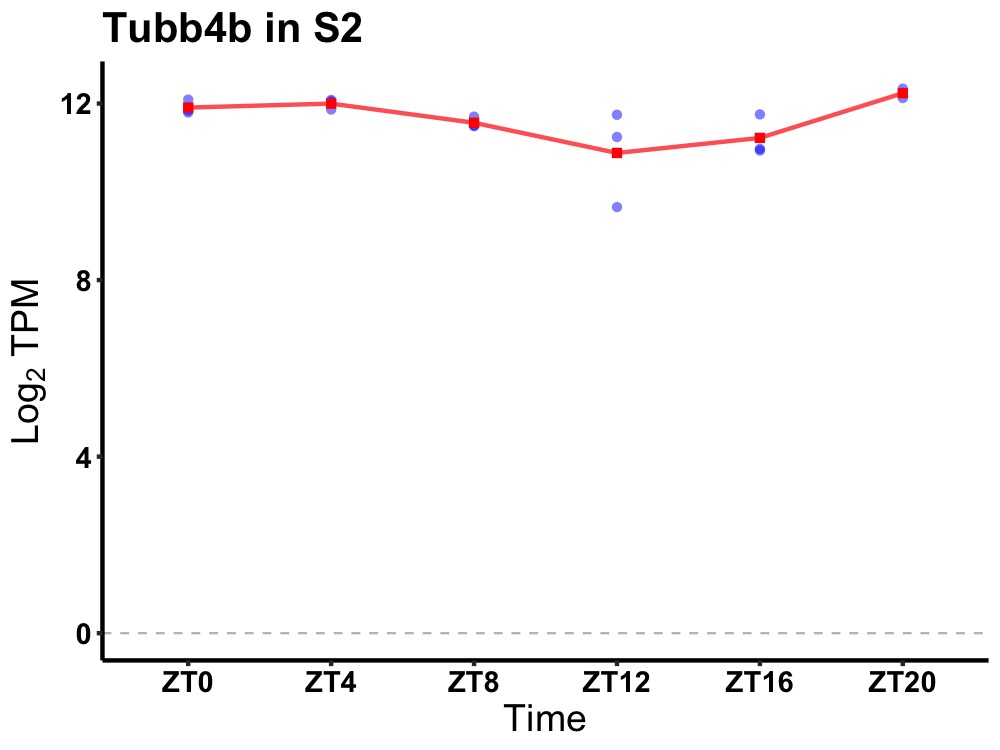 |
tubulin, beta 4B class IVB | 0.001 | 20 | 2 | 0.42 |
| ENSMUSG00000028085 | Ascc1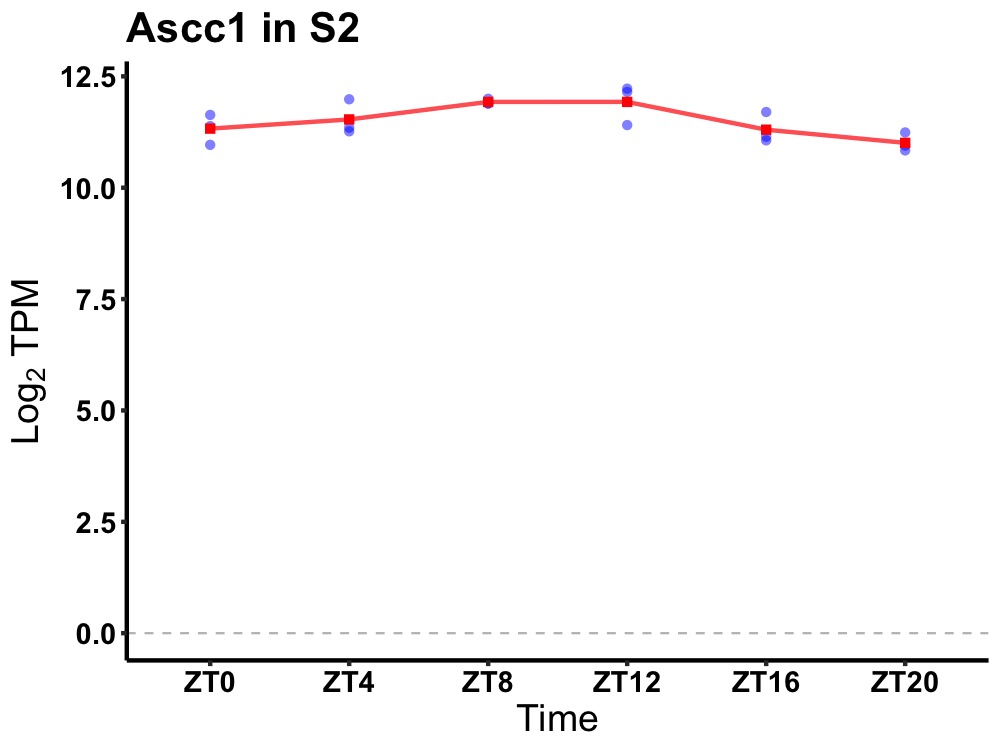 |
activating signal cointegrator 1 complex subunit 1 | 0.010 | 24 | 10 | 0.42 |
| ENSMUSG00000031483 | Lamp1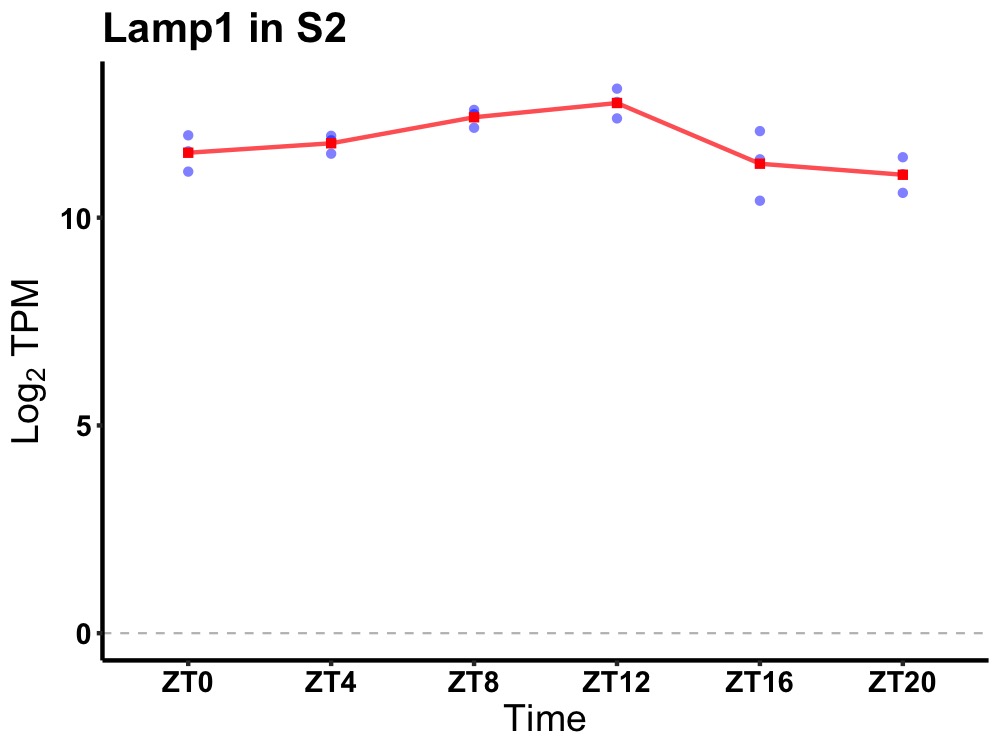 |
lysosomal-associated membrane protein 1 | 0.005 | 20 | 12 | 0.41 |
| ENSMUSG00000093548 | Gm6407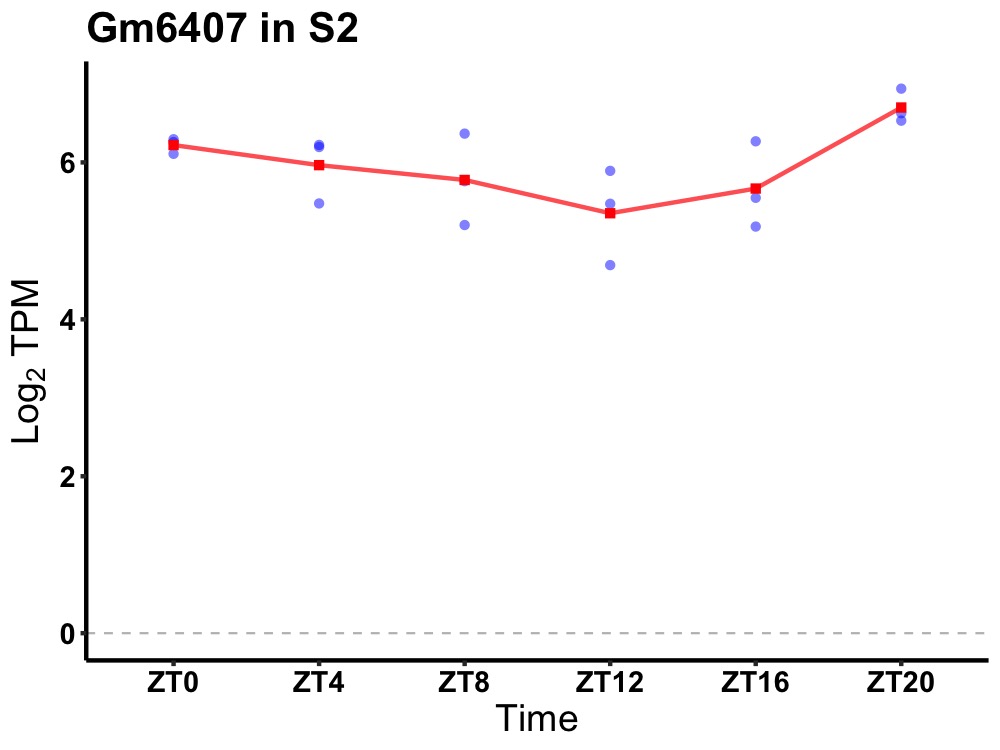 |
predicted gene 6407 | 0.036 | 20 | 2 | 0.41 |
| ENSMUSG00000083582 | Gm15393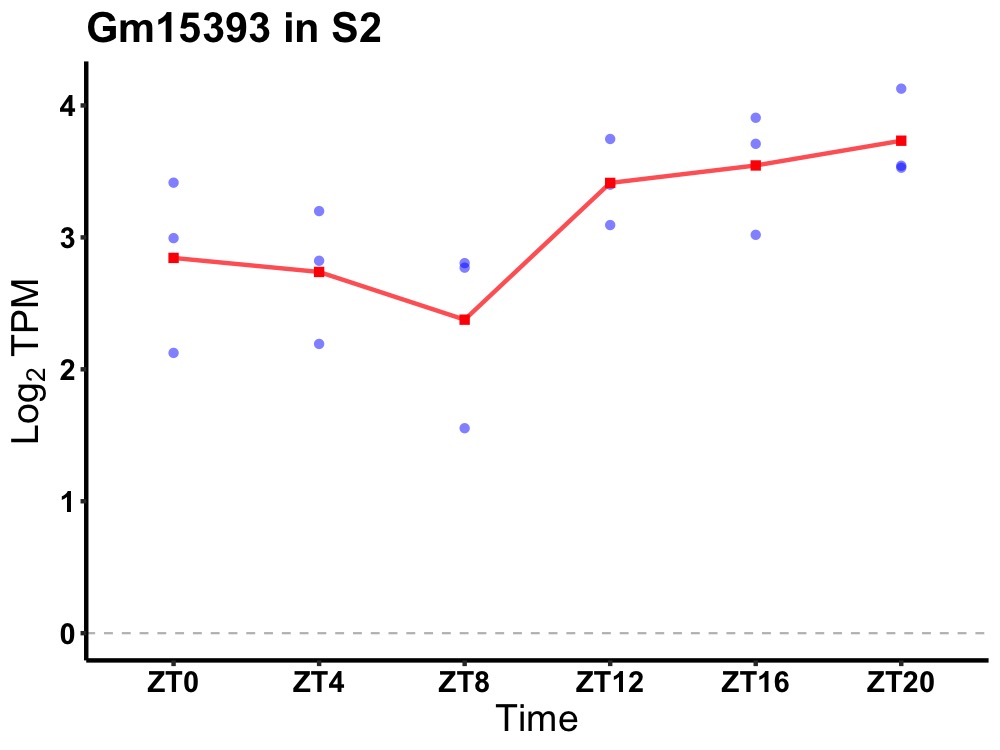 |
predicted gene 15393 | 0.036 | 24 | 20 | 0.41 |
| ENSMUSG00000075415 | Acp6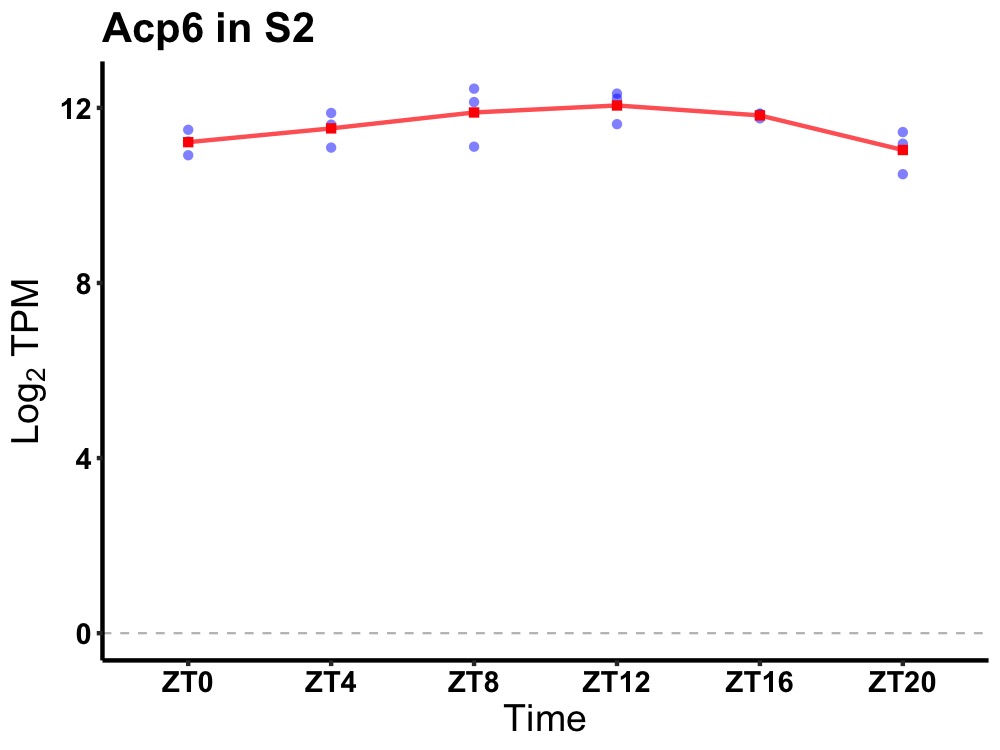 |
acid phosphatase 6, lysophosphatidic | 0.027 | 20 | 12 | 0.41 |
| ENSMUSG00000002769 | Impa1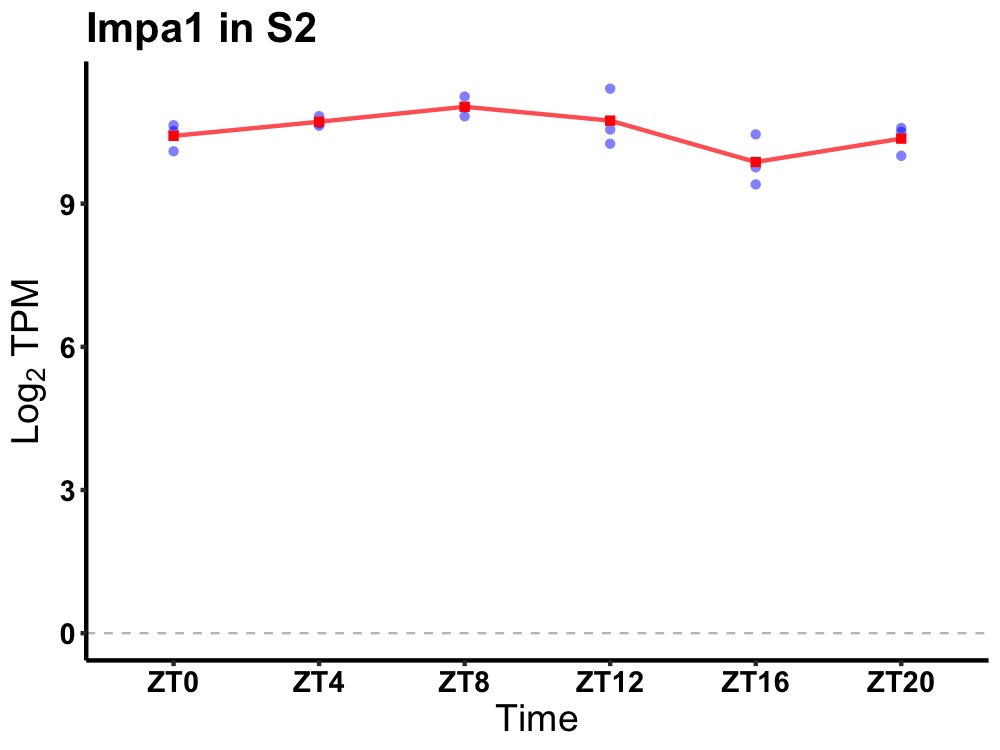 |
inositol (myo)-1(or 4)-monophosphatase 1 | 0.007 | 20 | 8 | 0.41 |
| ENSMUSG00000109171 | Spag7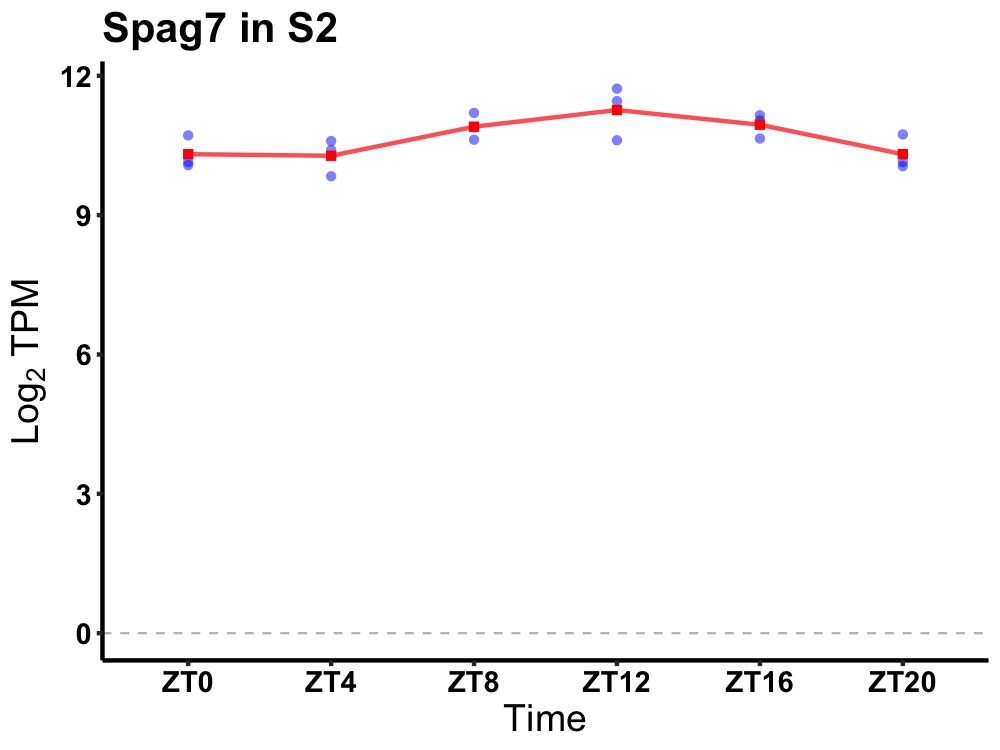 |
sperm associated antigen 7 | 0.036 | 20 | 14 | 0.40 |
| ENSMUSG00000036376 | mt-Nd2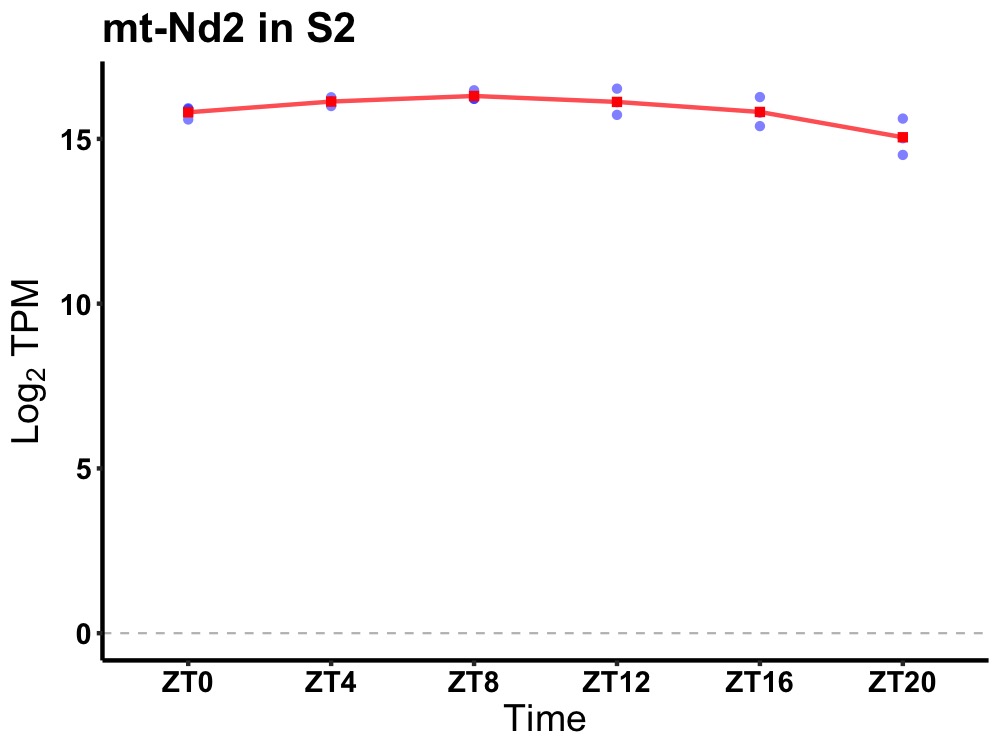 |
mitochondrially encoded NADH dehydrogenase 2 | 0.014 | 24 | 8 | 0.40 |
| ENSMUSG00000081731 | Calr-ps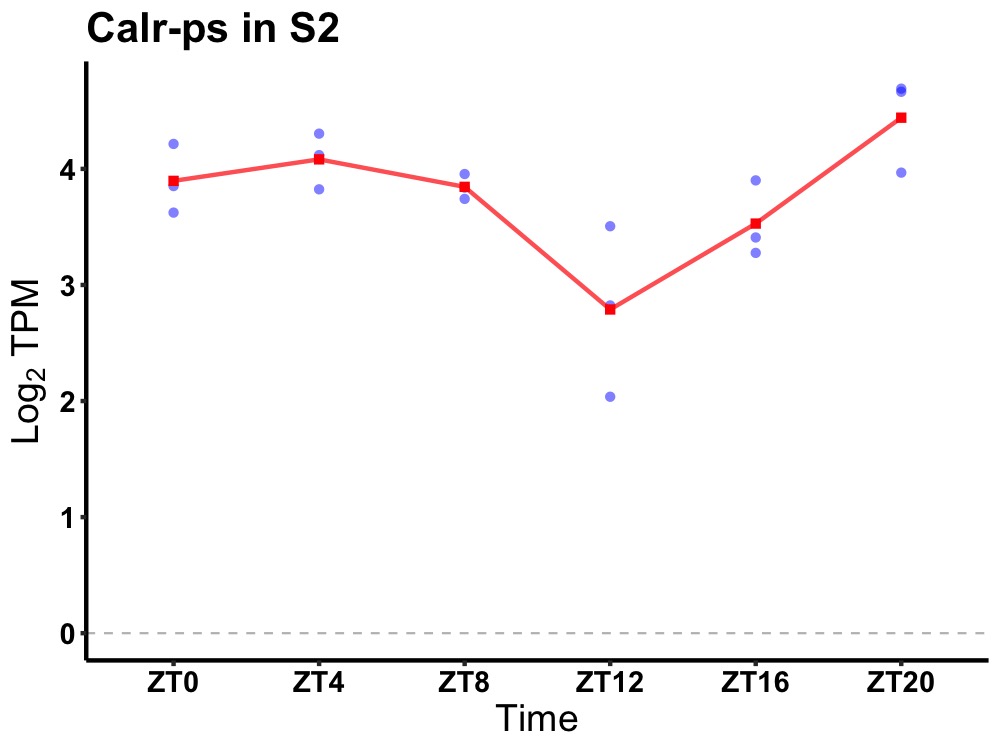 |
calreticulin, pseudogene | 0.010 | 20 | 4 | 0.40 |
| ENSMUSG00000081051 | Dnajc15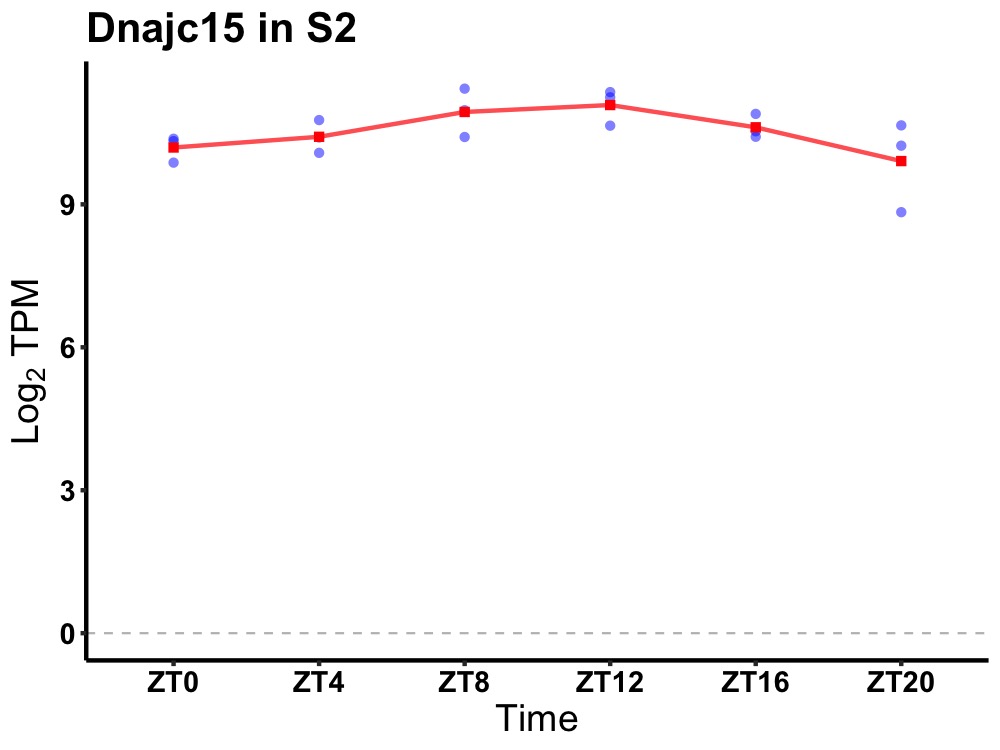 |
DnaJ heat shock protein family (Hsp40) member C15 | 0.014 | 20 | 12 | 0.40 |
| ENSMUSG00000035540 | Trappc3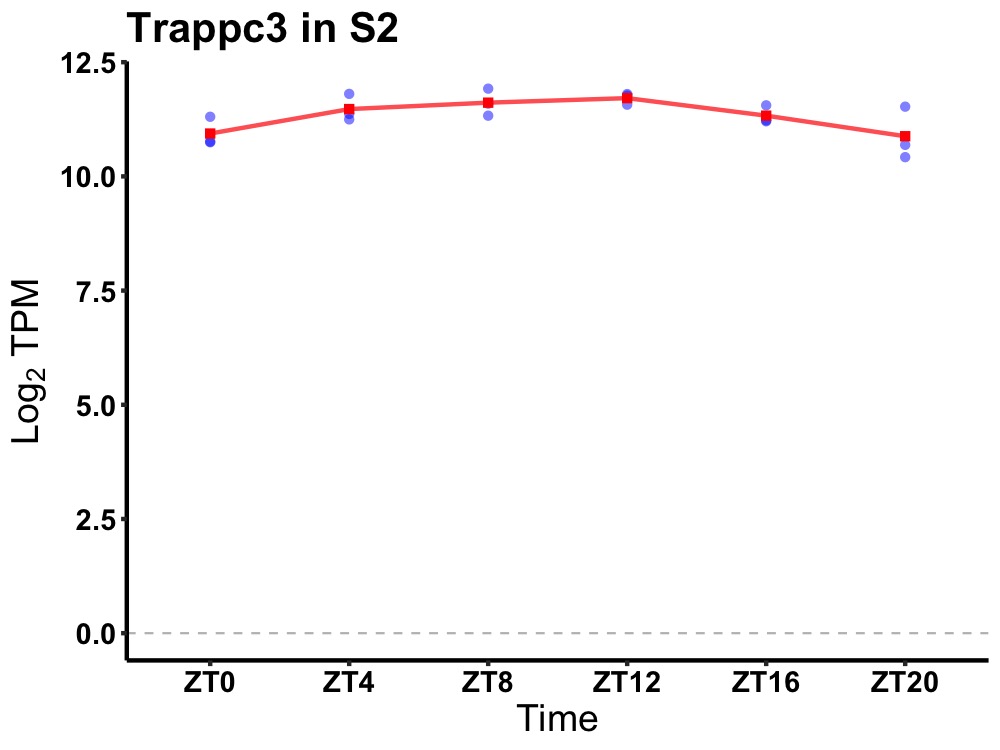 |
trafficking protein particle complex 3 | 0.014 | 24 | 10 | 0.39 |
| ENSMUSG00000021585 | Coq8a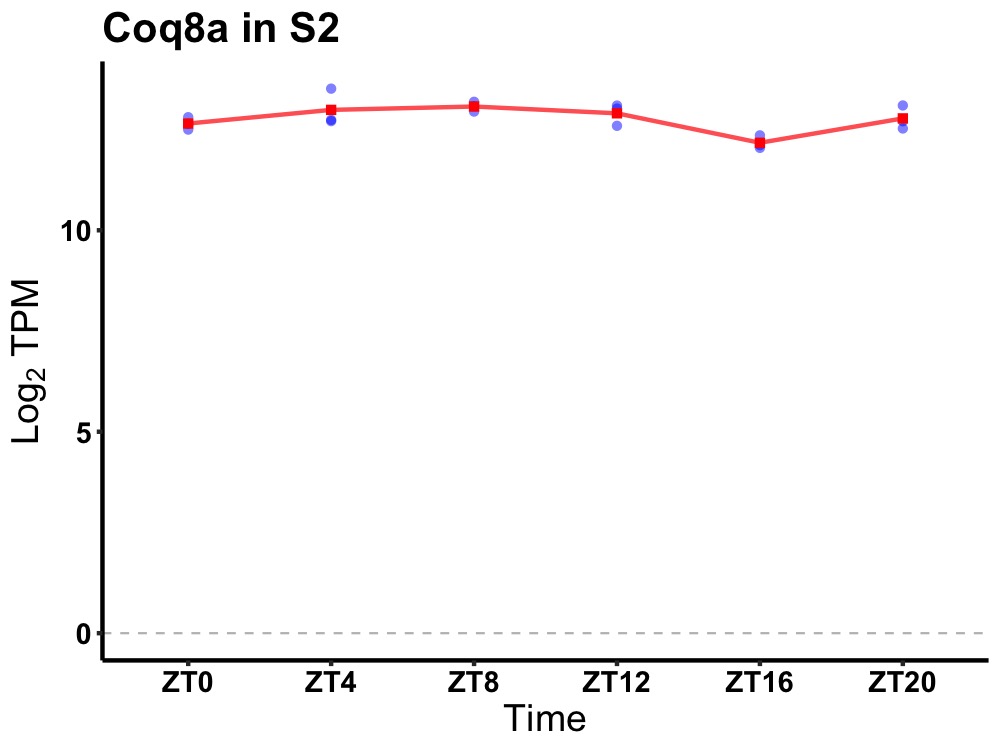 |
coenzyme Q8A | 0.014 | 20 | 8 | 0.39 |
| ENSMUSG00000106558 | Gm5552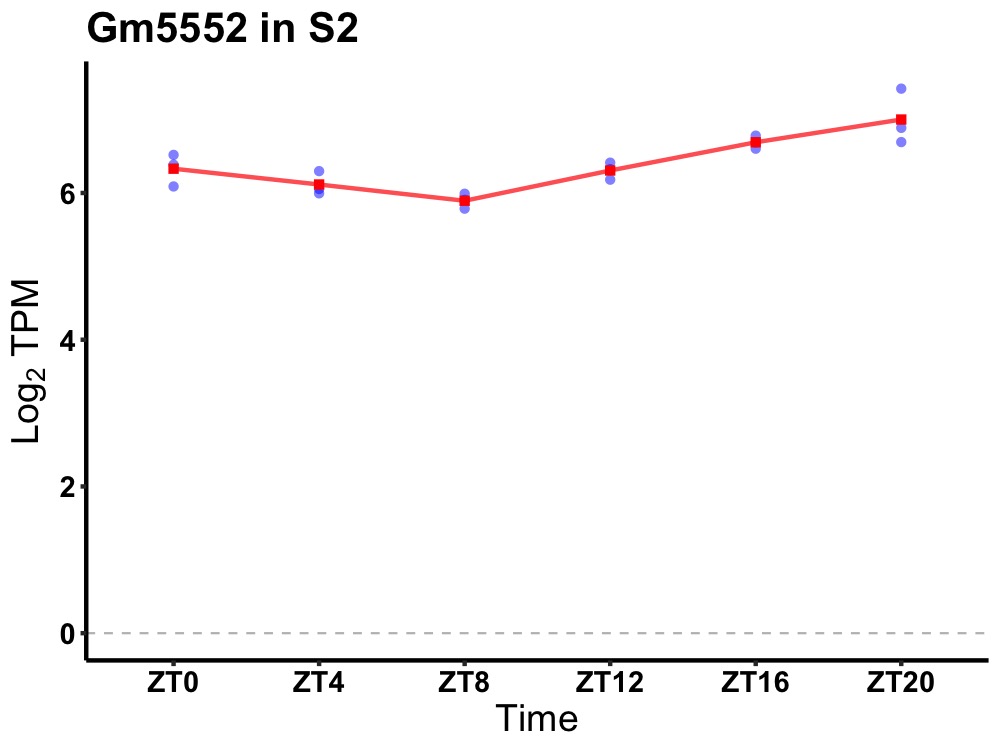 |
predicted gene 5552 | 0.000 | 24 | 20 | 0.39 |
| ENSMUSG00000067719 | Gm10221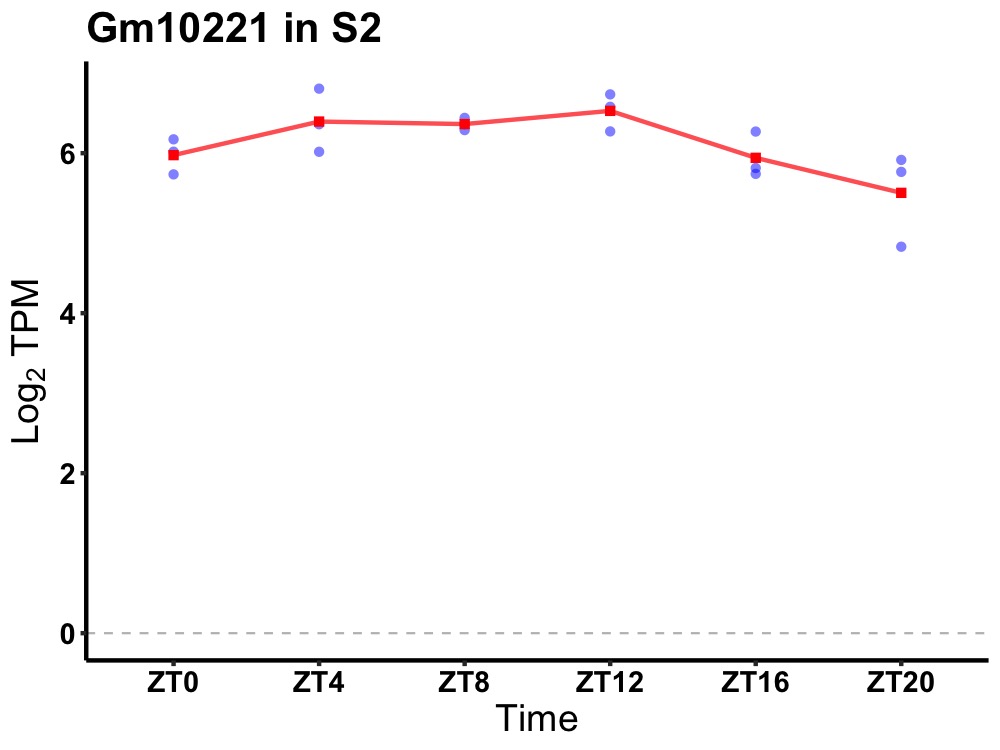 |
predicted gene 10221 | 0.019 | 24 | 10 | 0.39 |
| ENSMUSG00000081434 | Acot4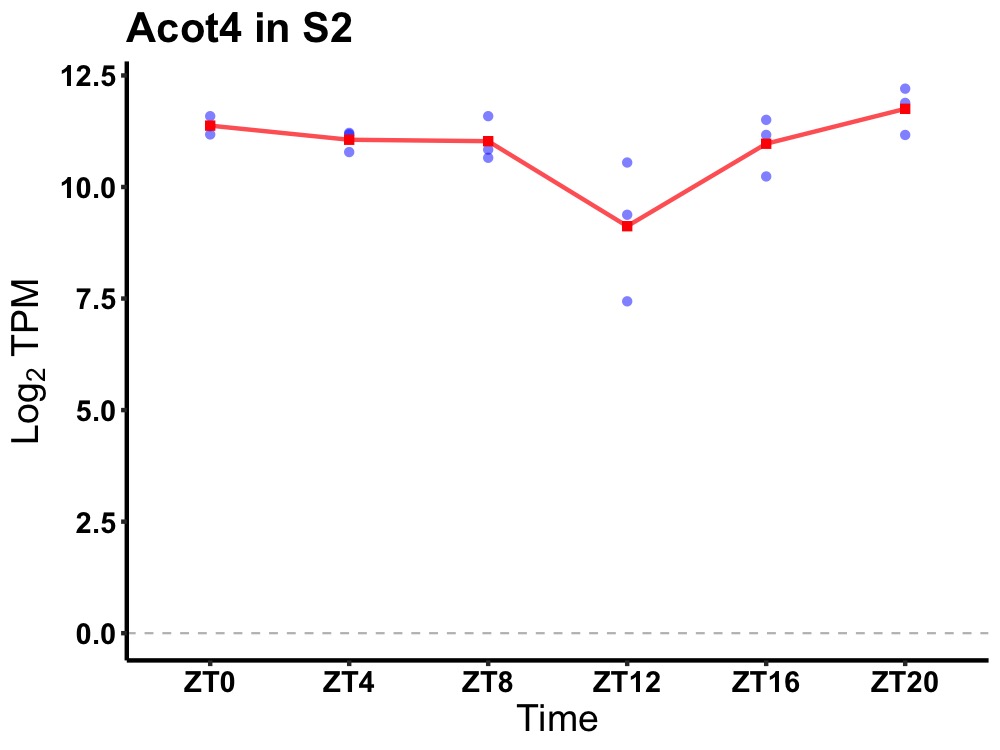 |
acyl-CoA thioesterase 4 | 0.049 | 20 | 2 | 0.38 |
| ENSMUSG00000081431 | Cfdp1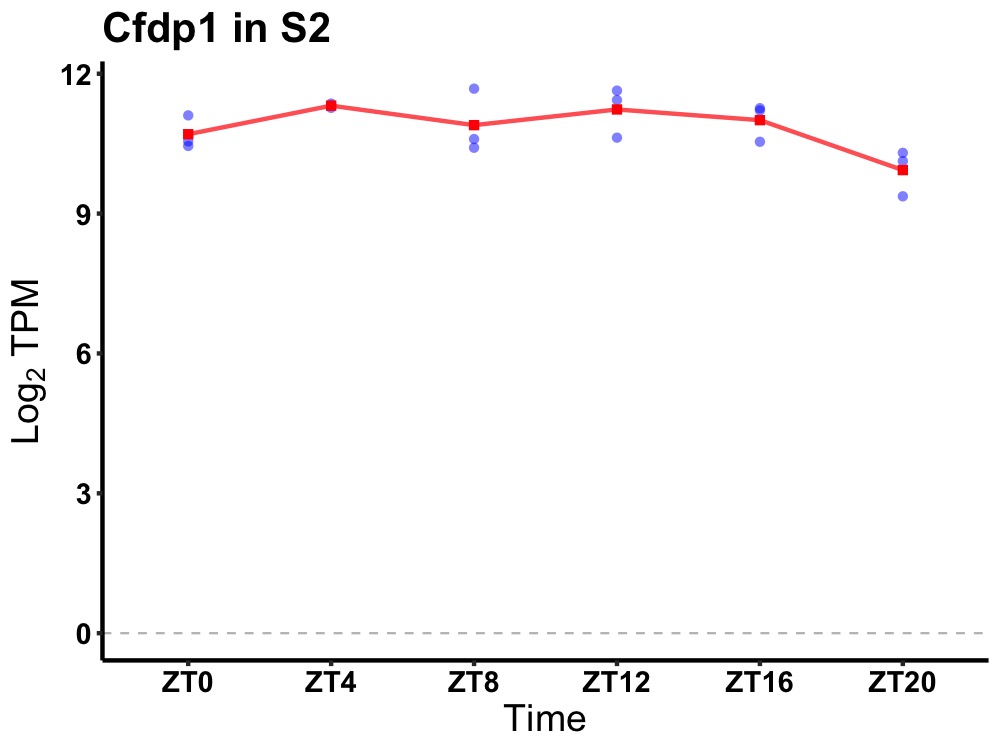 |
craniofacial development protein 1 | 0.049 | 24 | 10 | 0.38 |
| ENSMUSG00000000326 | Slc25a11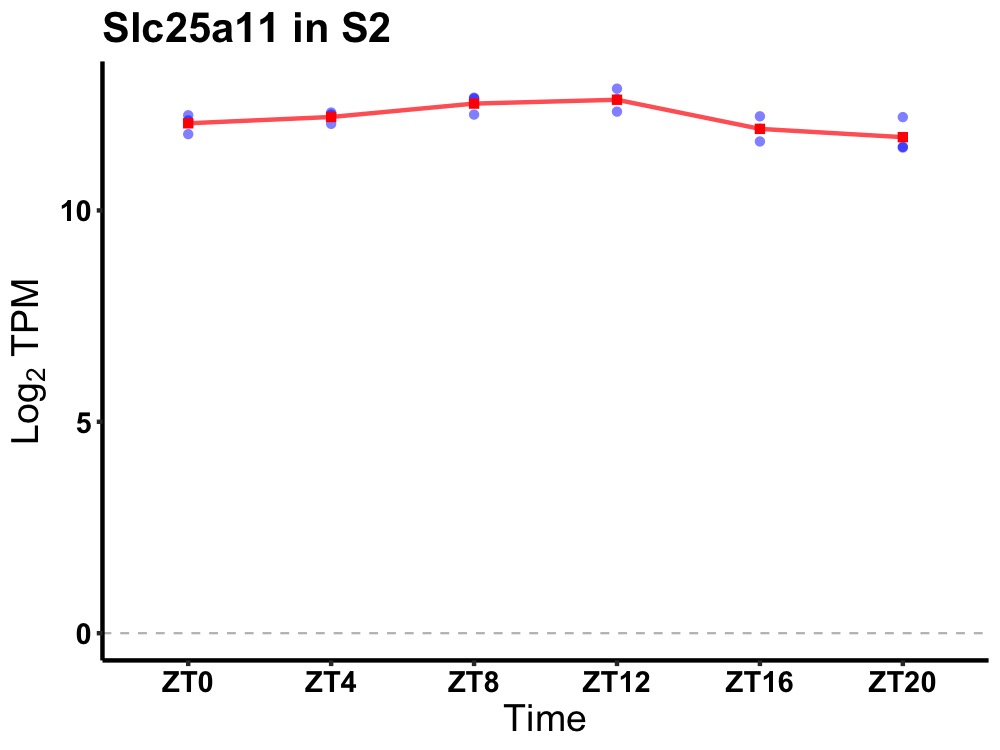 |
solute carrier family 25 (mitochondrial carrier oxoglutarate carrier), member 11 | 0.002 | 24 | 10 | 0.38 |
| ENSMUSG00000108652 | AC160336.1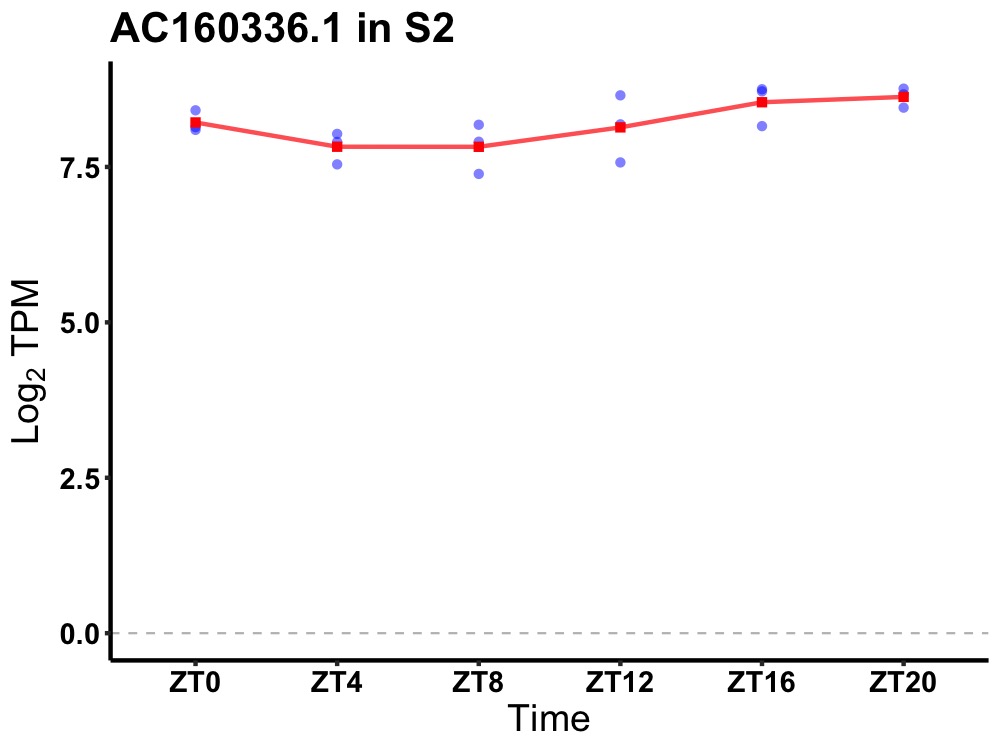 |
novel transcript | 0.010 | 24 | 18 | 0.38 |
| ENSMUSG00000031517 | Prps1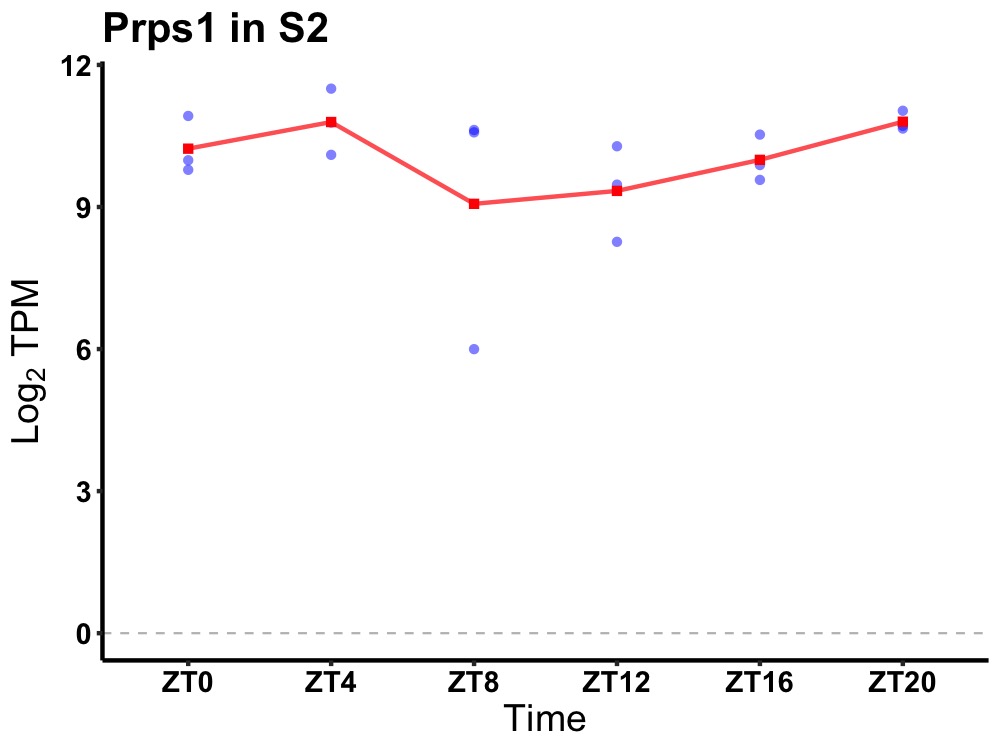 |
phosphoribosyl pyrophosphate synthetase 1 | 0.049 | 20 | 4 | 0.38 |
| ENSMUSG00000017493 | AW209491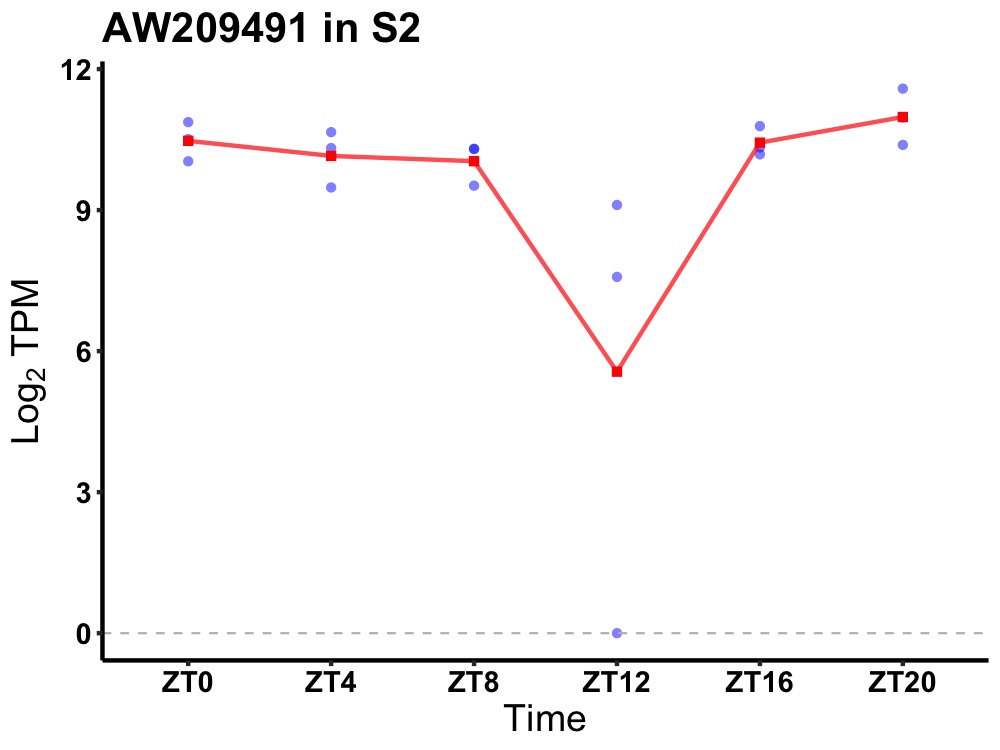 |
UPF0415 protein C7orf25 homolog | 0.010 | 20 | 2 | 0.38 |
| ENSMUSG00000084260 | Gm12844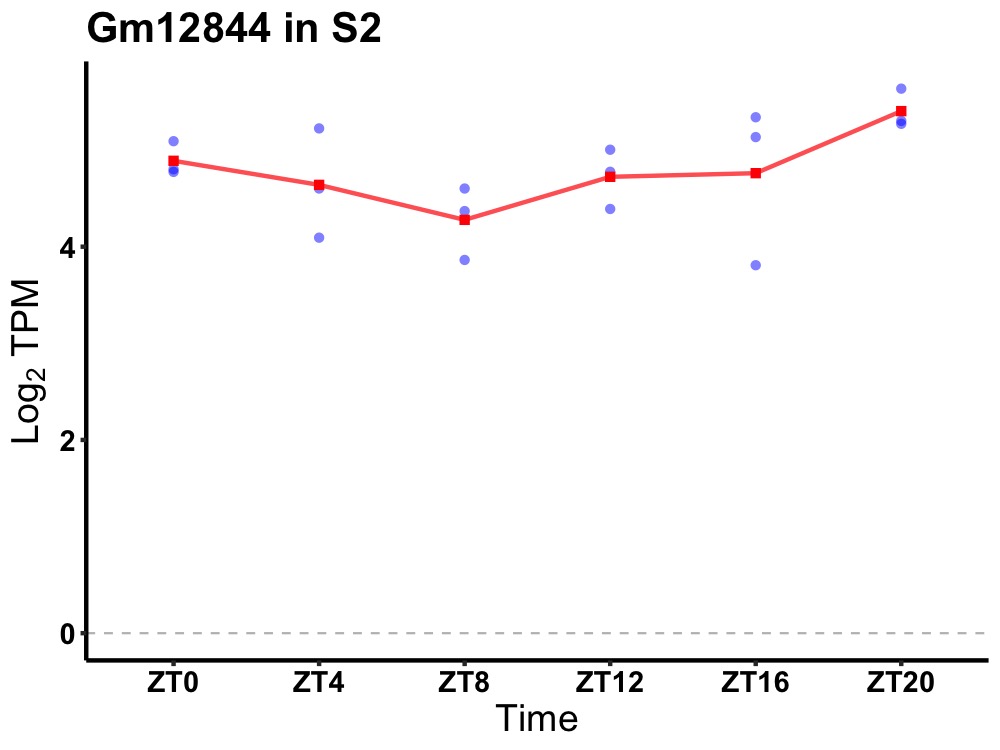 |
predicted gene 12844 | 0.036 | 24 | 20 | 0.38 |
| ENSMUSG00000026688 | Lonp1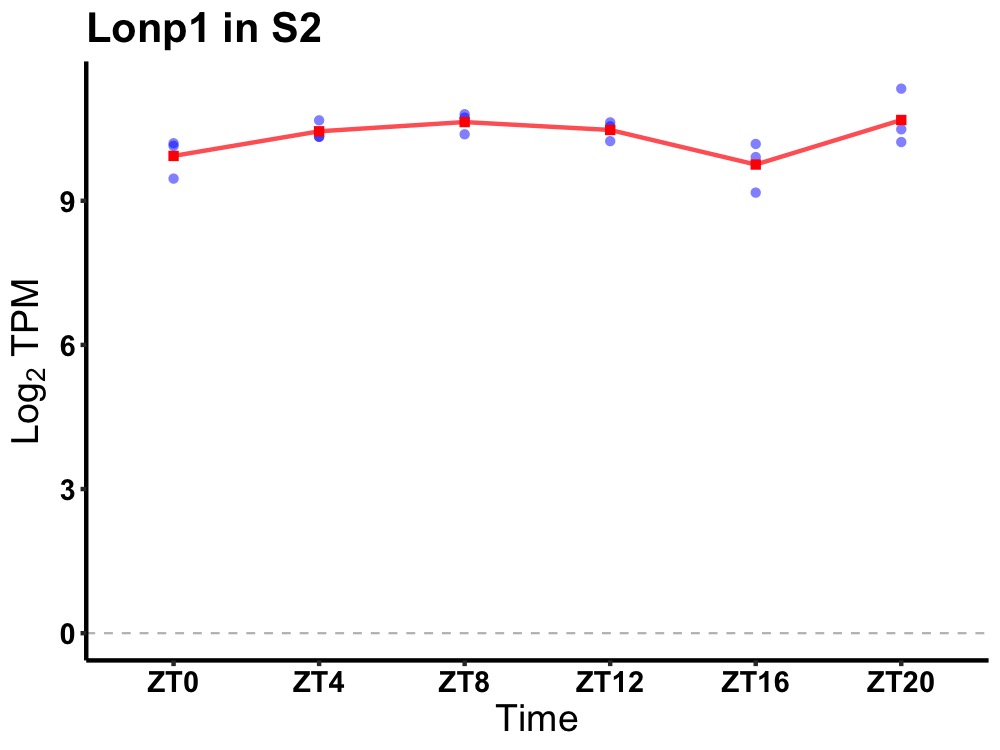 |
lon peptidase 1, mitochondrial | 0.007 | 20 | 8 | 0.38 |
| ENSMUSG00000115808 | Ubl5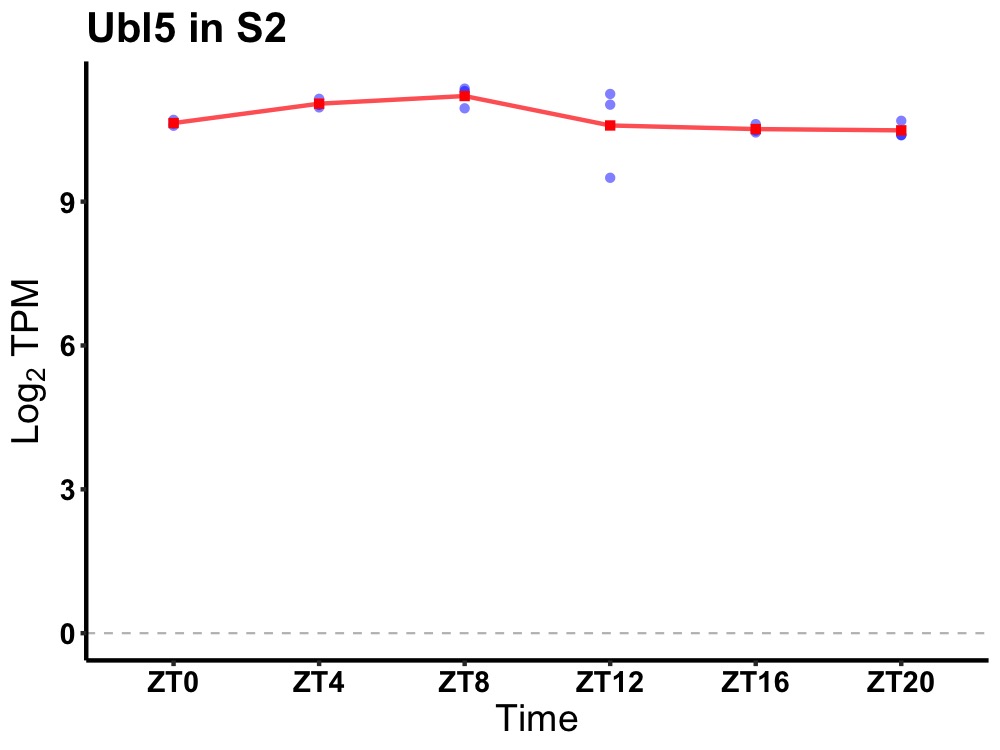 |
ubiquitin-like 5 | 0.014 | 24 | 8 | 0.37 |
| ENSMUSG00000024527 | Serpinf2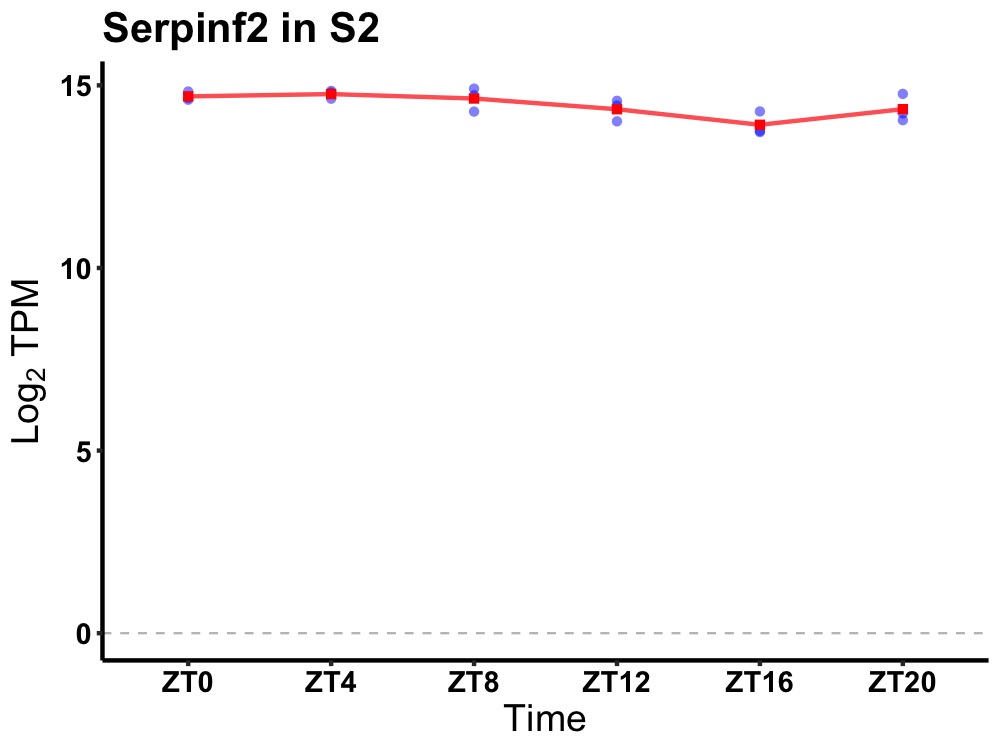 |
serine (or cysteine) peptidase inhibitor, clade F, member 2 | 0.027 | 24 | 6 | 0.37 |
| ENSMUSG00000072501 | Hscb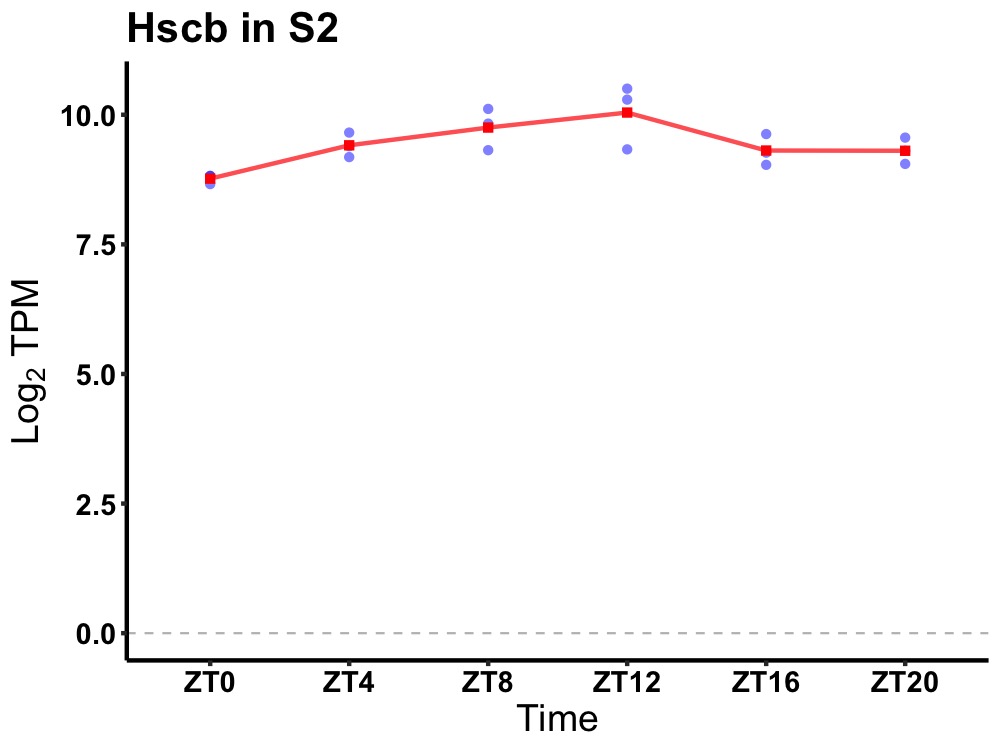 |
HscB iron-sulfur cluster co-chaperone | 0.010 | 24 | 12 | 0.36 |
| ENSMUSG00000025103 | Cox5b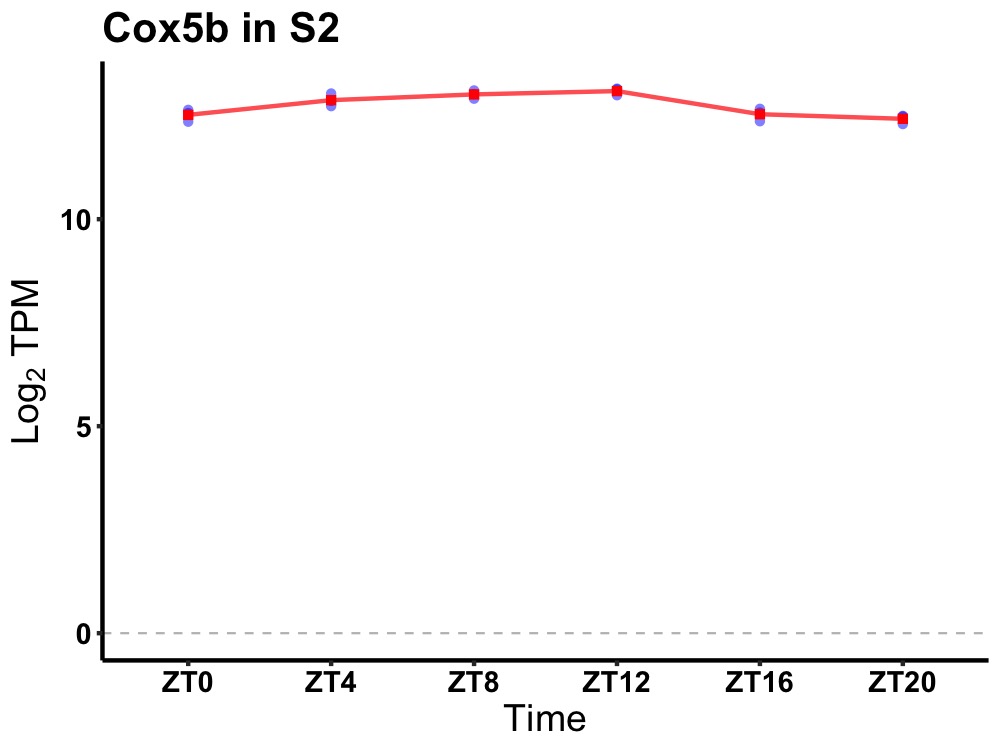 |
cytochrome c oxidase subunit 5B | 0.001 | 24 | 10 | 0.36 |
| ENSMUSG00000104664 | Aamdc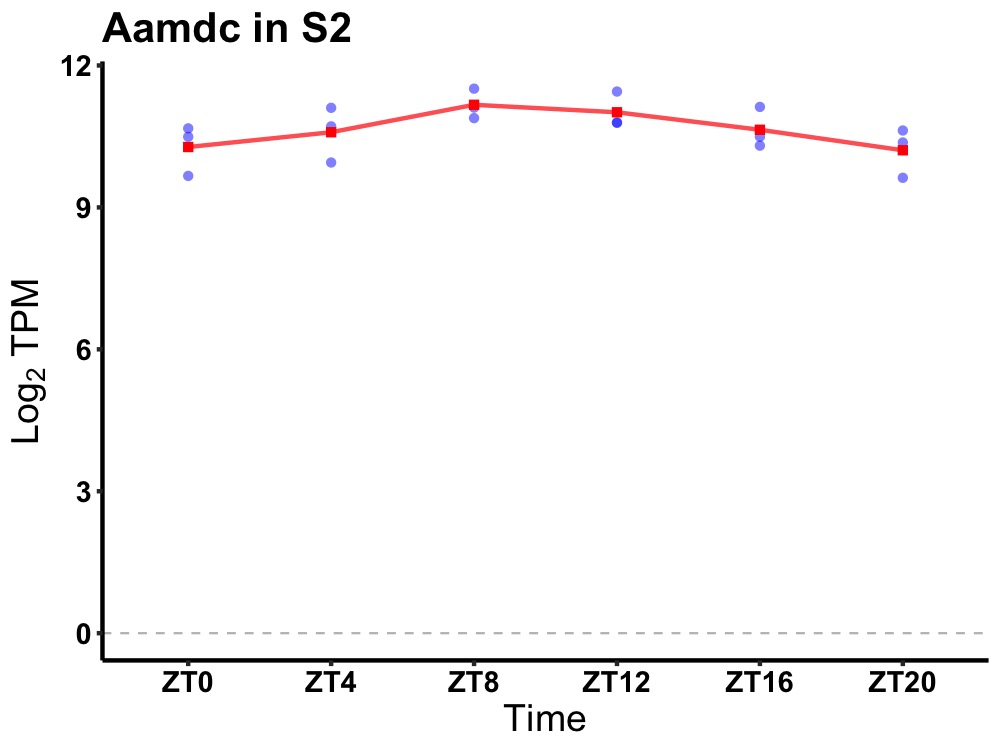 |
adipogenesis associated Mth938 domain containing | 0.027 | 24 | 10 | 0.36 |
| ENSMUSG00000078440 | Psmc5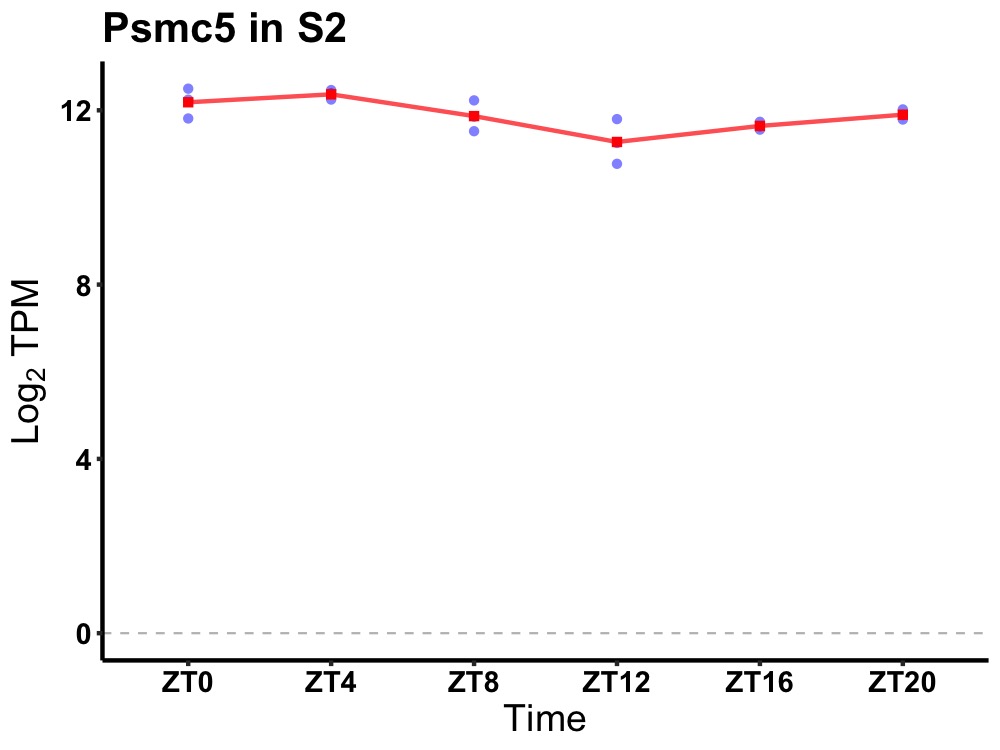 |
protease (prosome, macropain) 26S subunit, ATPase 5 | 0.005 | 24 | 2 | 0.36 |
| ENSMUSG00000021939 | Smdt1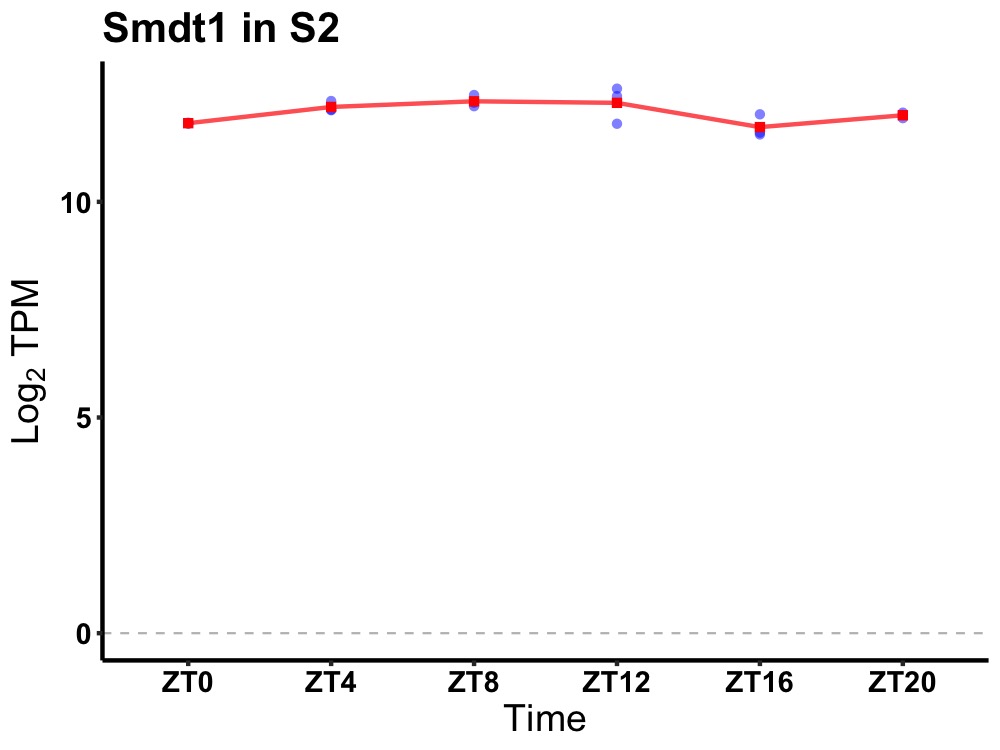 |
single-pass membrane protein with aspartate rich tail 1 | 0.014 | 20 | 8 | 0.36 |
| ENSMUSG00000084215 | Gm11814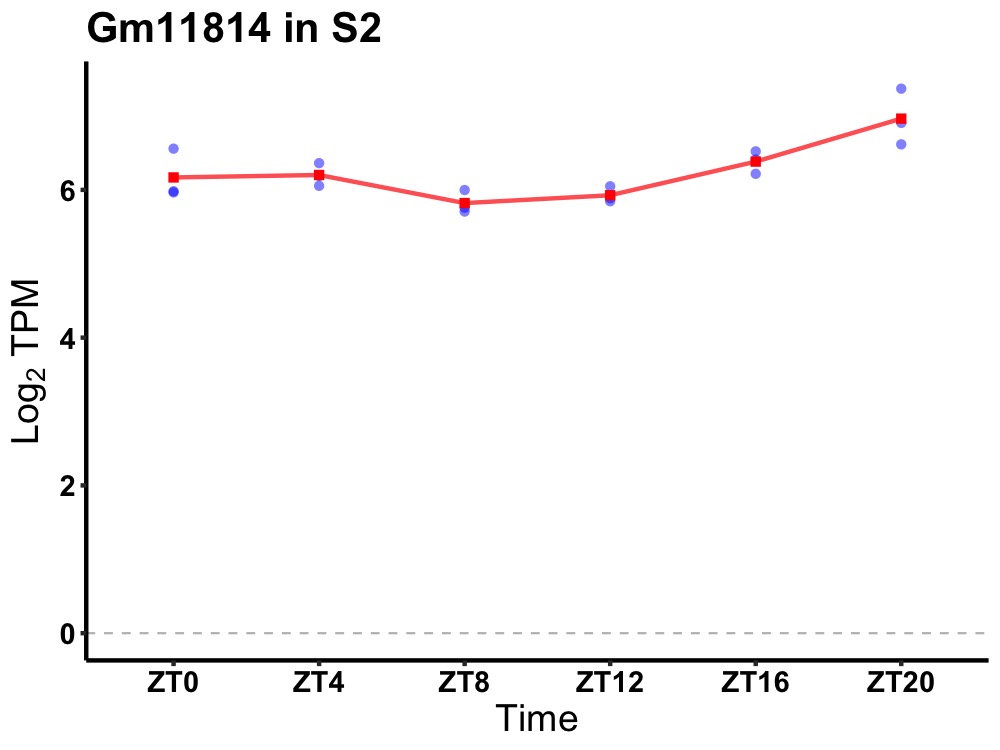 |
predicted gene 11814 | 0.001 | 24 | 22 | 0.36 |
| ENSMUSG00000022387 | Comt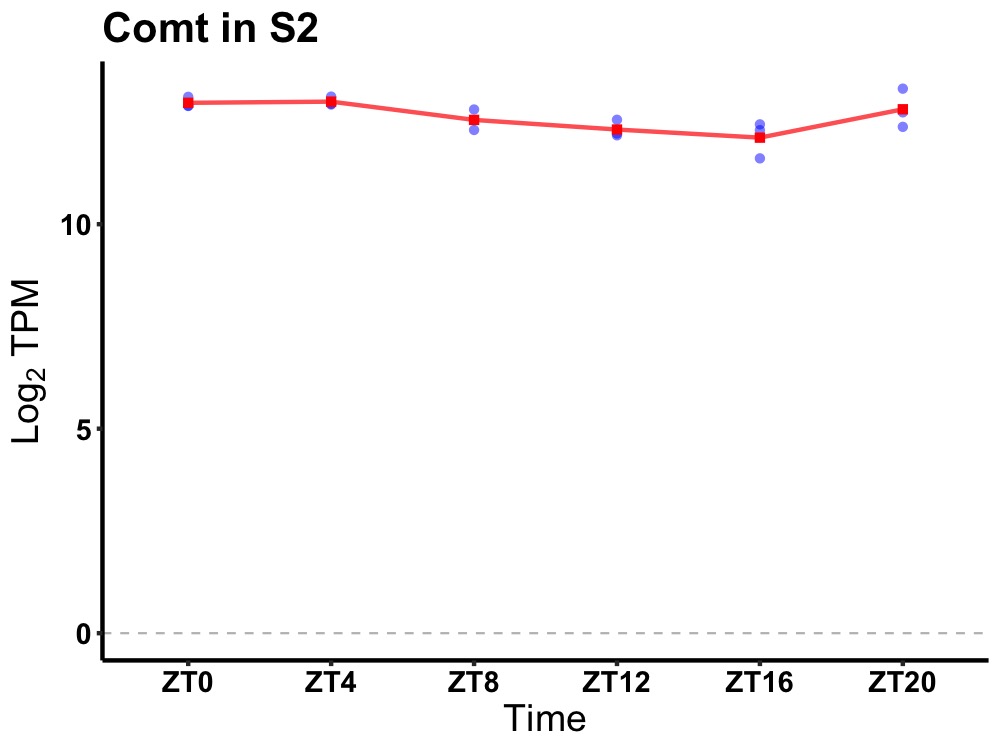 |
catechol-O-methyltransferase | 0.007 | 24 | 4 | 0.36 |
| ENSMUSG00000031751 | Folr1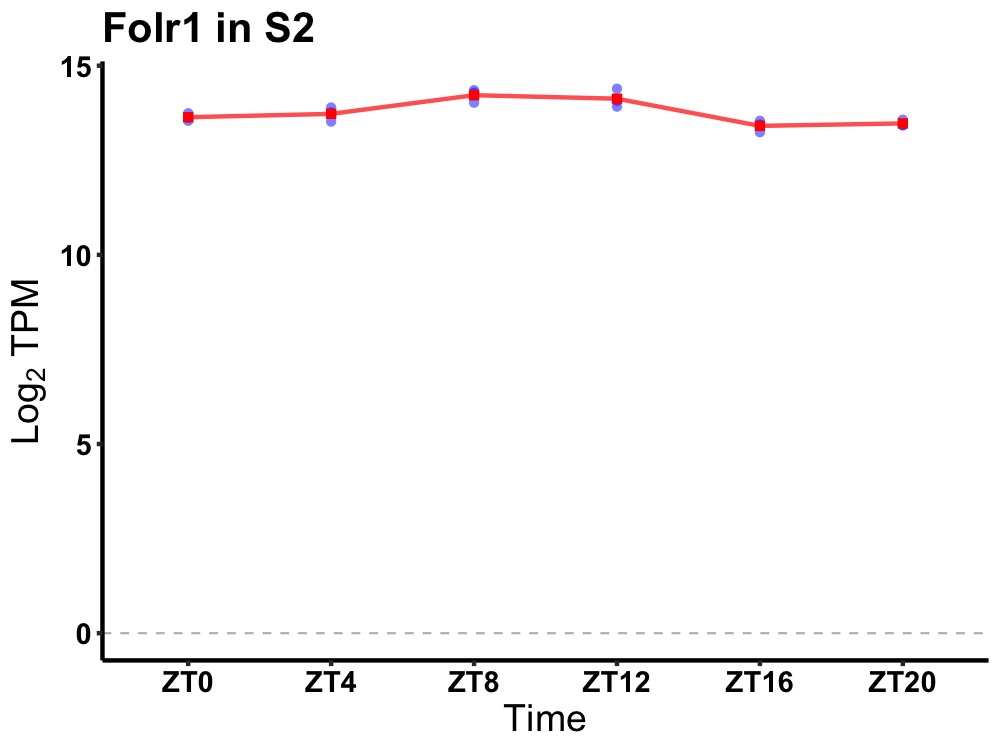 |
folate receptor 1 (adult) | 0.010 | 24 | 10 | 0.36 |
| ENSMUSG00000037458 | Cldn10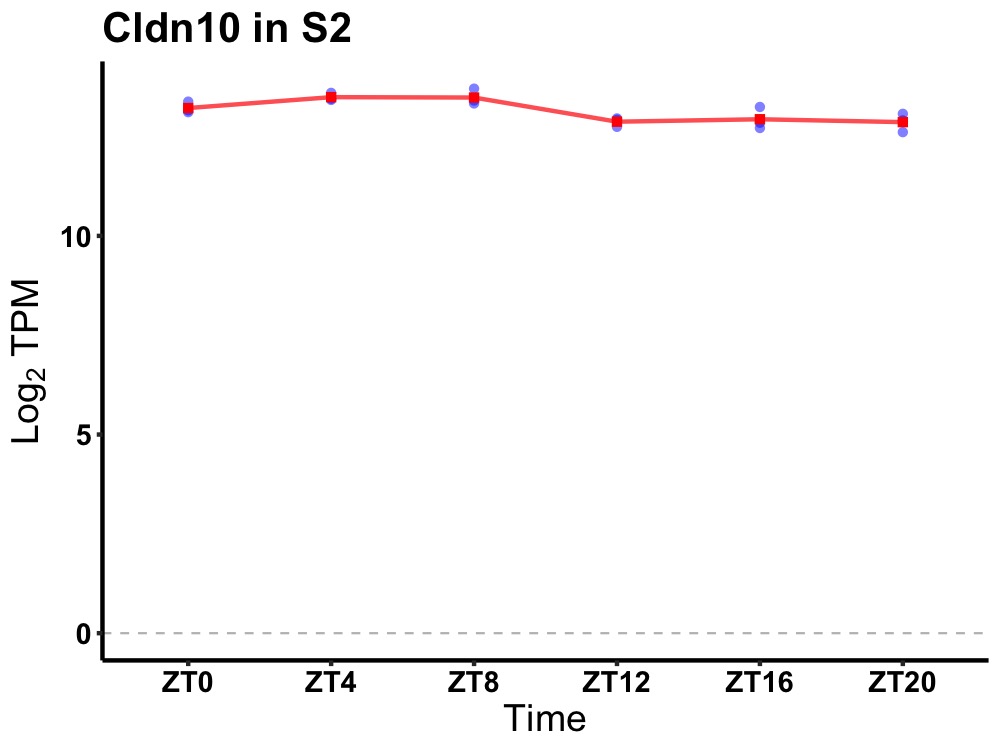 |
claudin 10 | 0.001 | 24 | 6 | 0.35 |
| ENSMUSG00000020109 | Klhl9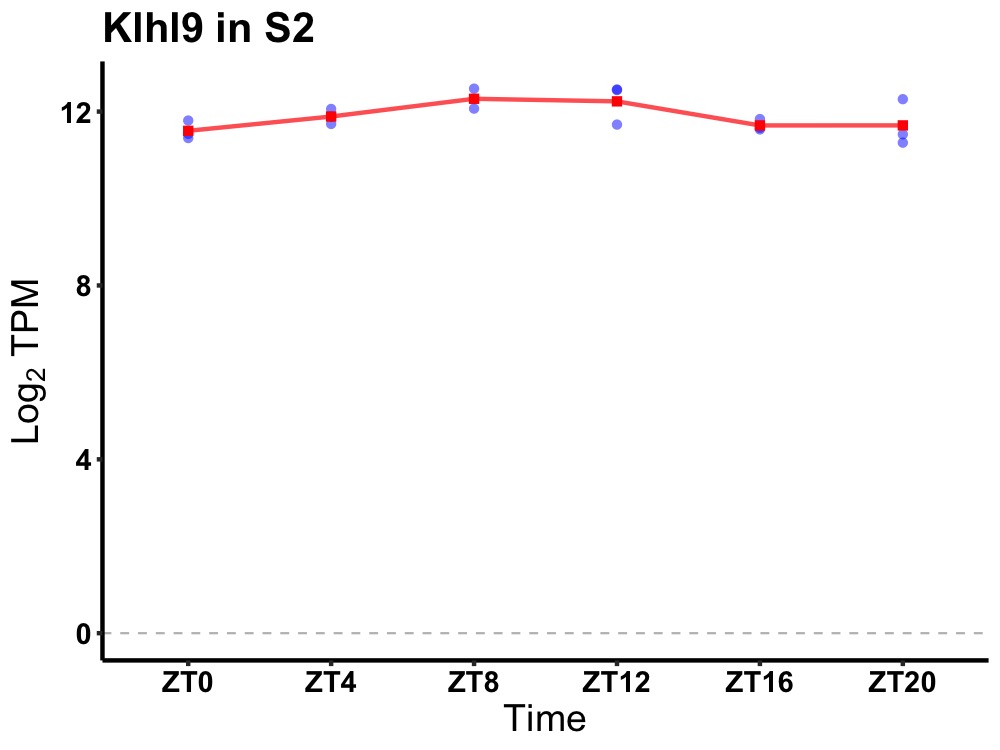 |
kelch-like 9 | 0.014 | 24 | 10 | 0.35 |
| ENSMUSG00000105965 | Cycs-ps2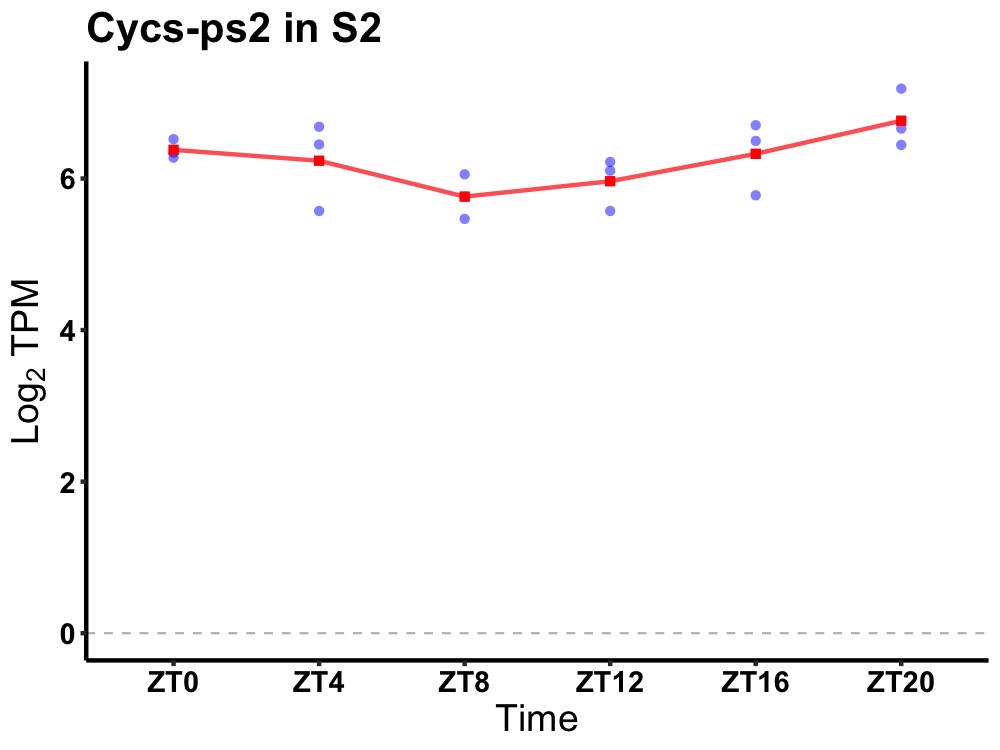 |
cytochrome c, pseudogene 2 | 0.049 | 24 | 22 | 0.35 |
| ENSMUSG00000096726 | Gm5558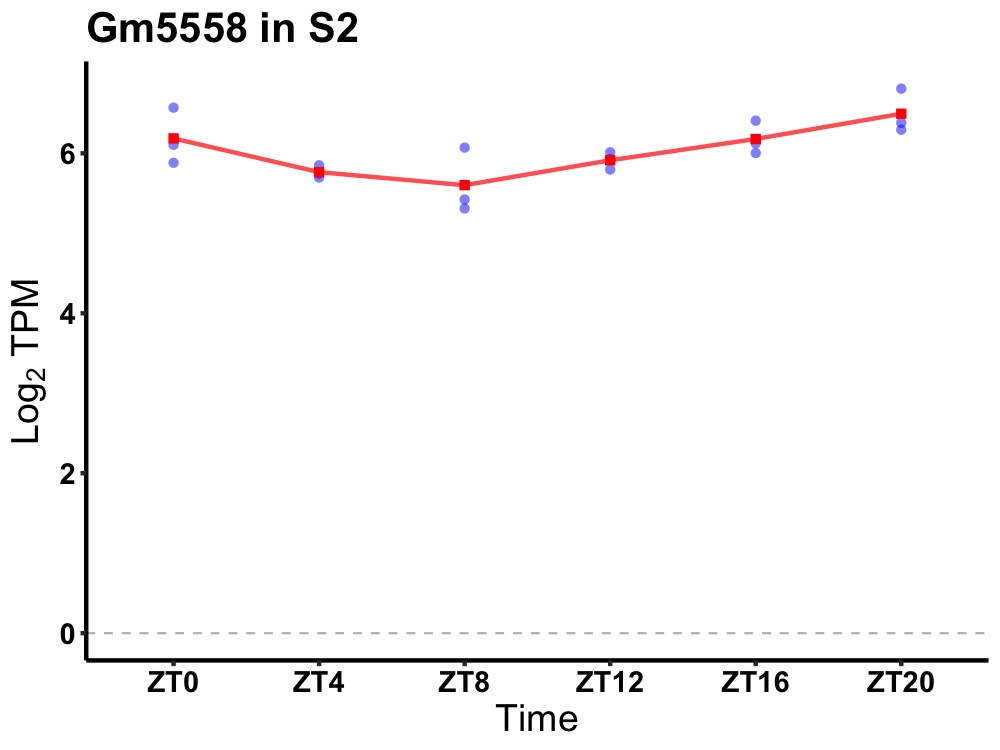 |
predicted gene 5558 | 0.002 | 24 | 20 | 0.35 |
| ENSMUSG00000060038 | Afap1l1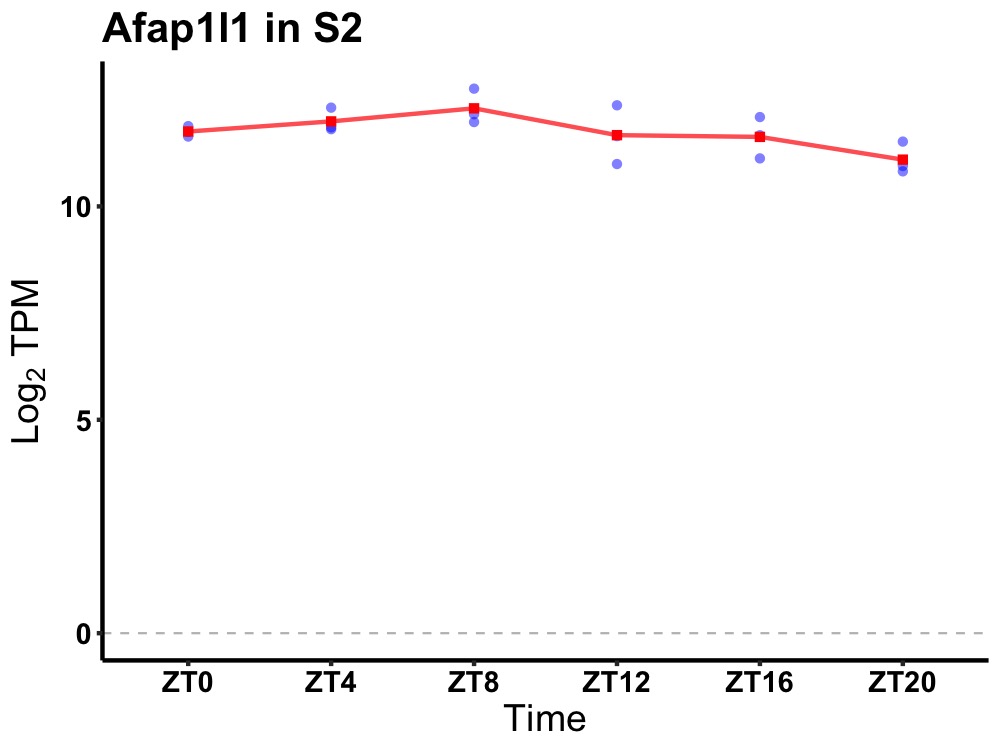 |
actin filament associated protein 1-like 1 | 0.036 | 24 | 8 | 0.35 |
| ENSMUSG00000116614 | Gm46610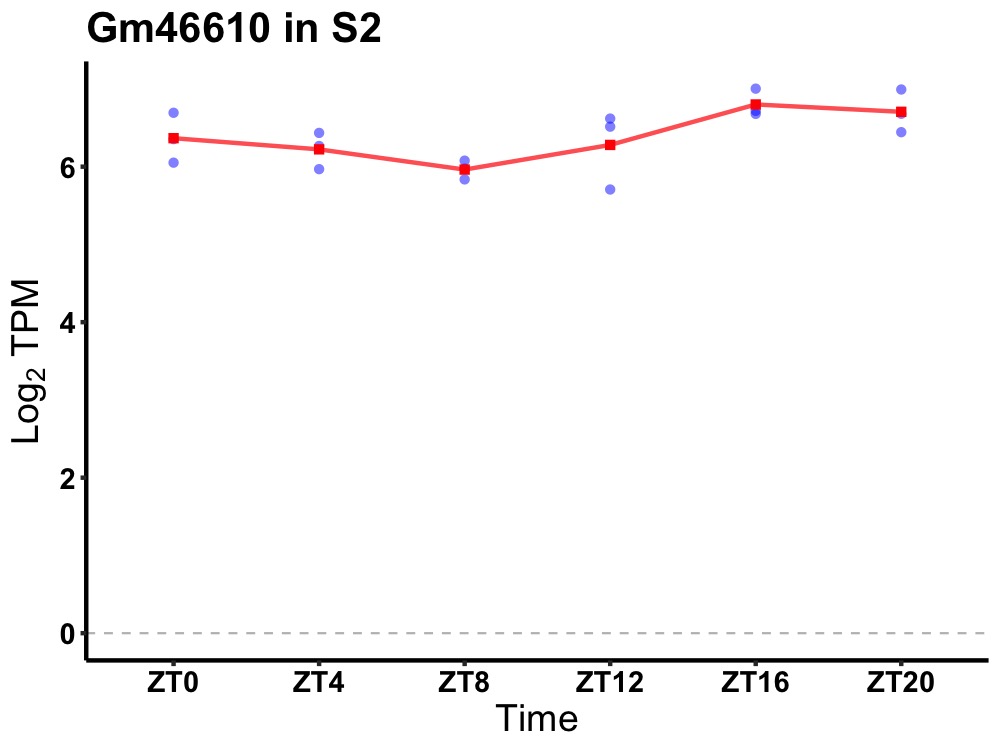 |
predicted gene, 46610 | 0.027 | 24 | 20 | 0.35 |
| ENSMUSG00000114579 | Gm4130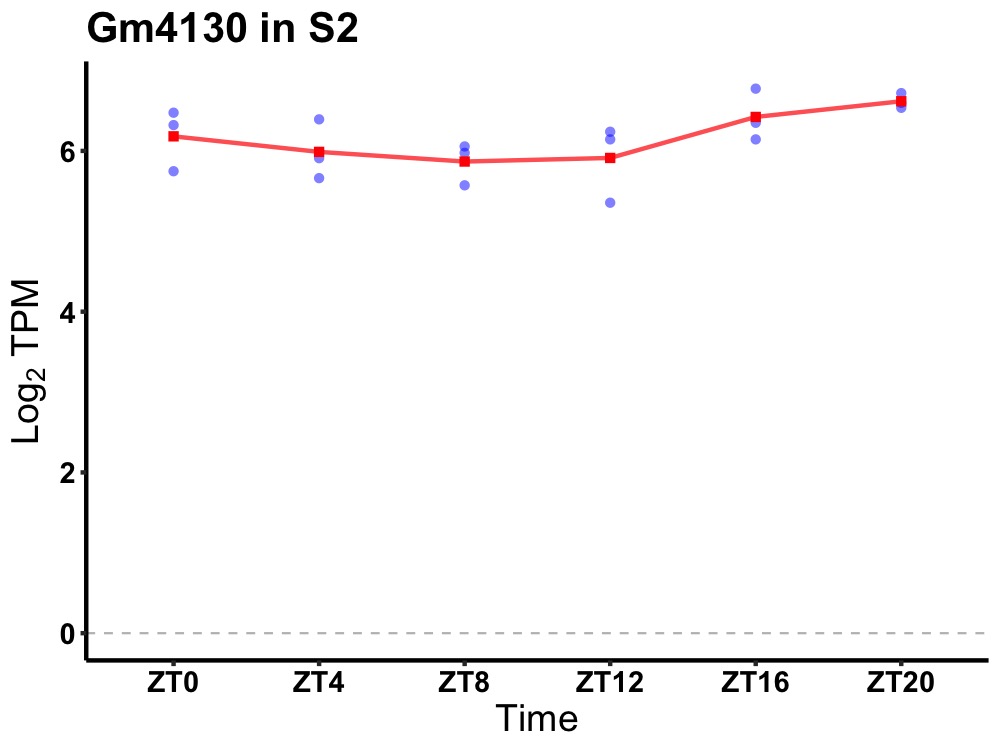 |
predicted gene 4130 | 0.049 | 24 | 20 | 0.35 |
| ENSMUSG00000024039 | Rps27l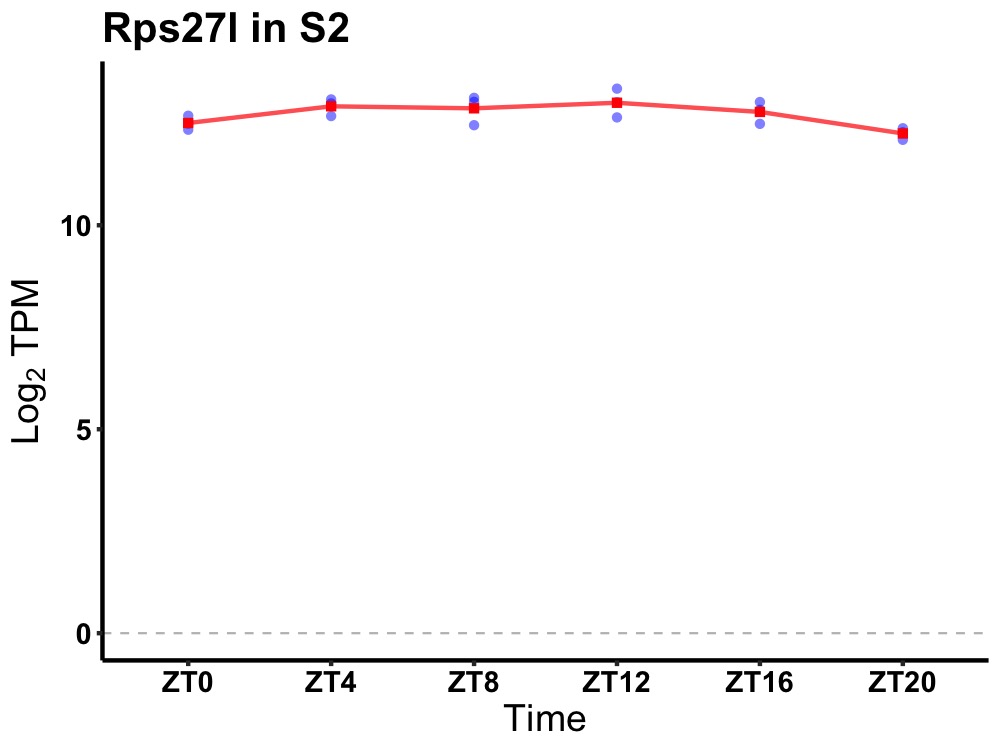 |
ribosomal protein S27-like | 0.049 | 24 | 10 | 0.34 |
| ENSMUSG00000117951 | CR555305.2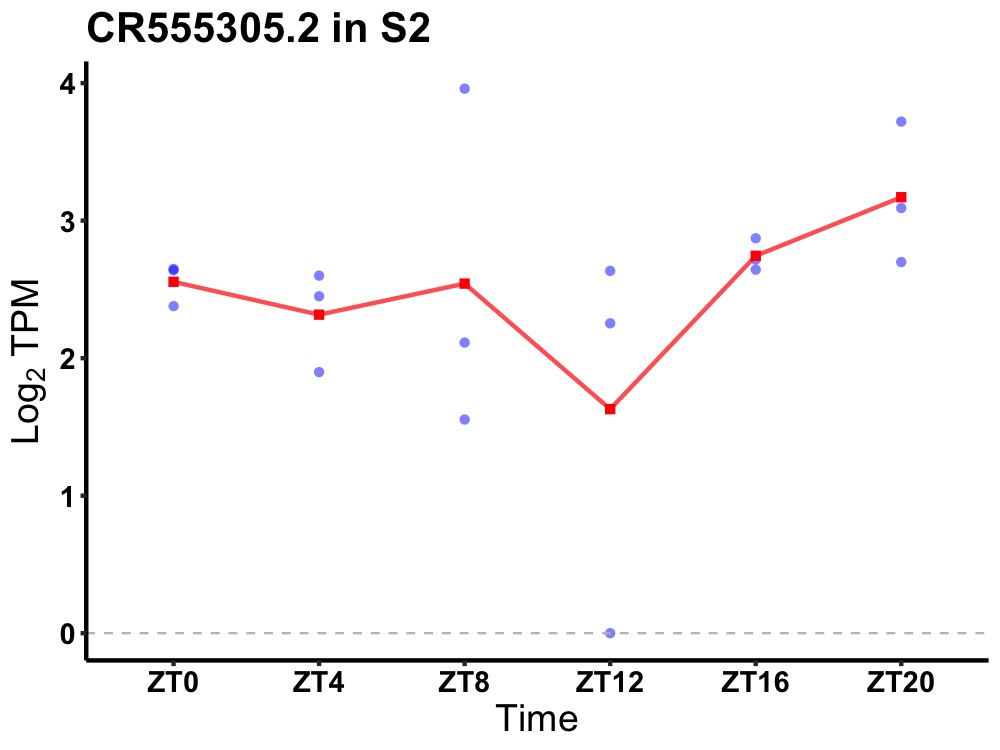 |
none | 0.049 | 24 | 20 | 0.34 |
| ENSMUSG00000035392 | Smpd2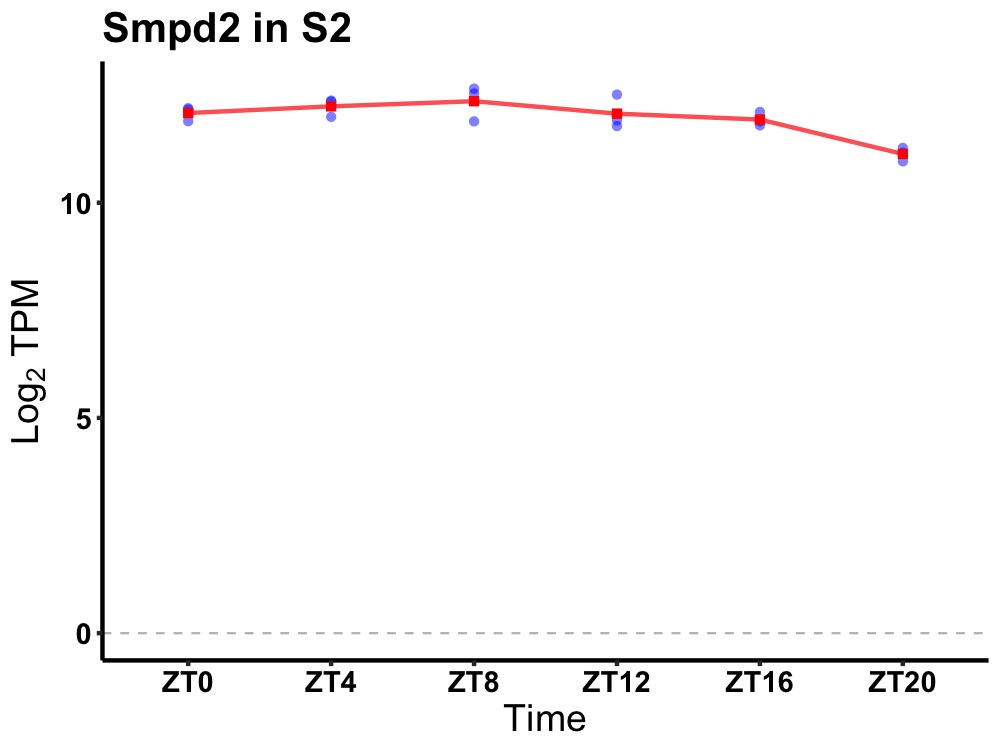 |
sphingomyelin phosphodiesterase 2, neutral | 0.019 | 24 | 8 | 0.34 |
| ENSMUSG00000038871 | Coq9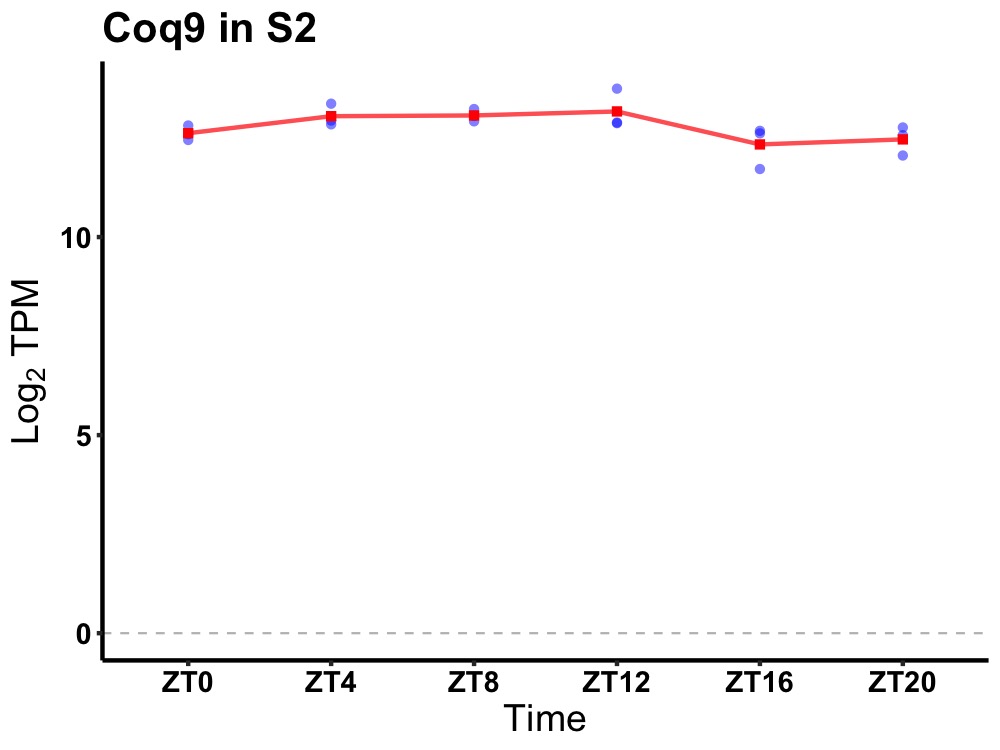 |
coenzyme Q9 | 0.010 | 24 | 10 | 0.34 |
| ENSMUSG00000066809 | Coa6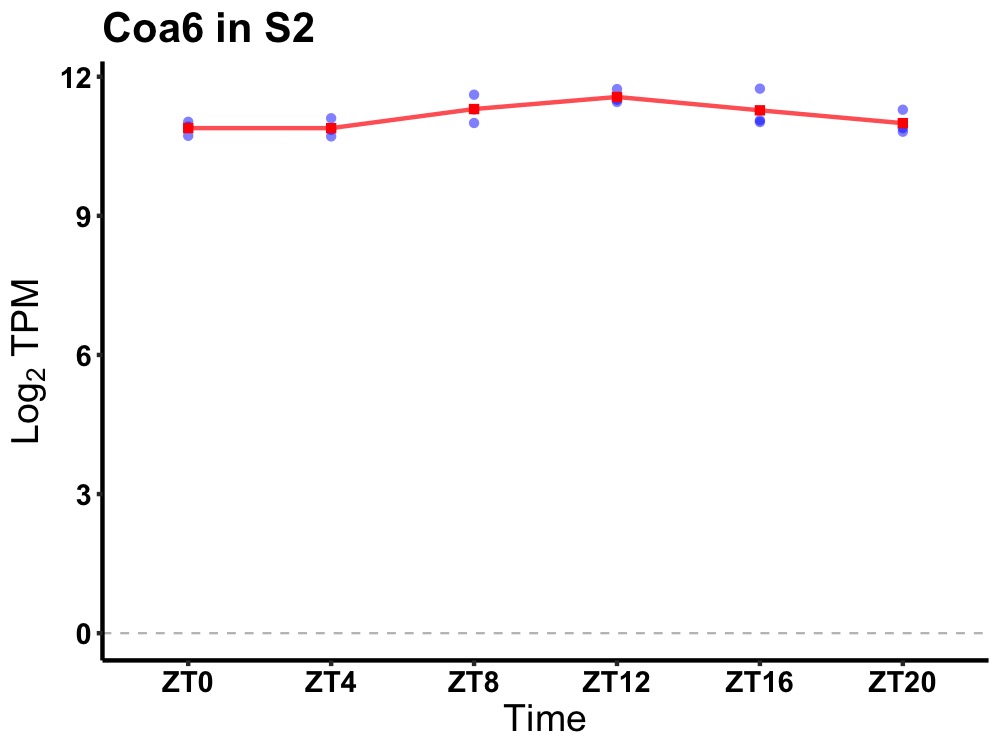 |
cytochrome c oxidase assembly factor 6 | 0.049 | 24 | 14 | 0.33 |
| ENSMUSG00000028973 | Acy3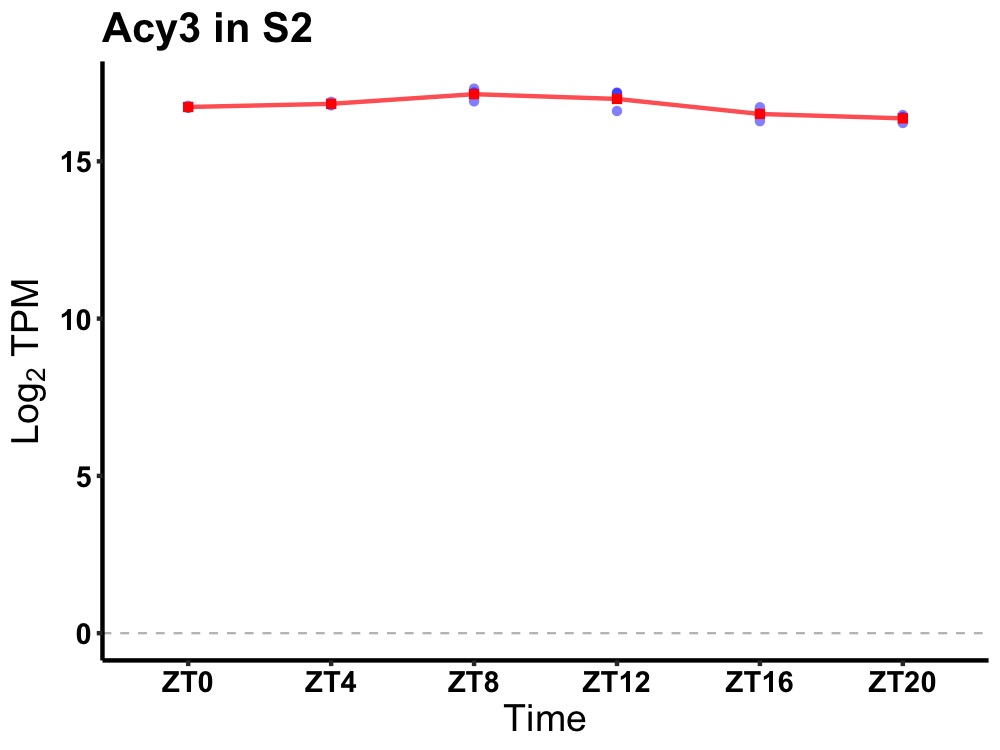 |
aspartoacylase (aminoacylase) 3 | 0.000 | 24 | 8 | 0.33 |
| ENSMUSG00000020122 | Dynlrb1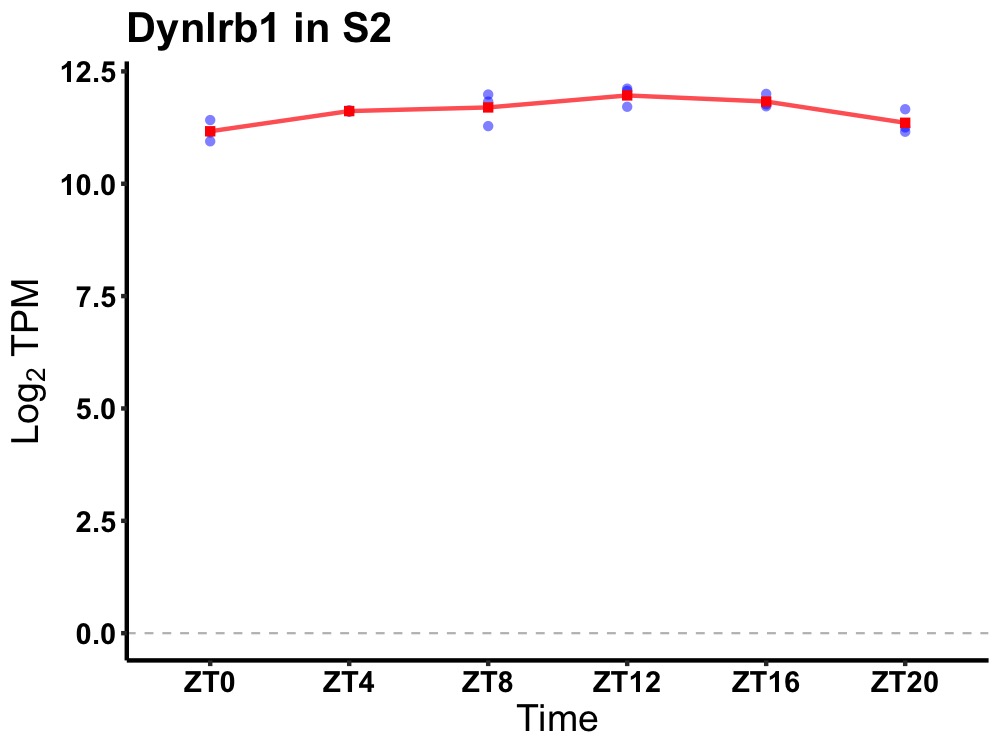 |
dynein light chain roadblock-type 1 | 0.002 | 24 | 12 | 0.33 |
| ENSMUSG00000100405 | Psme2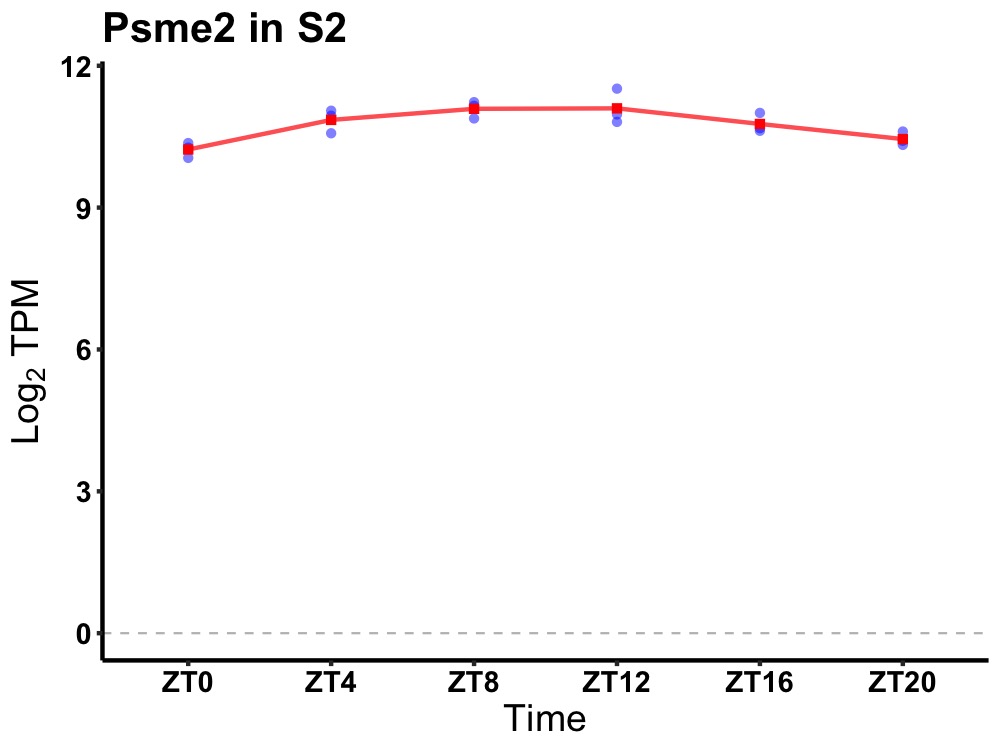 |
proteasome (prosome, macropain) activator subunit 2 (PA28 beta) | 0.001 | 20 | 10 | 0.32 |
| ENSMUSG00000031879 | Commd3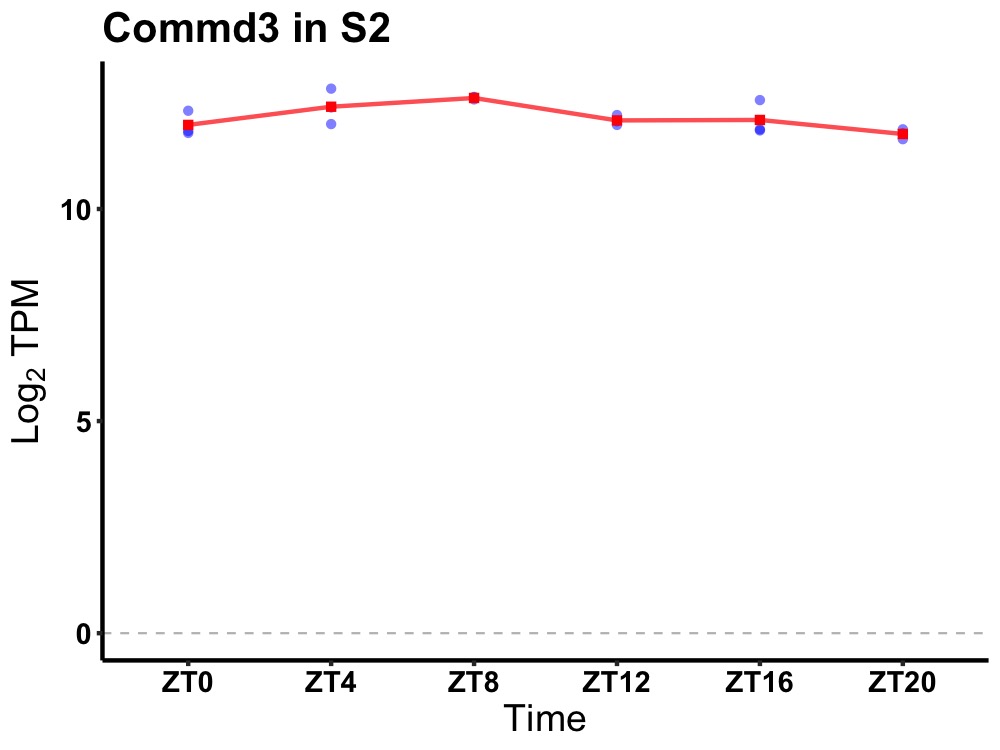 |
COMM domain containing 3 | 0.014 | 24 | 10 | 0.32 |
| ENSMUSG00000027193 | Hoga1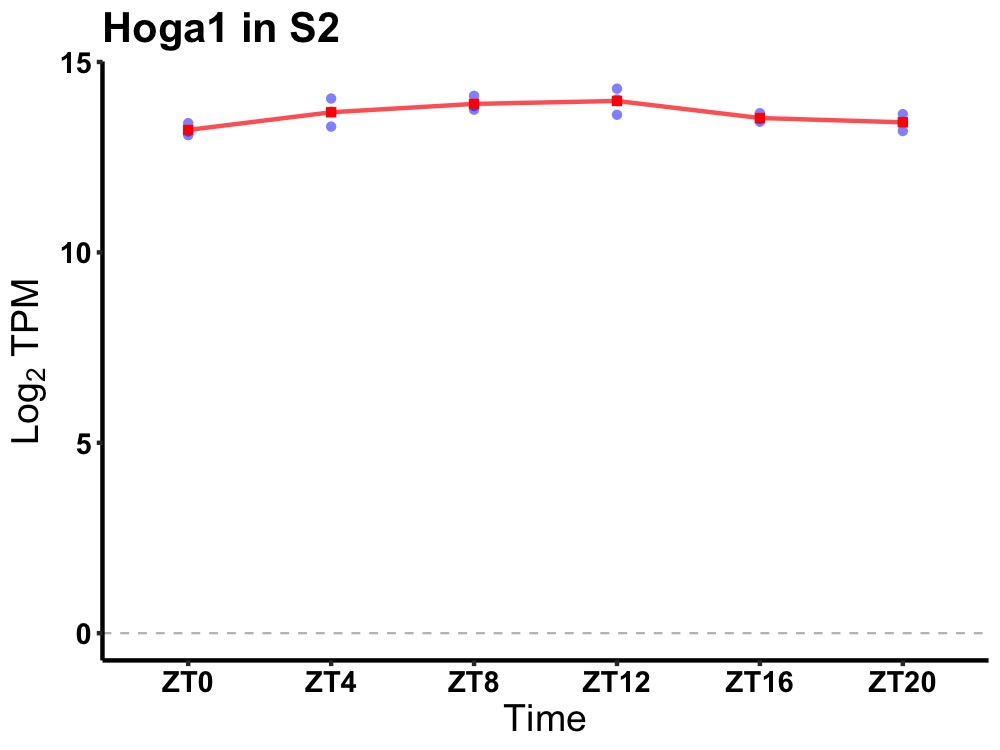 |
4-hydroxy-2-oxoglutarate aldolase 1 | 0.003 | 20 | 10 | 0.32 |
| ENSMUSG00000052520 | Krt8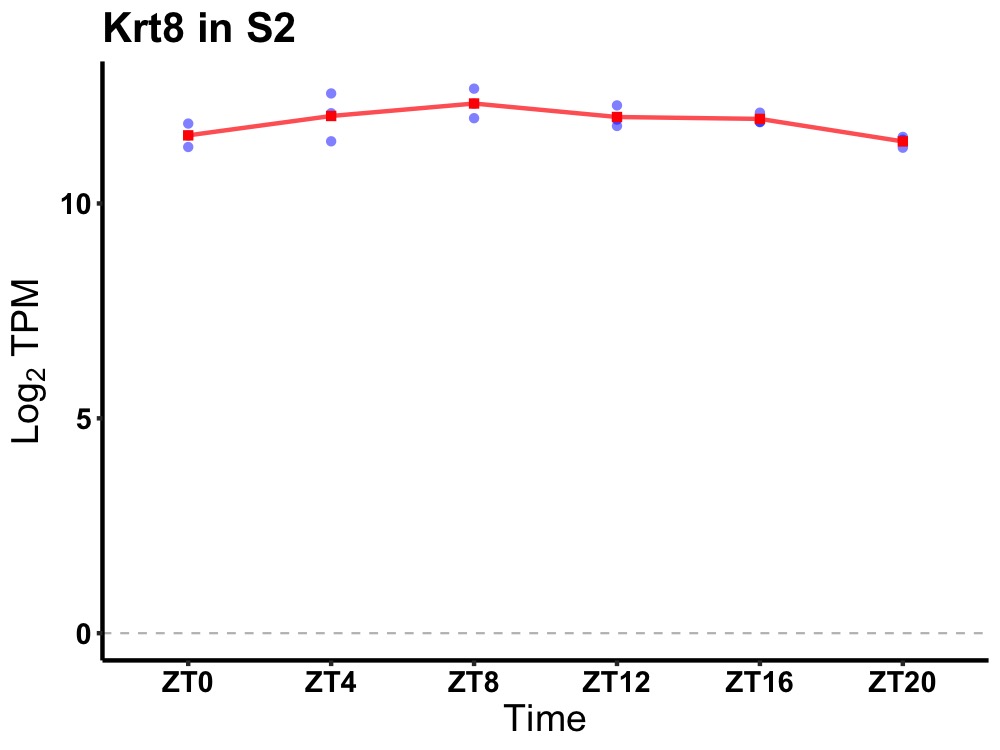 |
keratin 8 | 0.014 | 24 | 10 | 0.32 |
| ENSMUSG00000029681 | Glul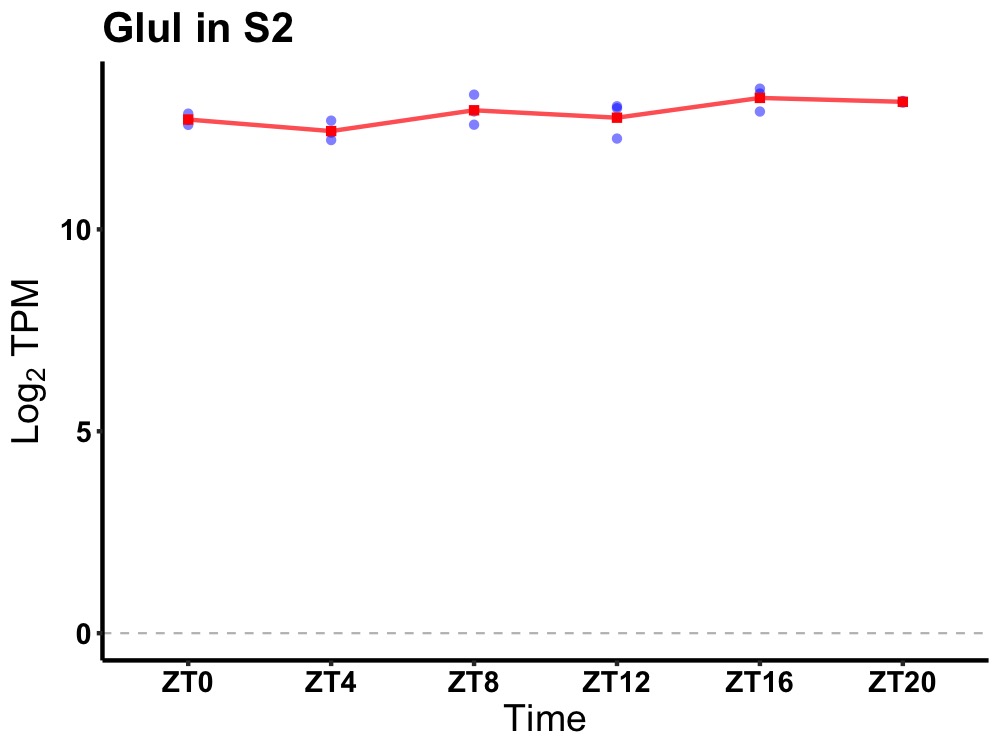 |
glutamate-ammonia ligase (glutamine synthetase) | 0.049 | 24 | 18 | 0.31 |
| ENSMUSG00000080935 | Got2-ps1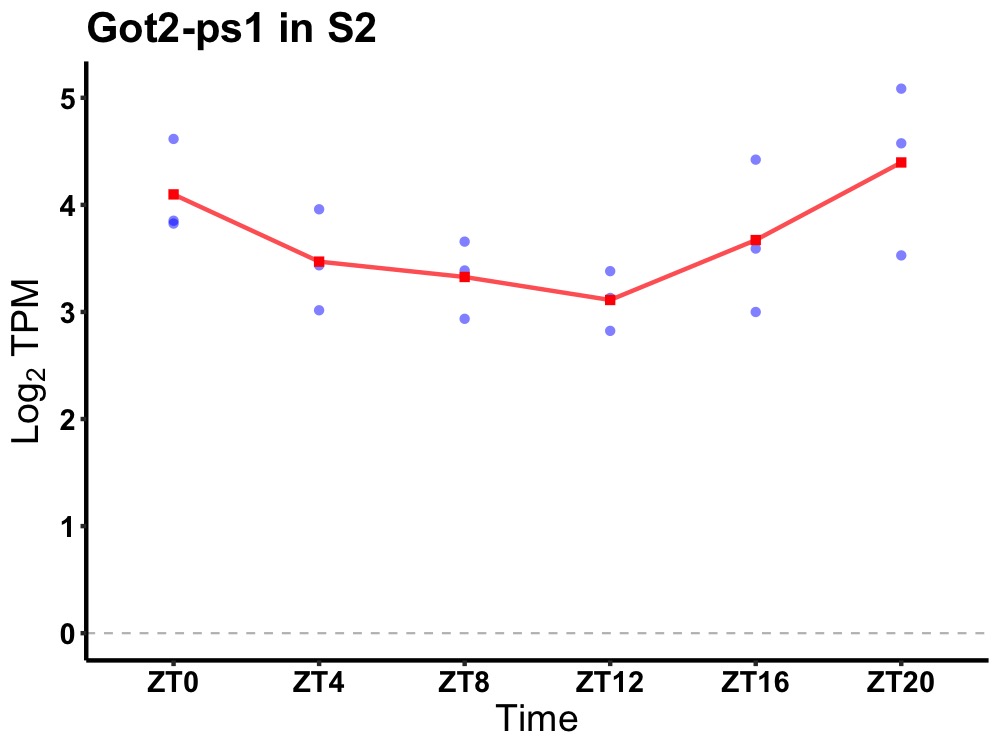 |
glutamatic-oxaloacetic transaminase 2, mitochondrial, pseudogene 1 | 0.049 | 20 | 2 | 0.31 |
| ENSMUSG00000114809 | Ptma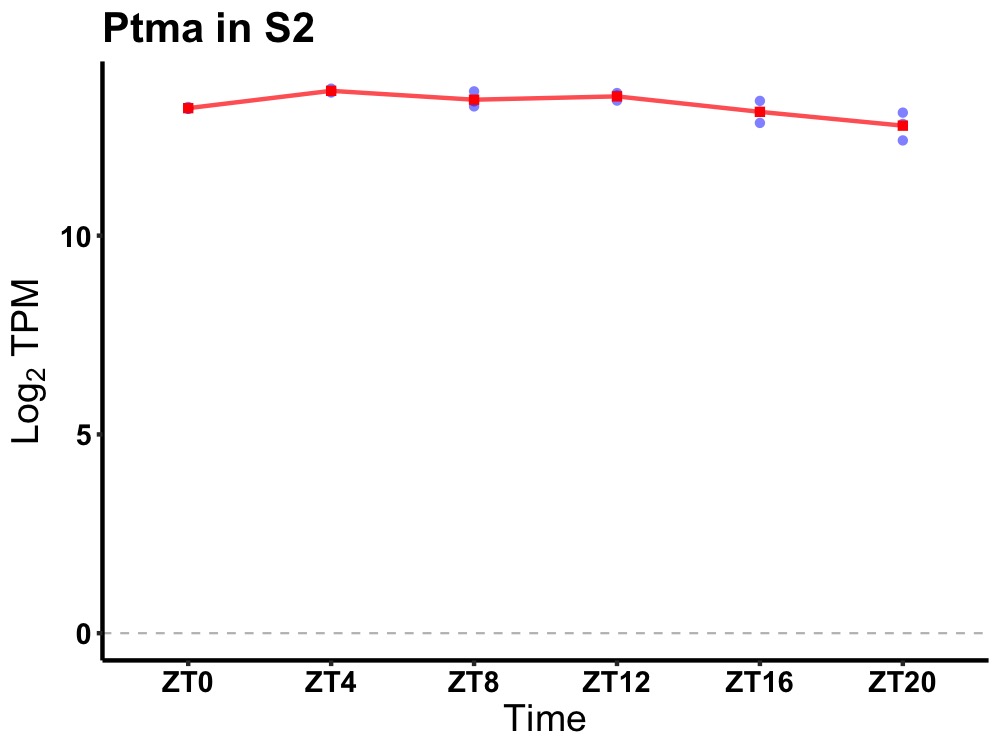 |
prothymosin alpha | 0.002 | 24 | 8 | 0.31 |
| ENSMUSG00000106956 | Gm19617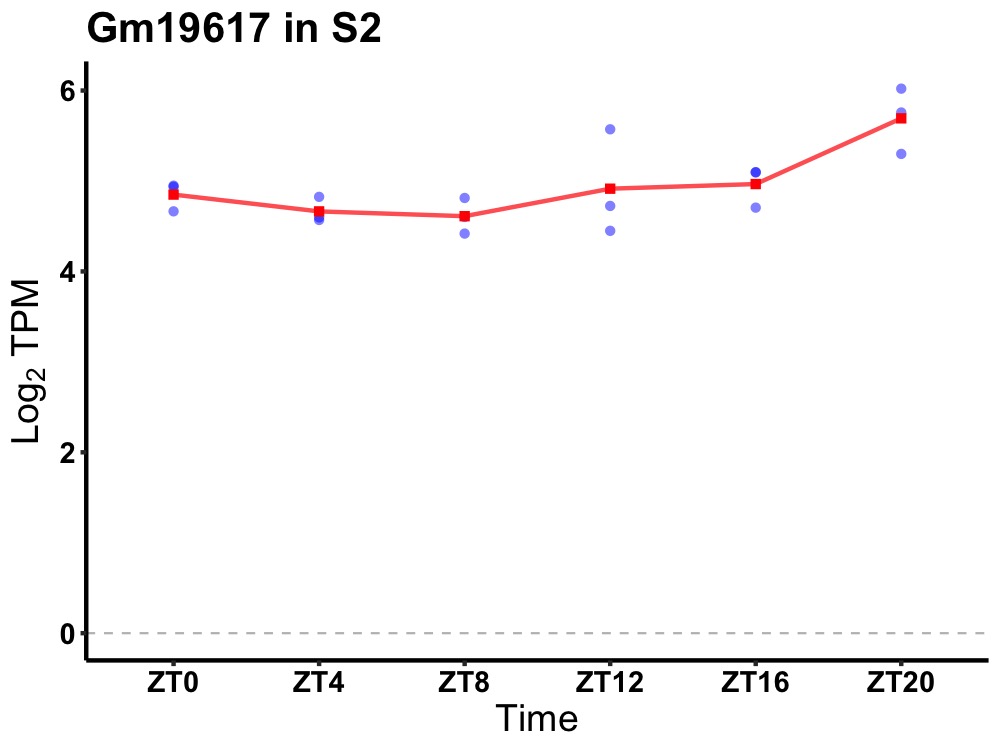 |
predicted gene, 19617 | 0.019 | 24 | 20 | 0.31 |
| ENSMUSG00000025922 | Gm5045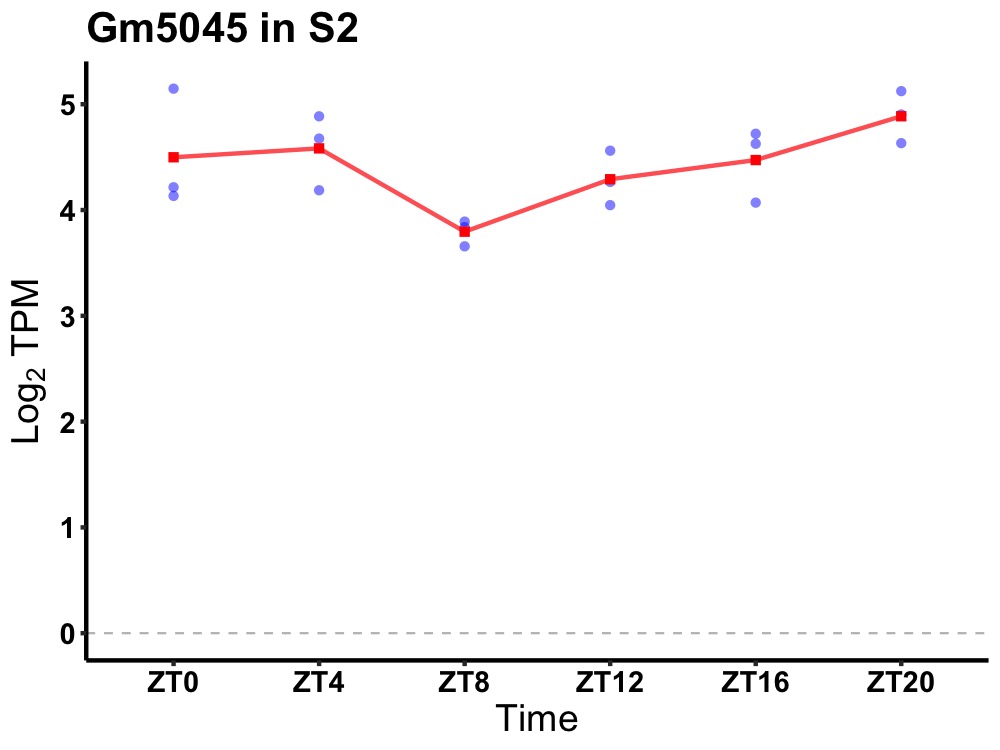 |
predicted gene 5045 | 0.049 | 24 | 22 | 0.31 |
| ENSMUSG00000107596 | Hmgn2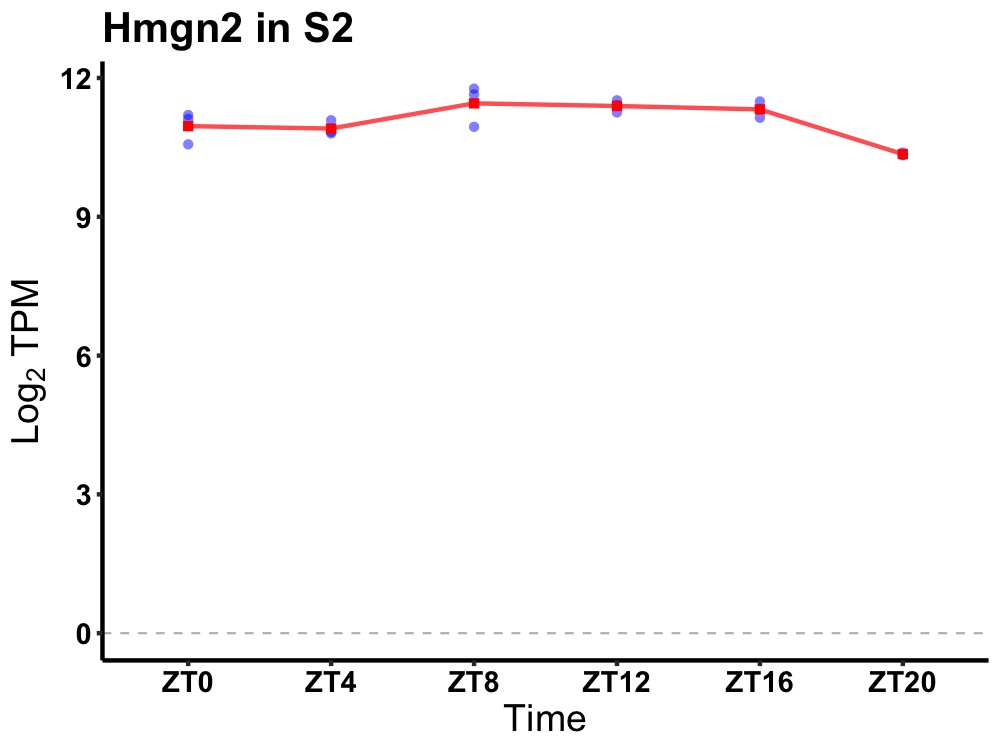 |
high mobility group nucleosomal binding domain 2 | 0.005 | 20 | 12 | 0.30 |
| ENSMUSG00000082709 | Rps15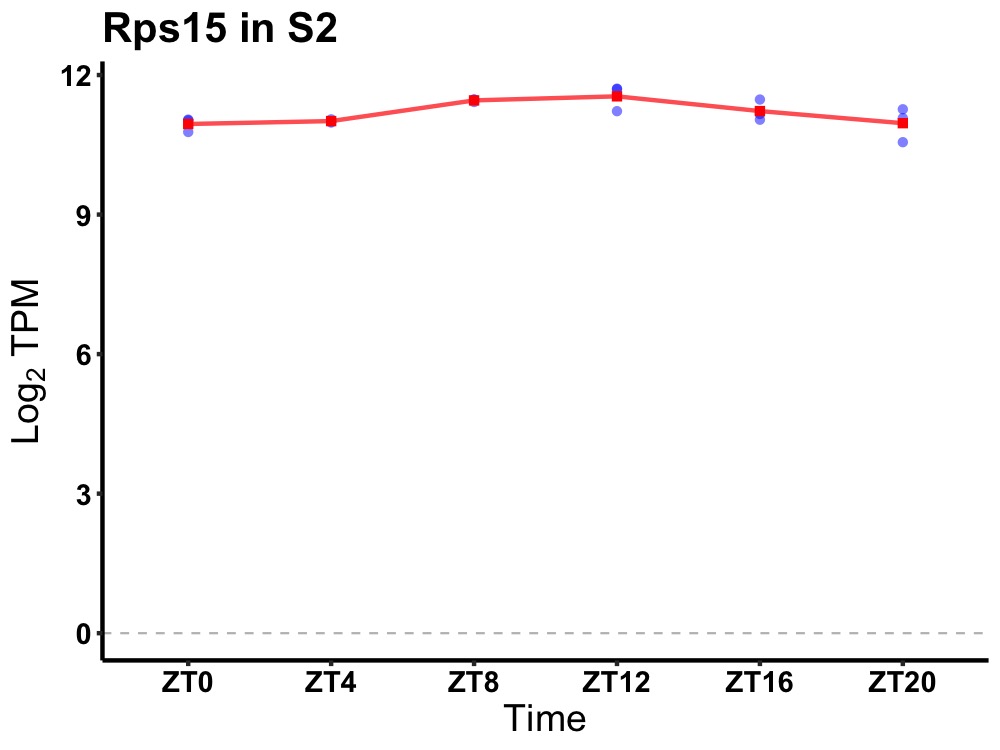 |
ribosomal protein S15 | 0.019 | 24 | 12 | 0.30 |
| ENSMUSG00000030861 | Hsp90ab1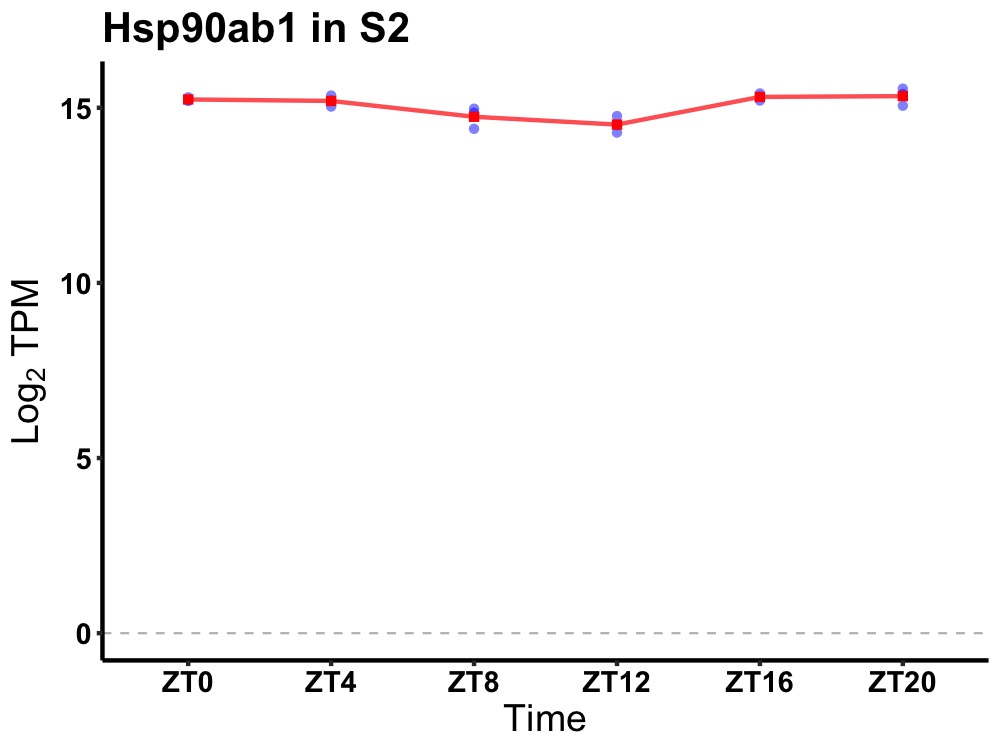 |
heat shock protein 90 alpha (cytosolic), class B member 1 | 0.049 | 24 | 22 | 0.30 |
| ENSMUSG00000105600 | Gm5987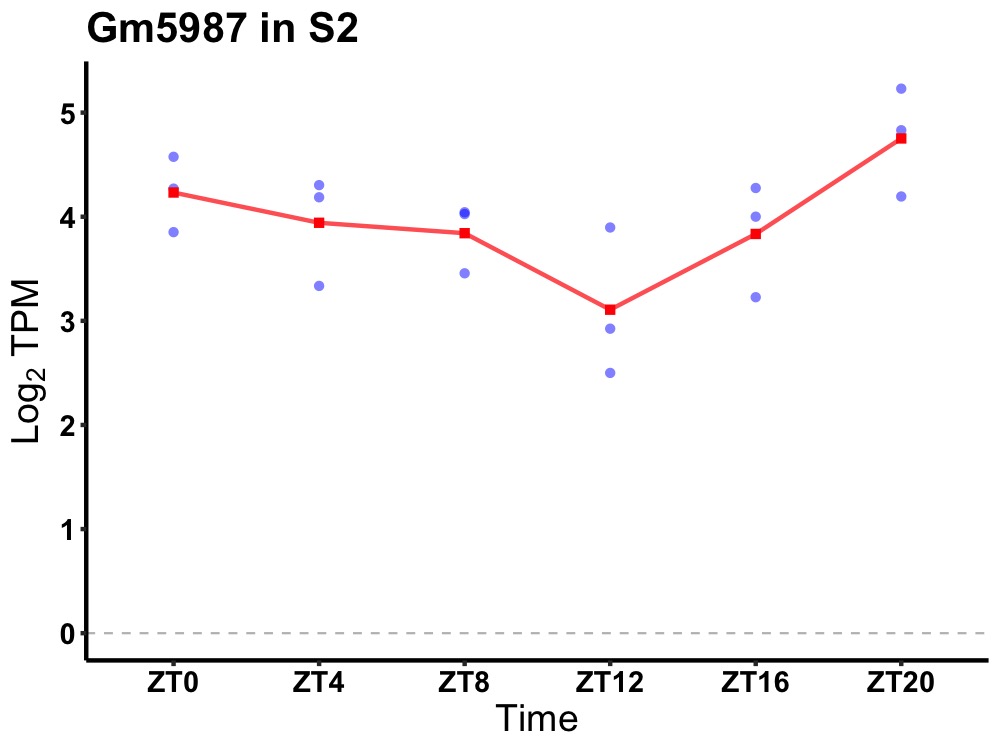 |
predicted gene 5987 | 0.027 | 20 | 2 | 0.30 |
| ENSMUSG00000083270 | Gm13498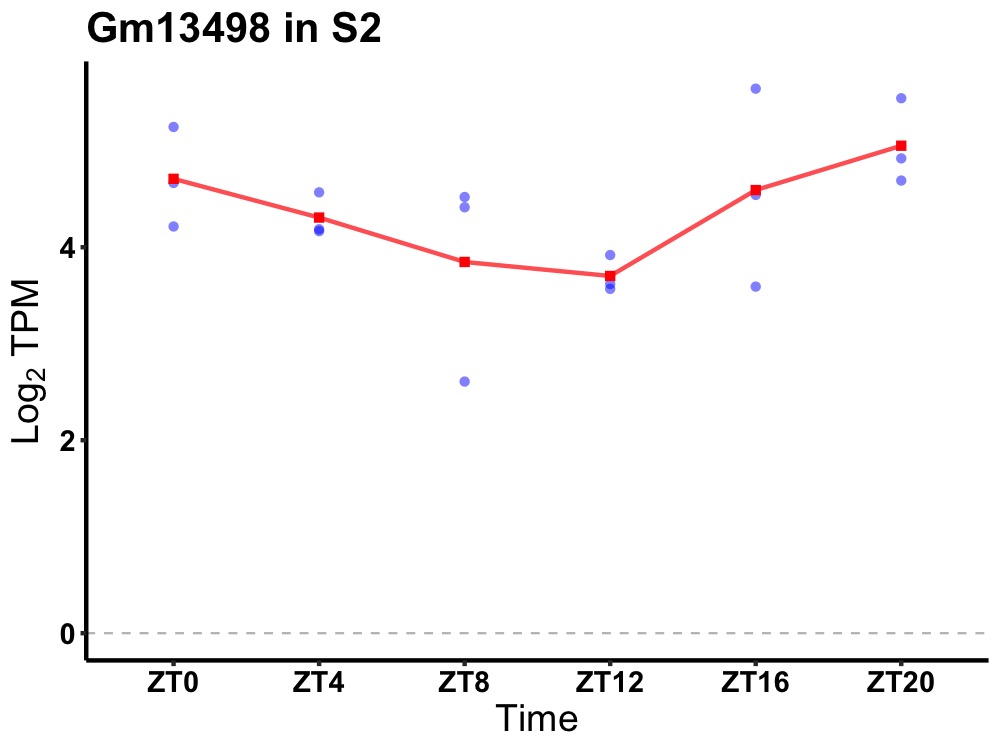 |
predicted gene 13498 | 0.036 | 20 | 2 | 0.30 |
| ENSMUSG00000021681 | Tmem174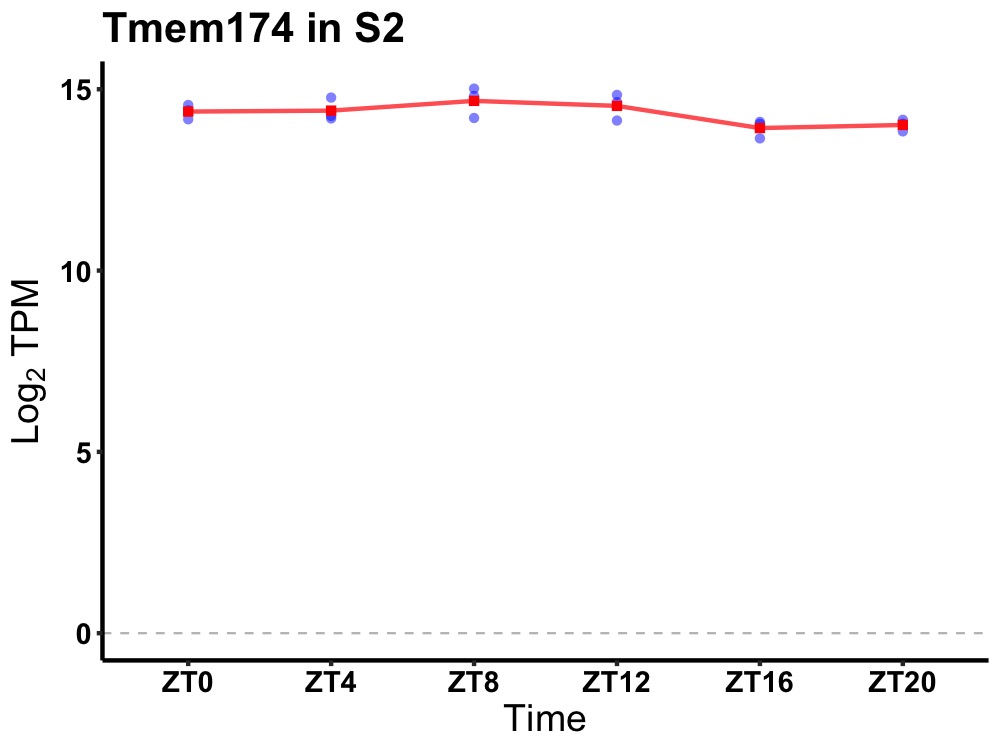 |
transmembrane protein 174 | 0.019 | 24 | 8 | 0.30 |
| ENSMUSG00000025410 | Rpl9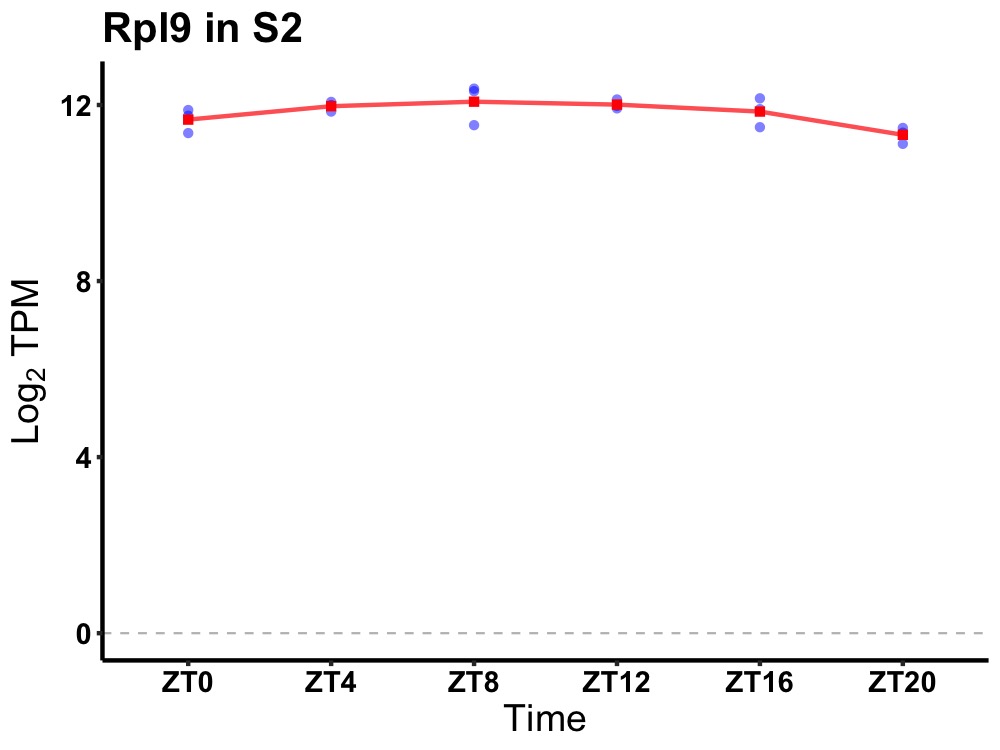 |
ribosomal protein L9 | 0.019 | 24 | 10 | 0.30 |
| ENSMUSG00000074656 | Ndufb3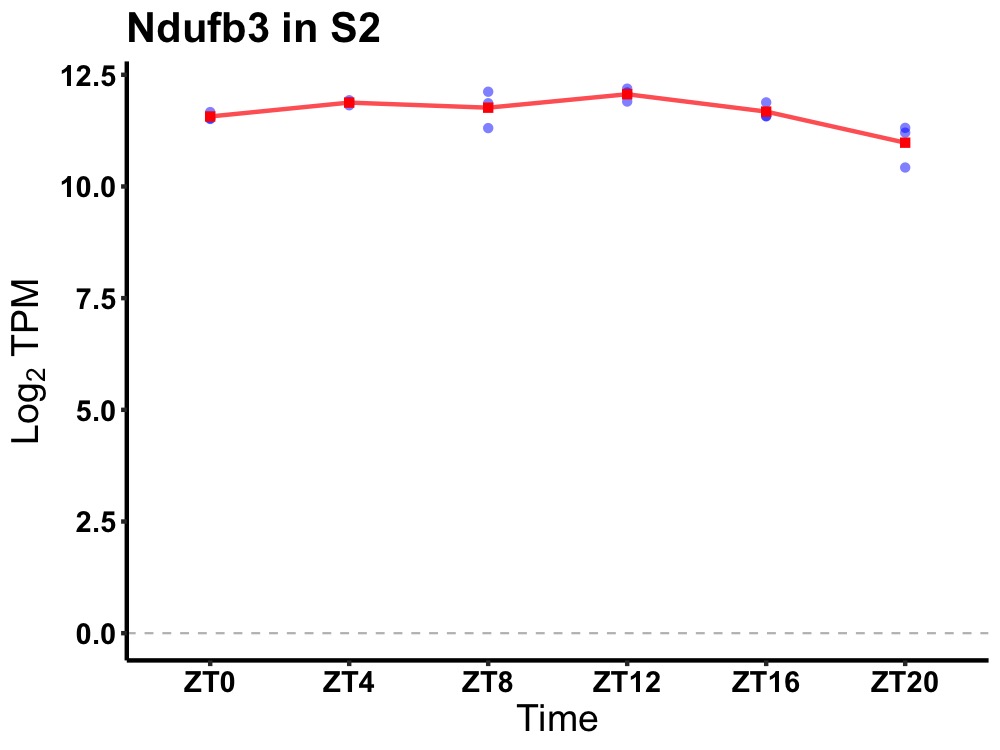 |
NADH:ubiquinone oxidoreductase subunit B3 | 0.010 | 24 | 10 | 0.30 |
| ENSMUSG00000001786 | Ap2a2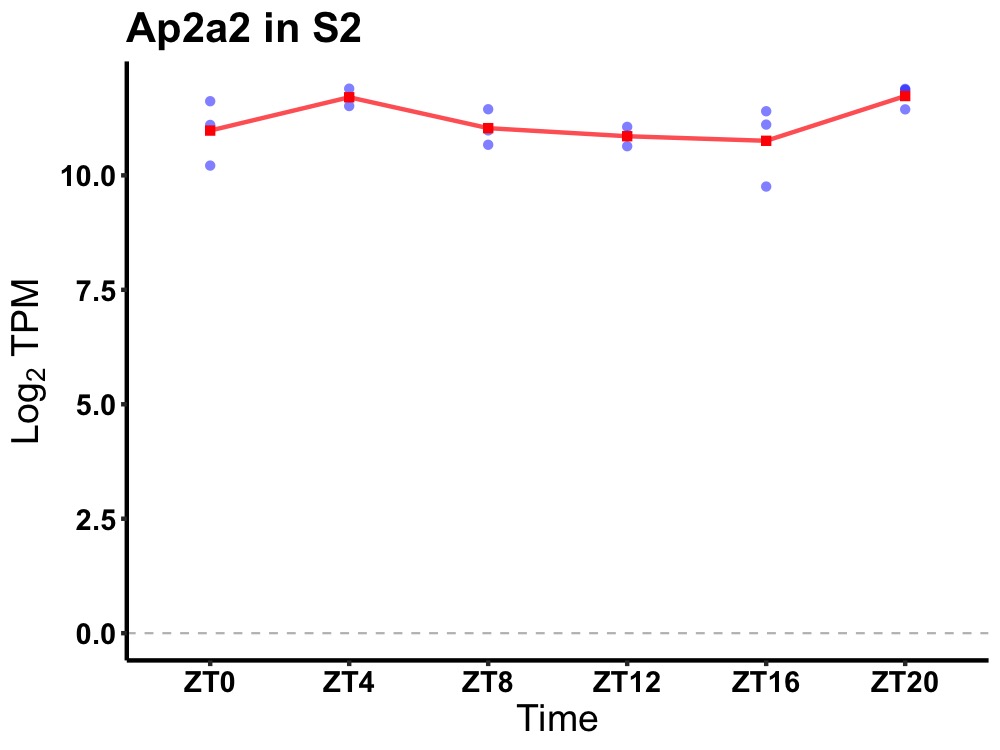 |
adaptor-related protein complex 2, alpha 2 subunit | 0.036 | 20 | 4 | 0.29 |
| ENSMUSG00000022339 | Sec13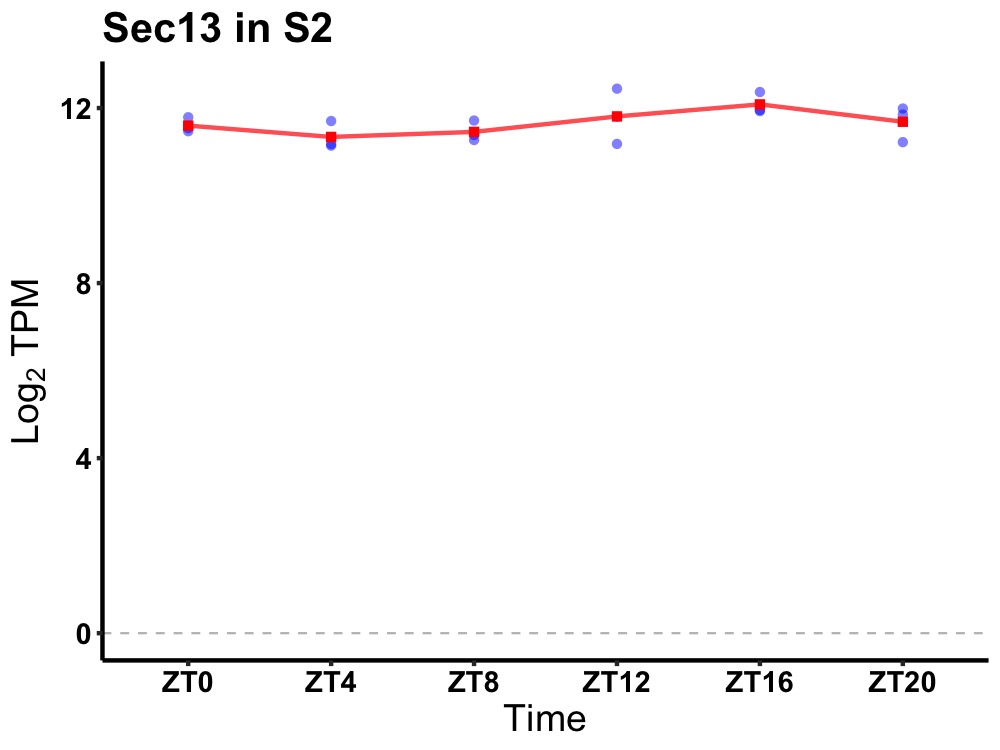 |
SEC13 homolog, nuclear pore and COPII coat complex component | 0.049 | 24 | 18 | 0.29 |
| ENSMUSG00000082062 | Ftl2-ps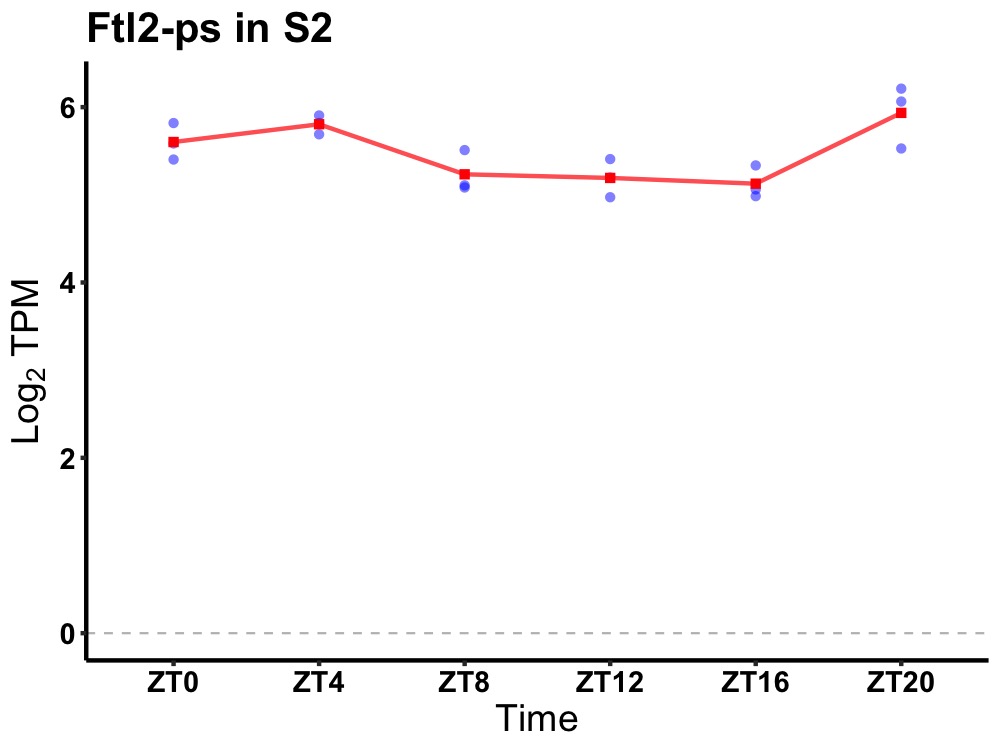 |
ferritin light polypeptide 2, pseudogene | 0.005 | 20 | 4 | 0.29 |
| ENSMUSG00000025545 | Gstp1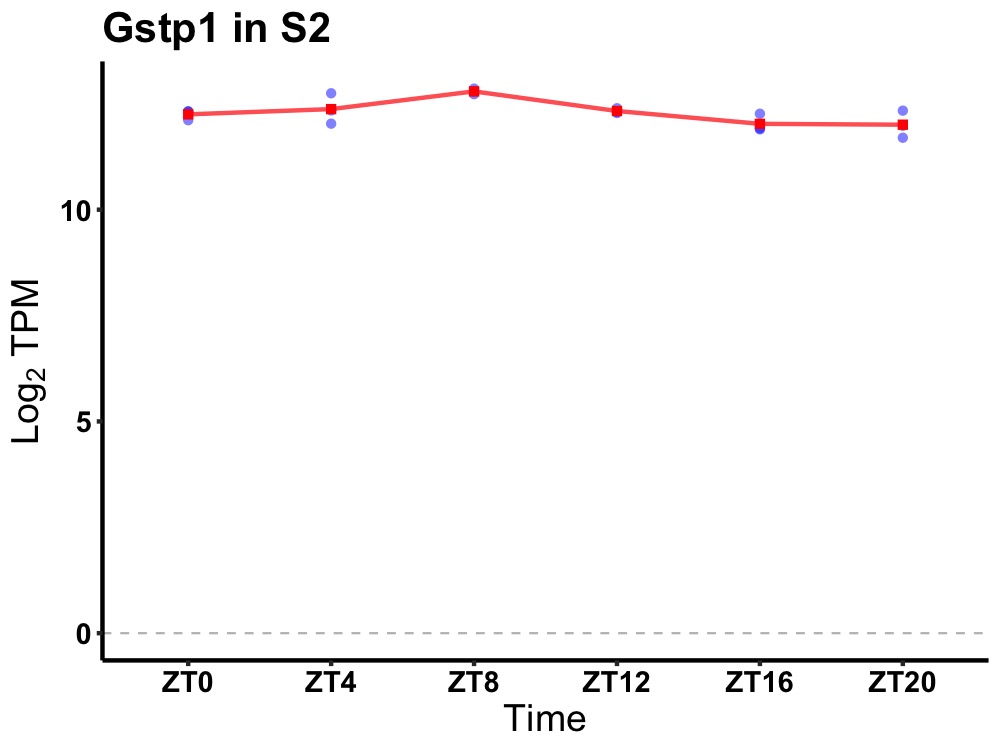 |
glutathione S-transferase, pi 1 | 0.014 | 24 | 8 | 0.29 |
| ENSMUSG00000085172 | Banf1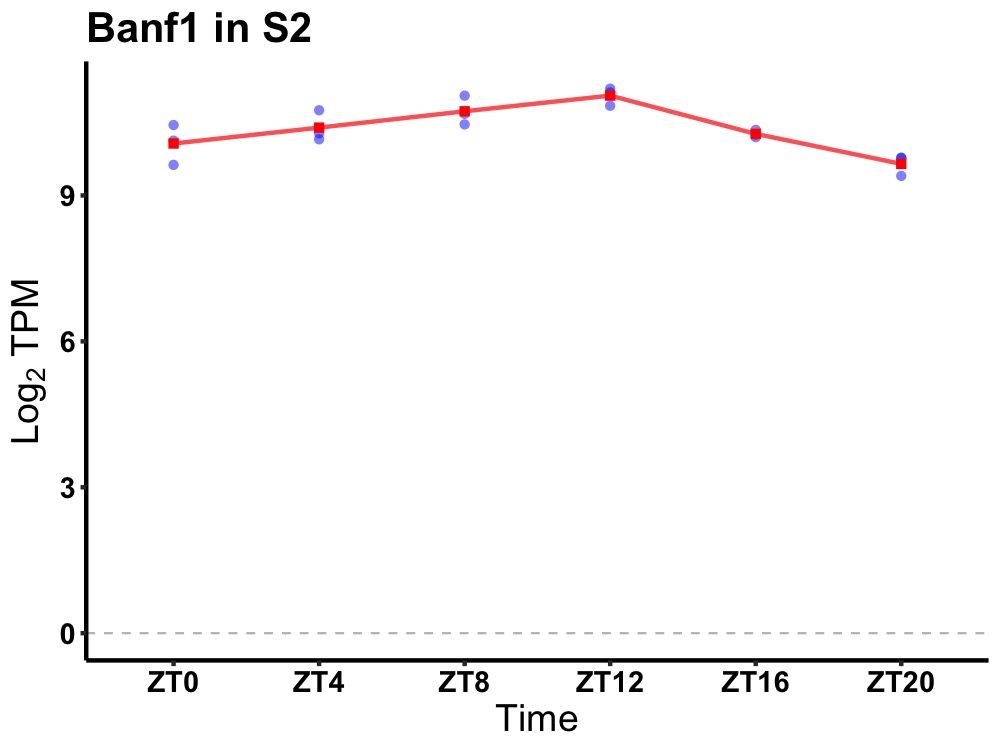 |
barrier to autointegration factor 1 | 0.000 | 20 | 12 | 0.29 |
| ENSMUSG00000114349 | Mrps16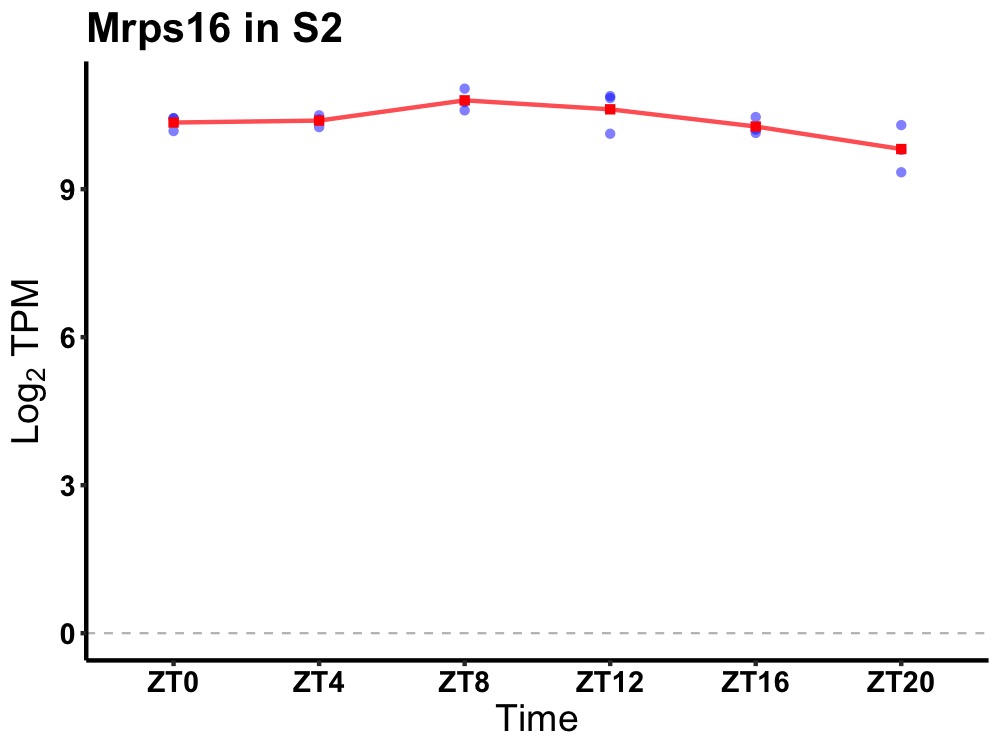 |
mitochondrial ribosomal protein S16 | 0.049 | 24 | 10 | 0.29 |
| ENSMUSG00000004285 | Pmpcb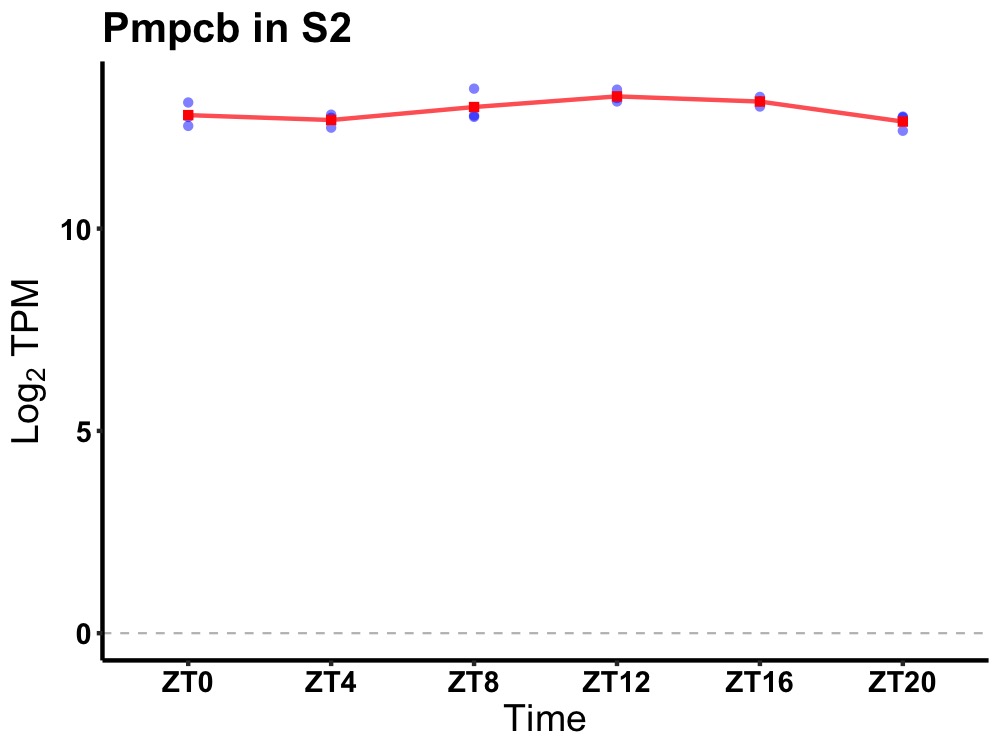 |
peptidase (mitochondrial processing) beta | 0.019 | 20 | 14 | 0.29 |
| ENSMUSG00000067017 | Gm3608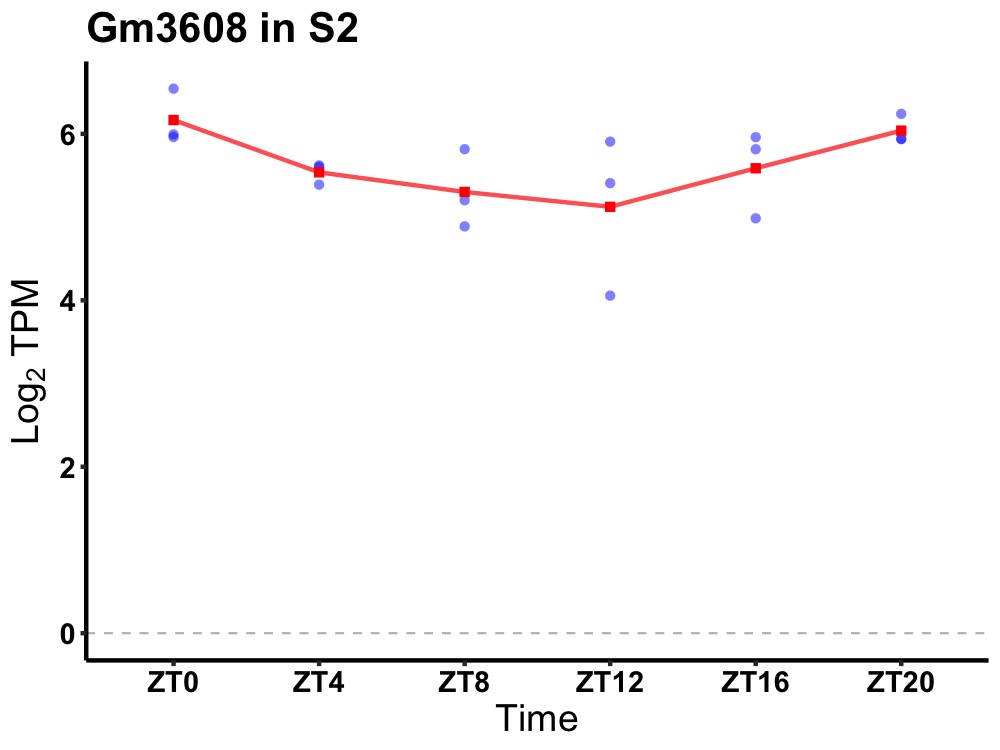 |
predicted gene 3608 | 0.010 | 20 | 0 | 0.29 |
| ENSMUSG00000000355 | Hipk2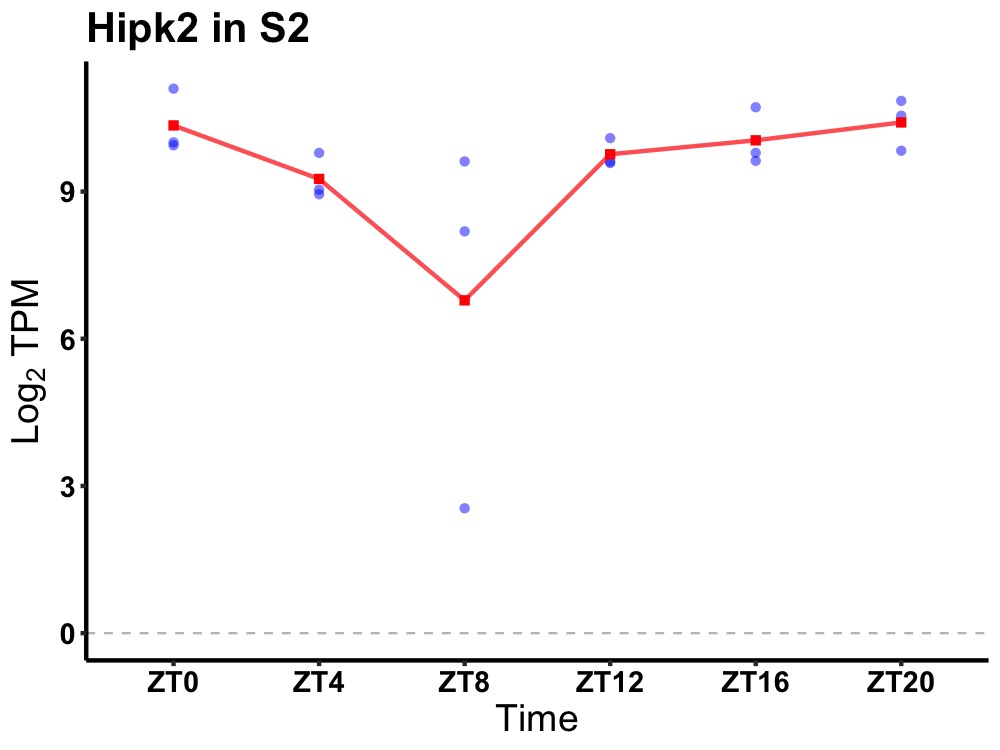 |
homeodomain interacting protein kinase 2 | 0.014 | 20 | 0 | 0.28 |
| ENSMUSG00000061559 | AC124578.3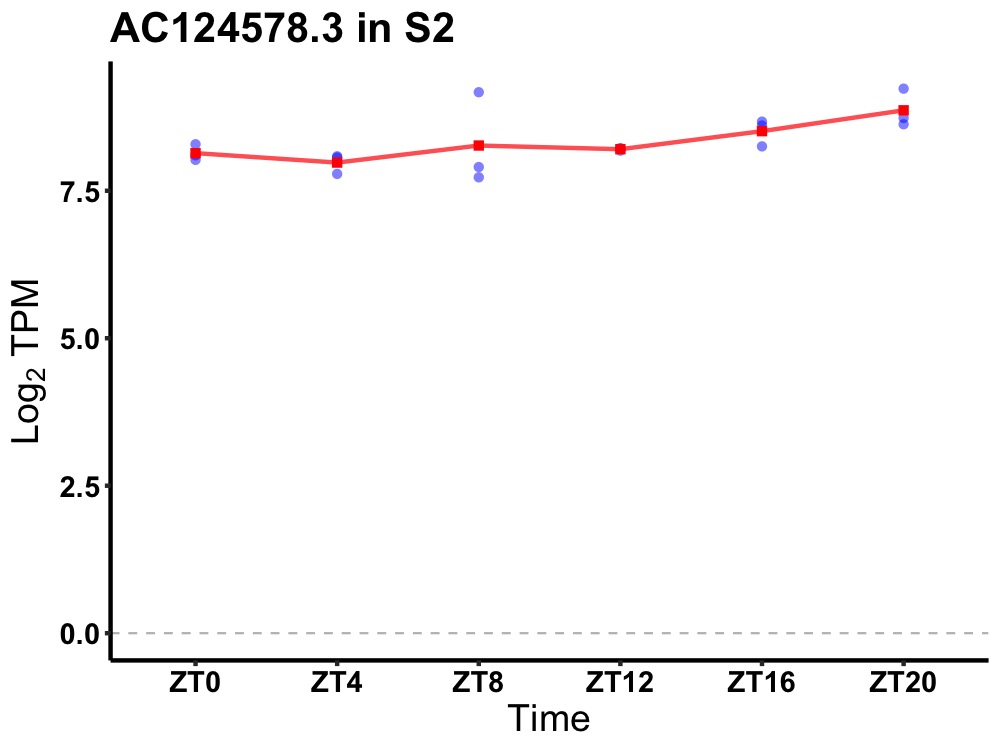 |
none | 0.014 | 24 | 20 | 0.28 |
| ENSMUSG00000090197 | Rps25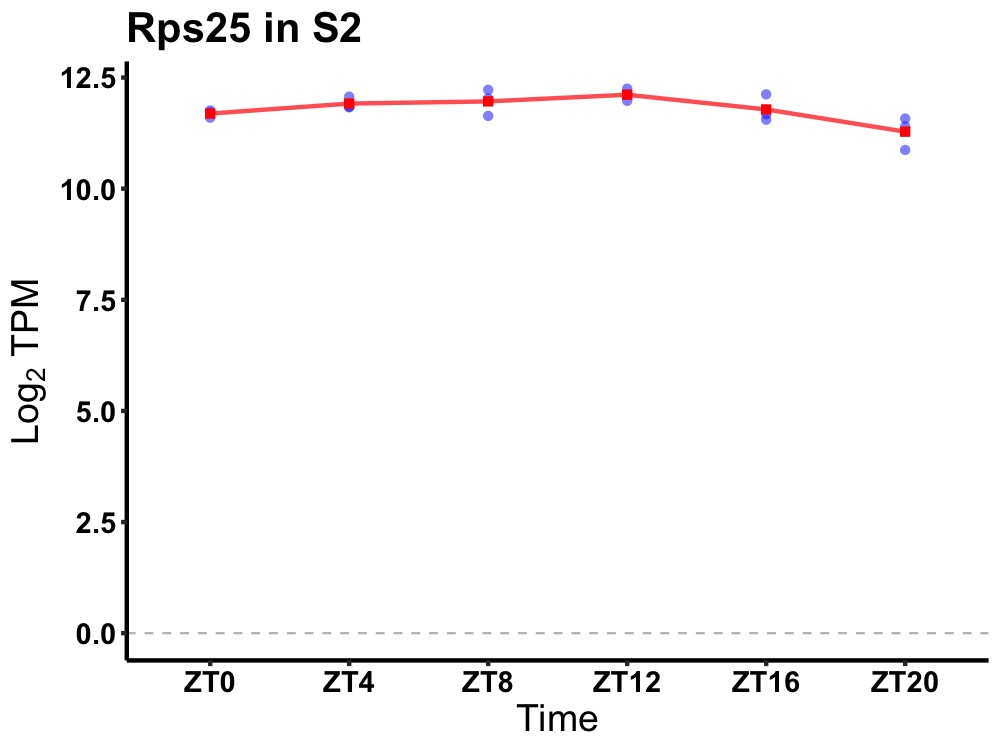 |
ribosomal protein S25 | 0.027 | 24 | 10 | 0.28 |
| ENSMUSG00000029098 | Gpx4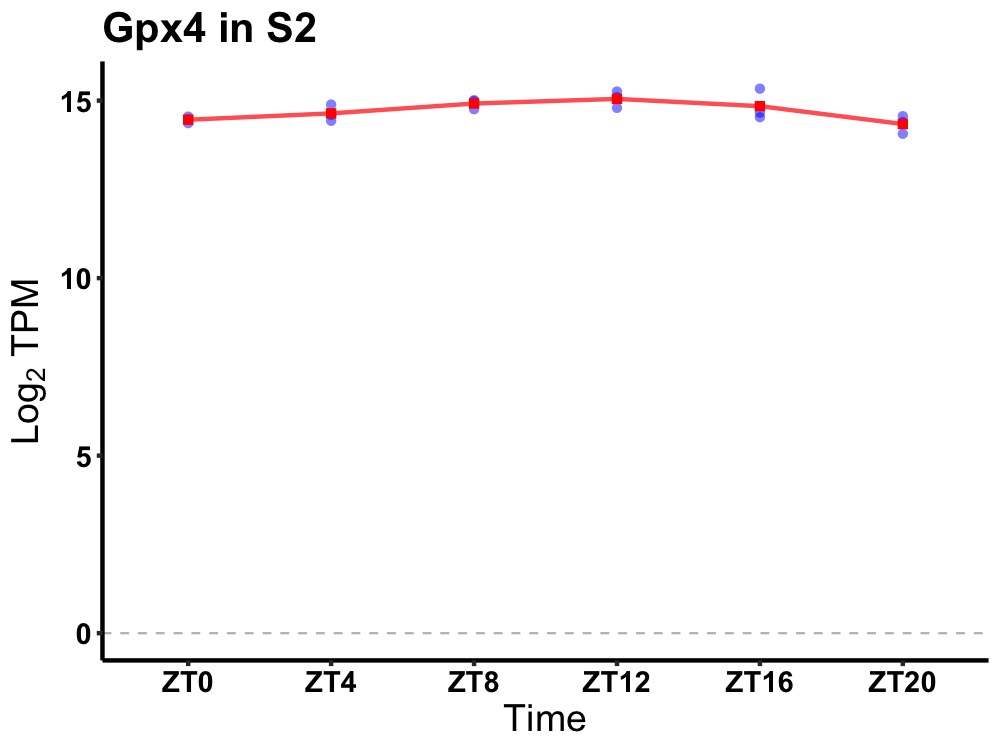 |
glutathione peroxidase 4 | 0.005 | 20 | 12 | 0.28 |
| ENSMUSG00000021696 | Tma7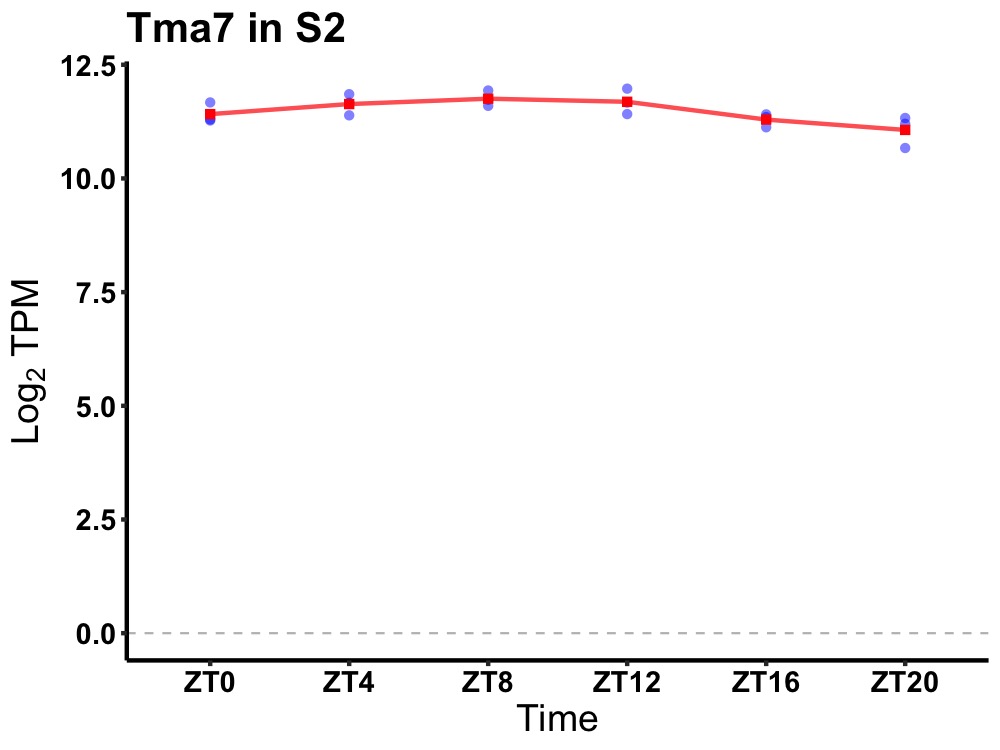 |
translational machinery associated 7 | 0.014 | 24 | 10 | 0.28 |
| ENSMUSG00000115593 | Gm4824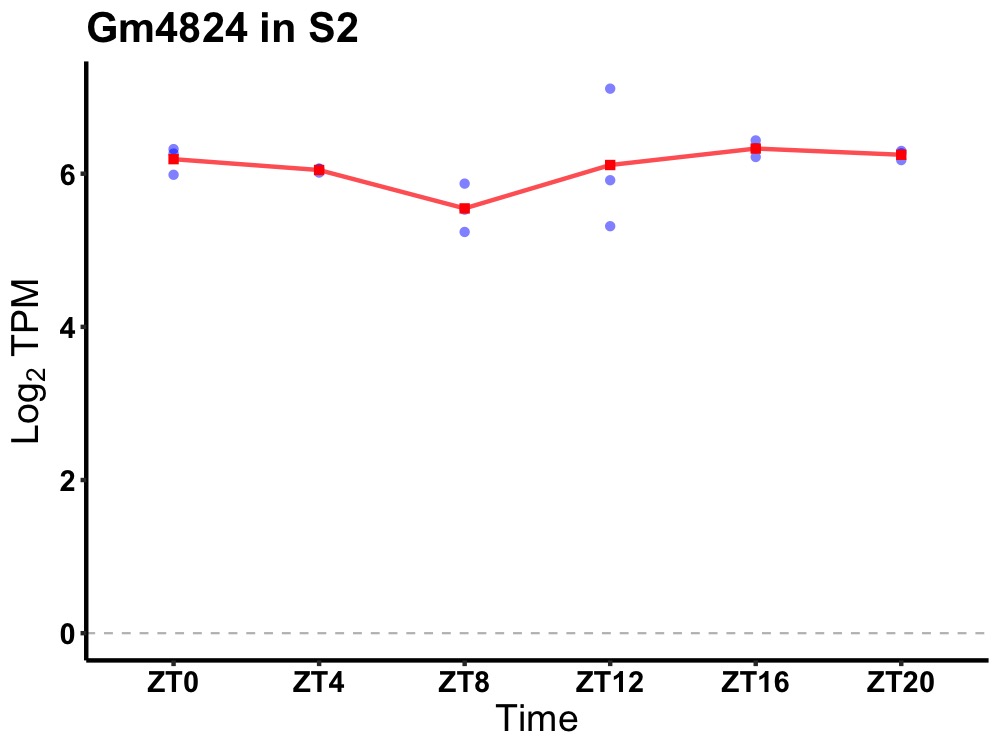 |
predicted gene 4824 | 0.049 | 20 | 18 | 0.28 |
| ENSMUSG00000112239 | Gm17823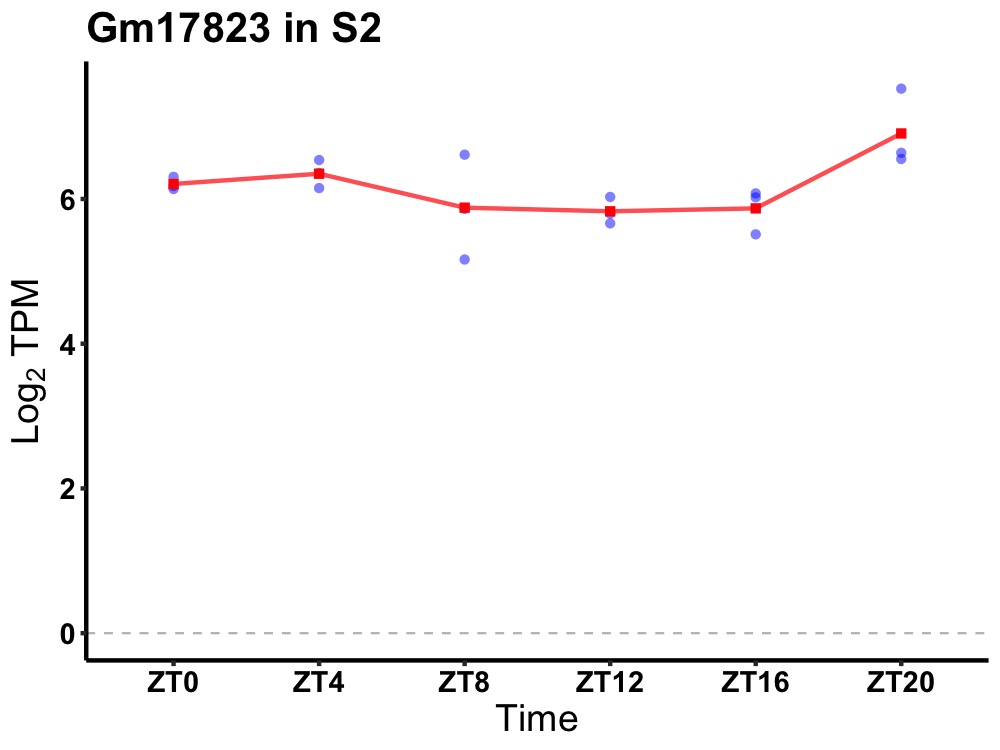 |
predicted gene, 17823 | 0.019 | 20 | 2 | 0.28 |
| ENSMUSG00000055013 | Cox6a1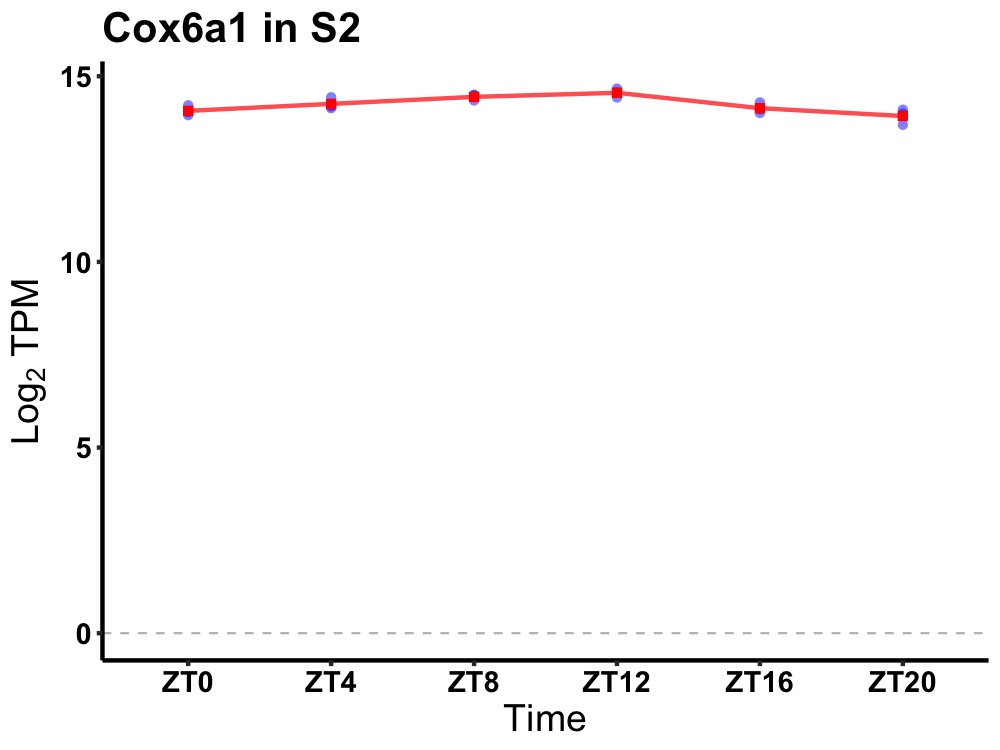 |
cytochrome c oxidase subunit 6A1 | 0.002 | 24 | 10 | 0.28 |
| ENSMUSG00000078153 | Psme2b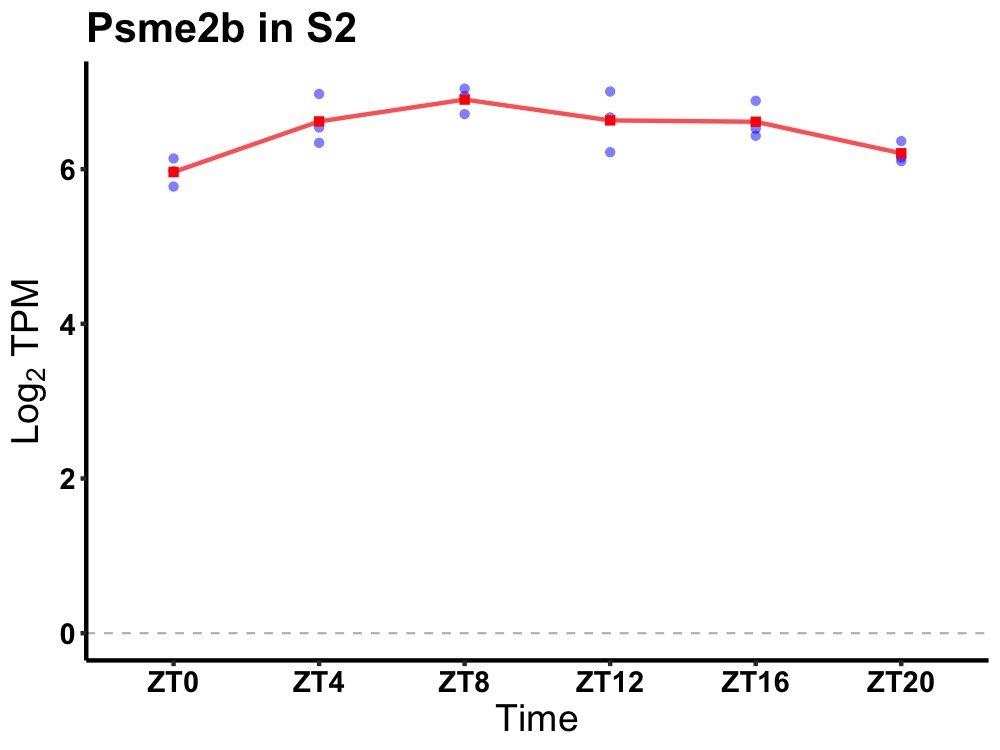 |
protease (prosome, macropain) activator subunit 2B | 0.002 | 20 | 10 | 0.28 |
| ENSMUSG00000103034 | Gm8797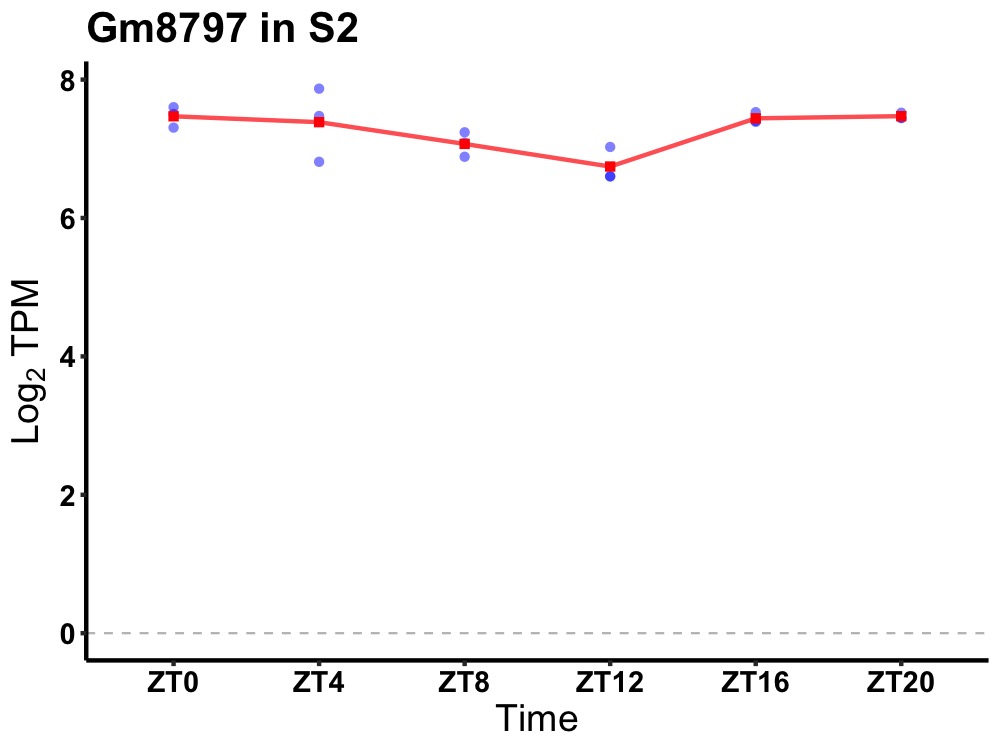 |
predicted pseudogene 8797 | 0.049 | 24 | 0 | 0.27 |
| ENSMUSG00000062382 | Ftl1-ps1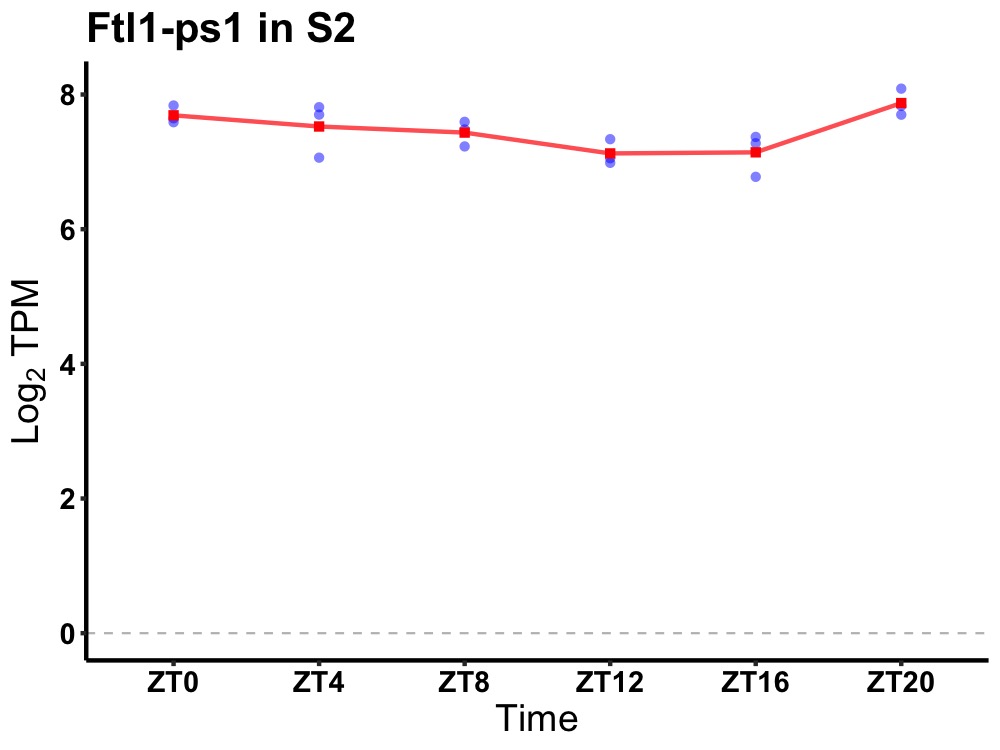 |
ferritin light polypeptide 1, pseudogene 1 | 0.027 | 20 | 2 | 0.27 |
| ENSMUSG00000029695 | mt-Co1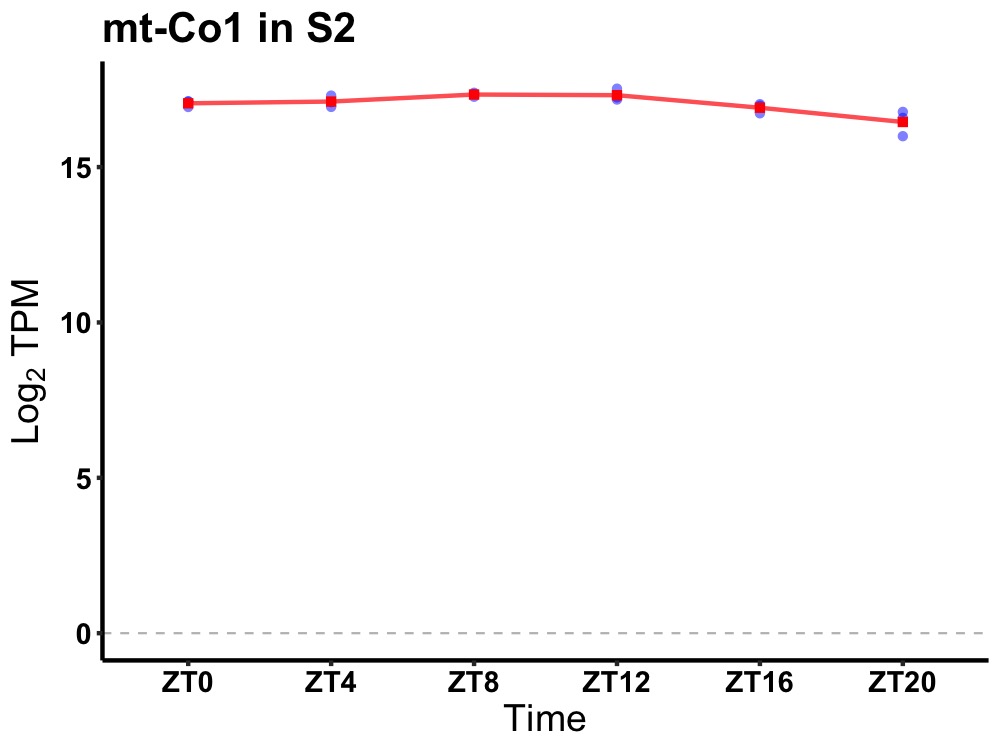 |
mitochondrially encoded cytochrome c oxidase I | 0.002 | 24 | 10 | 0.27 |
| ENSMUSG00000032299 | Ppa2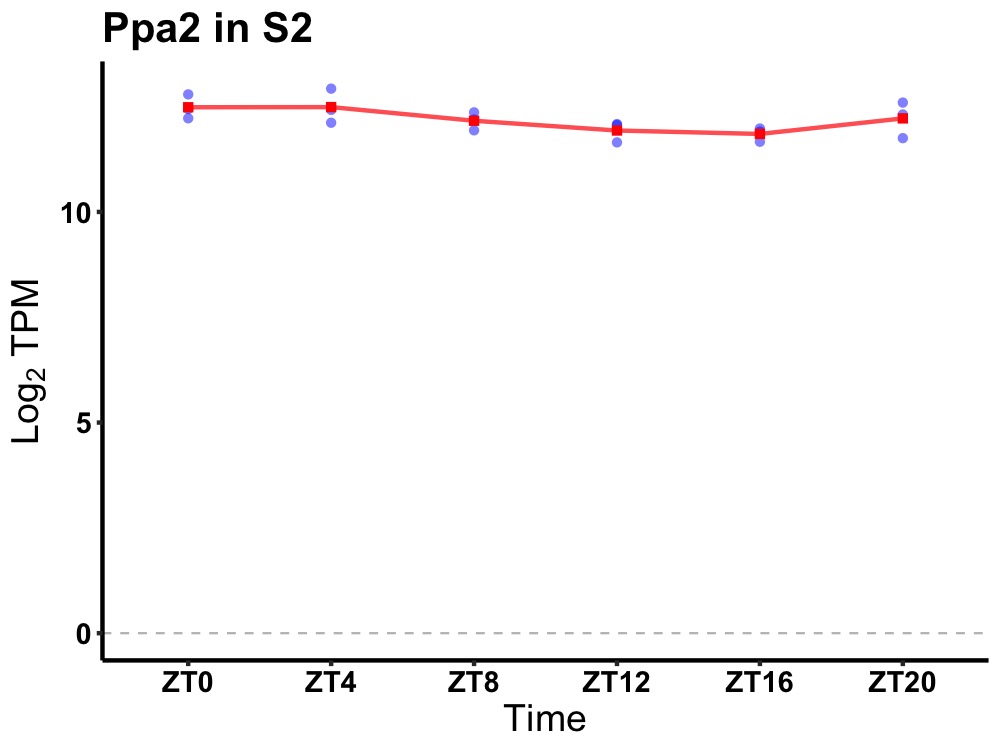 |
pyrophosphatase (inorganic) 2 | 0.049 | 24 | 4 | 0.27 |
| ENSMUSG00000084230 | Gpx4-ps2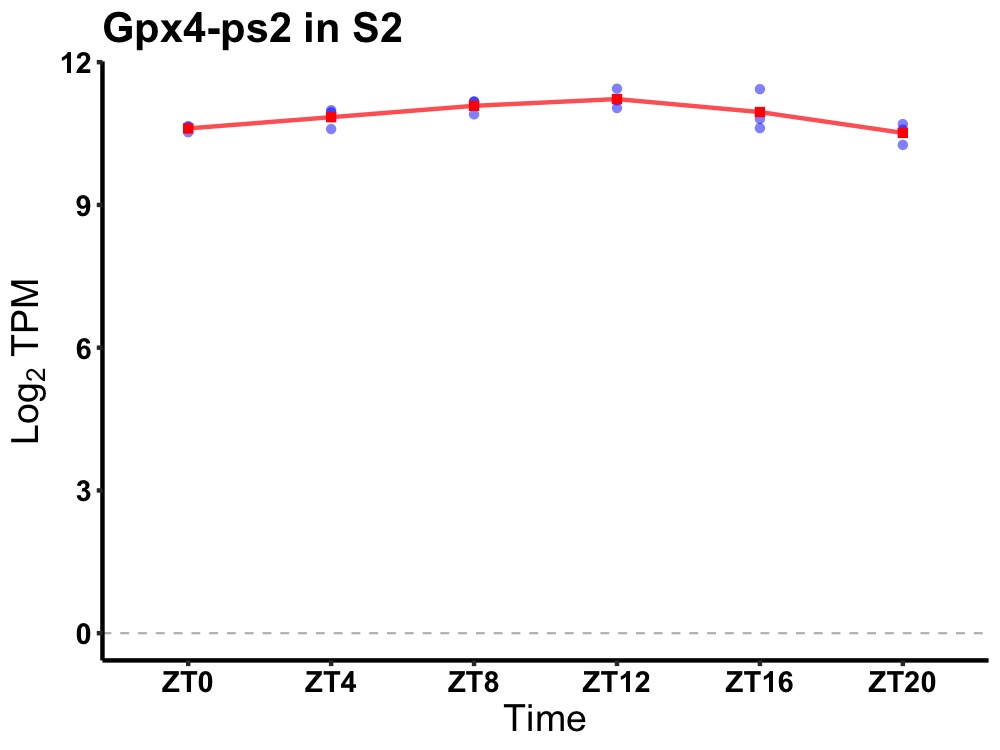 |
glutathione peroxidase 4, pseudogene 2 | 0.007 | 20 | 12 | 0.27 |
| ENSMUSG00000082453 | Gm11502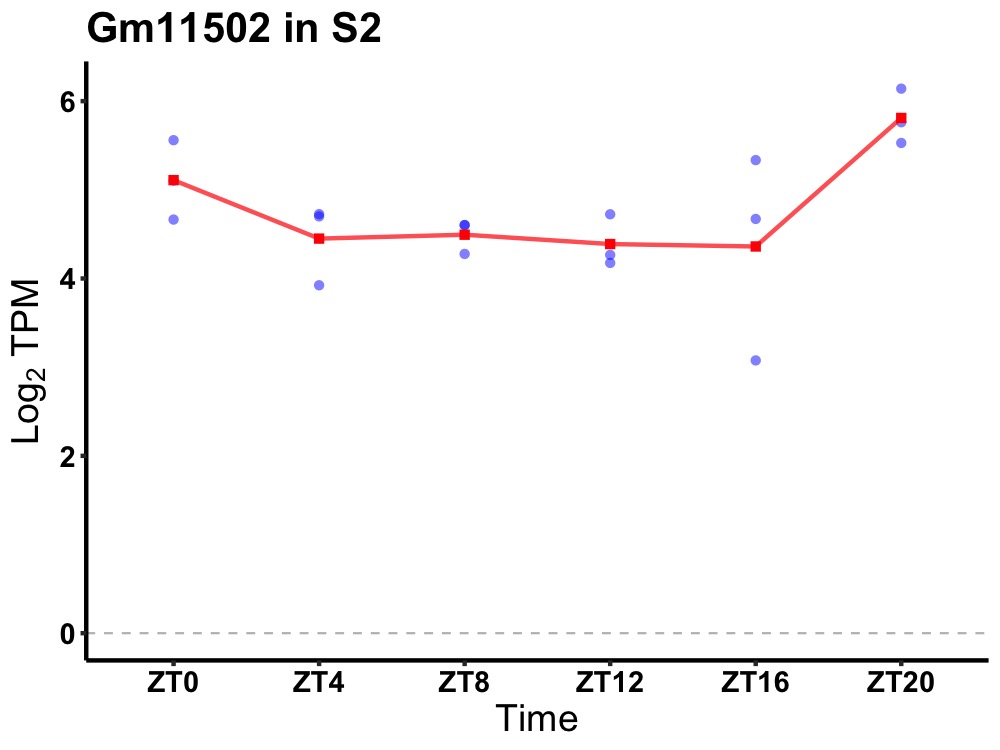 |
predicted gene 11502 | 0.036 | 20 | 2 | 0.27 |
| ENSMUSG00000066491 | Tmem250-ps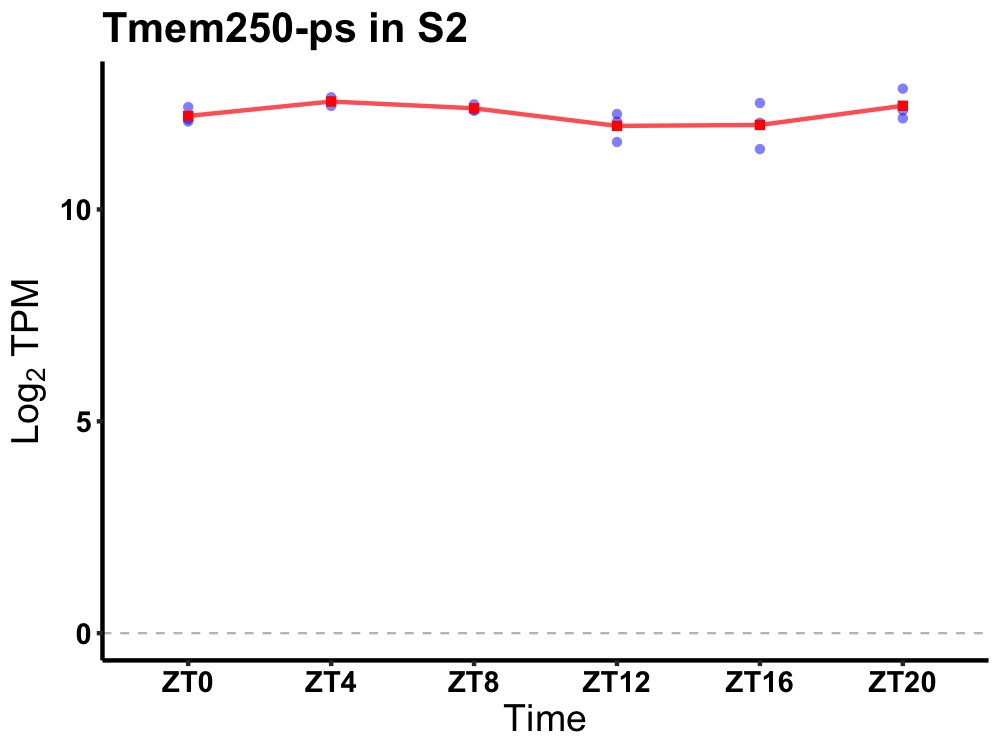 |
transmembrane protein 250, pseudogene | 0.036 | 20 | 6 | 0.26 |
| ENSMUSG00000104649 | Gm43712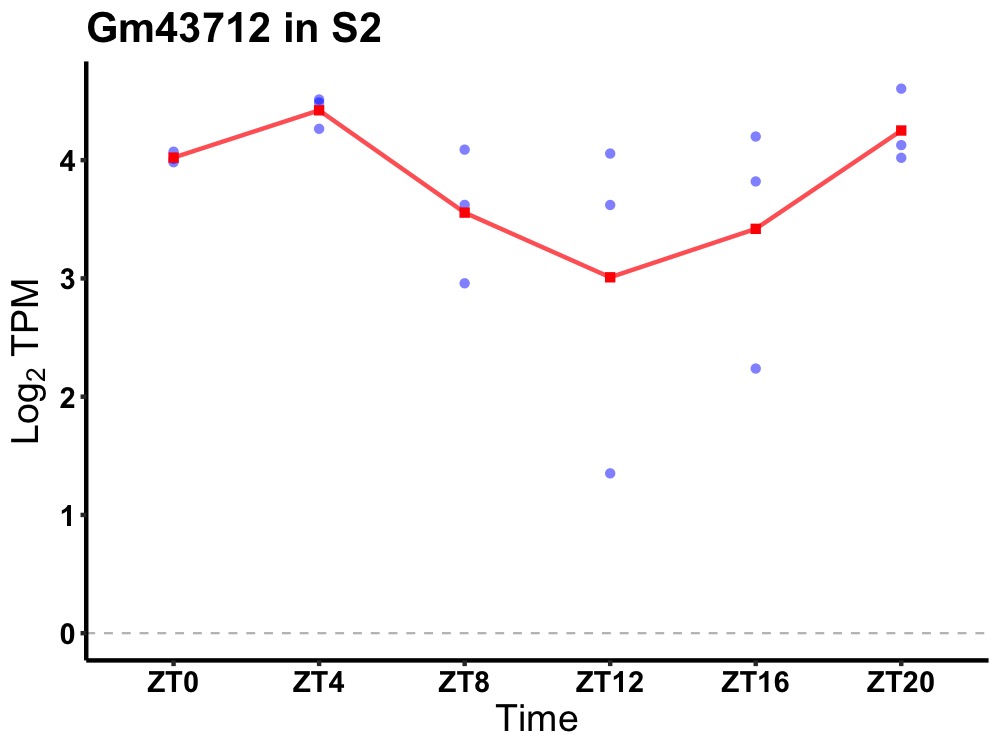 |
predicted gene 43712 | 0.027 | 20 | 4 | 0.26 |
| ENSMUSG00000082632 | Gm13365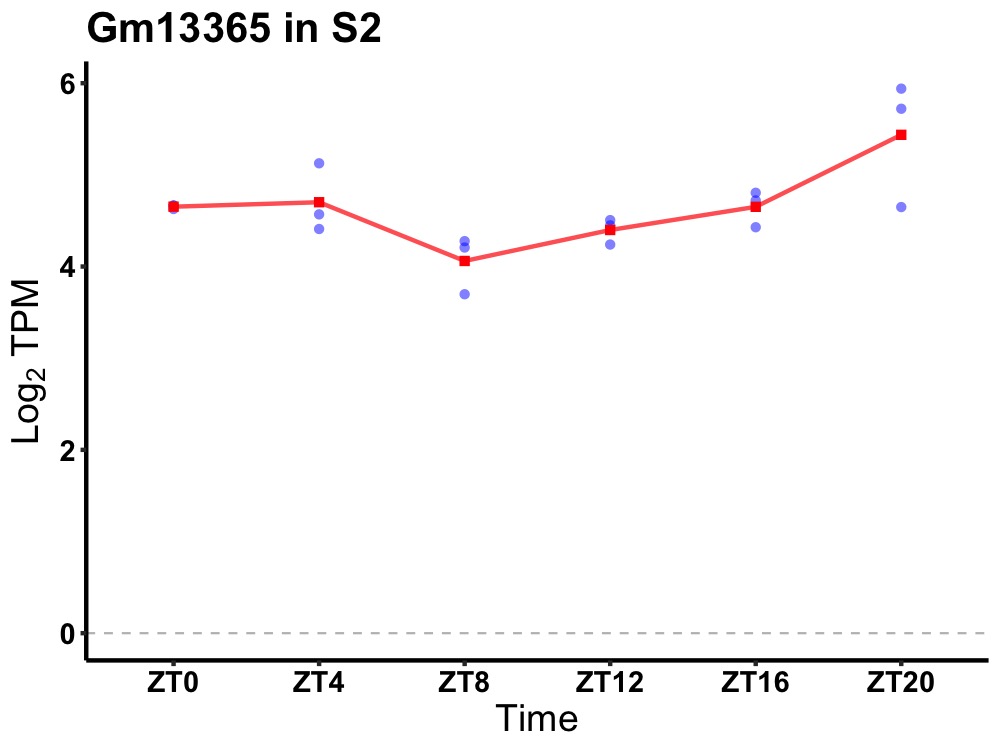 |
predicted gene 13365 | 0.007 | 24 | 22 | 0.26 |
| ENSMUSG00000029456 | Aldh8a1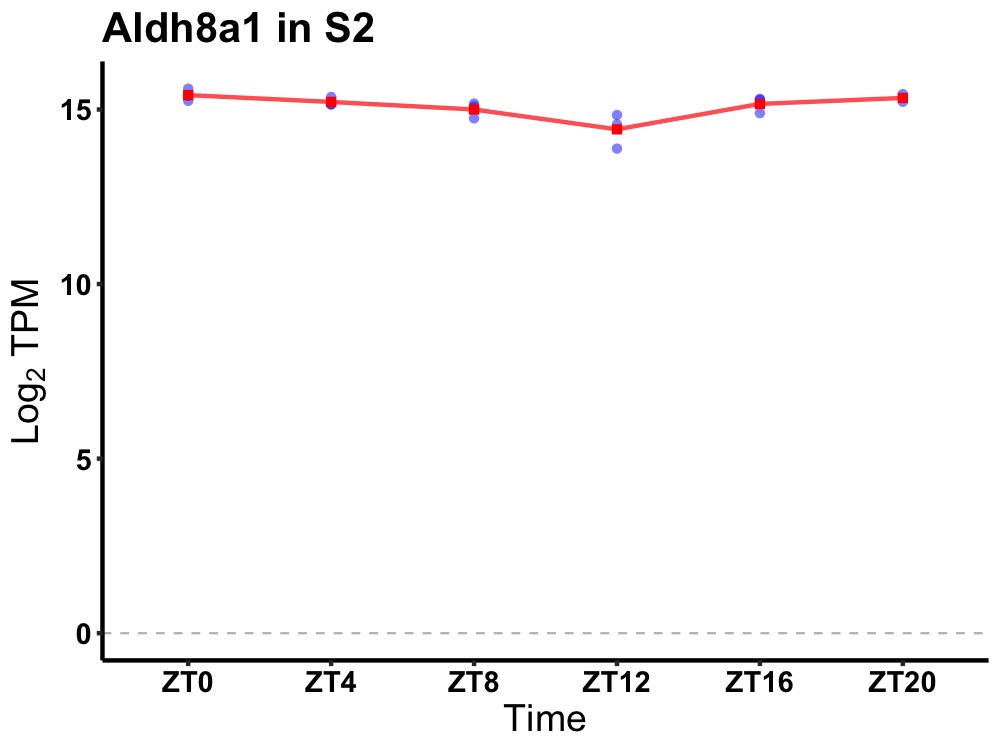 |
aldehyde dehydrogenase 8 family, member A1 | 0.001 | 24 | 0 | 0.26 |
| ENSMUSG00000085175 | Atp5g2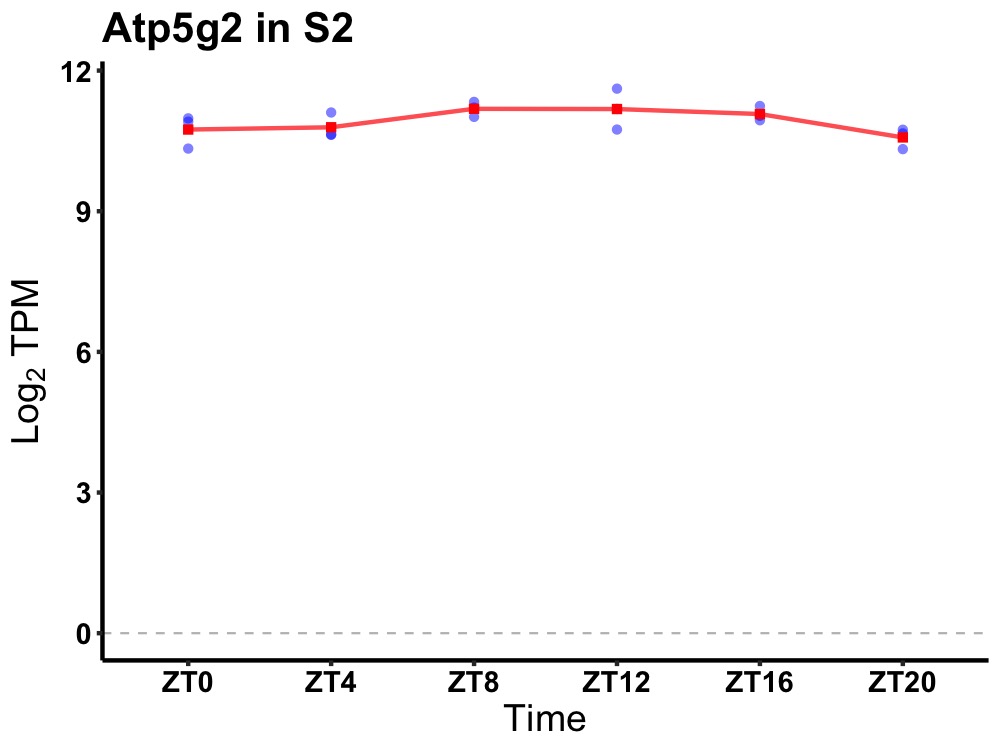 |
ATP synthase, H+ transporting, mitochondrial F0 complex, subunit C2 (subunit 9) | 0.049 | 20 | 12 | 0.25 |
| ENSMUSG00000020917 | Slc22a12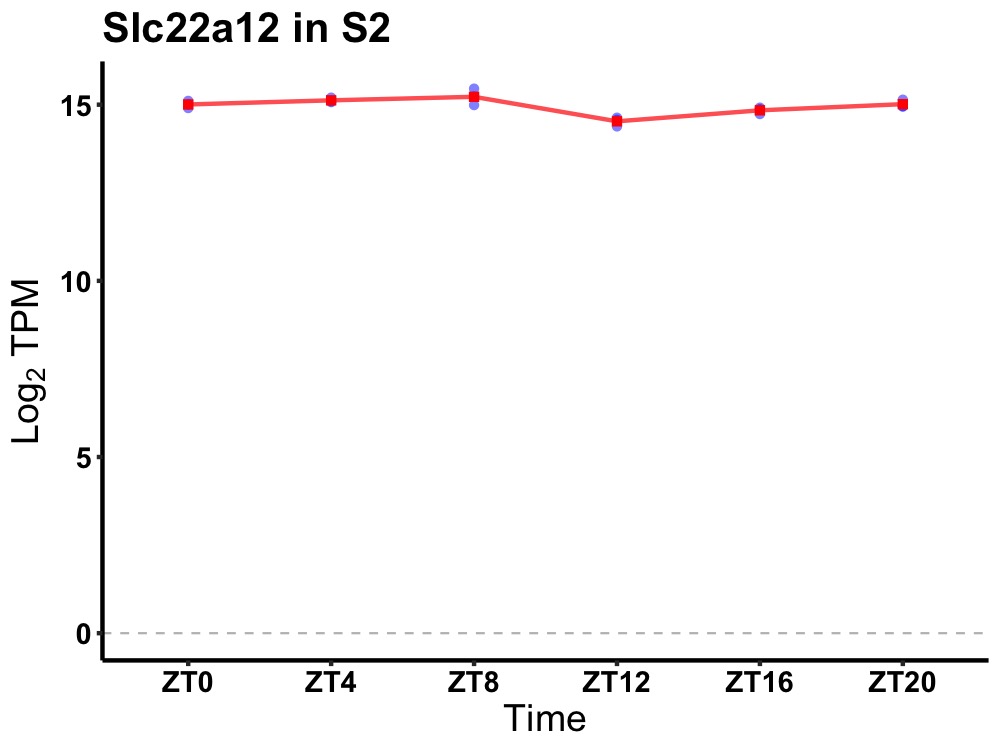 |
solute carrier family 22 (organic anion/cation transporter), member 12 | 0.027 | 20 | 6 | 0.25 |
| ENSMUSG00000082044 | Snrpert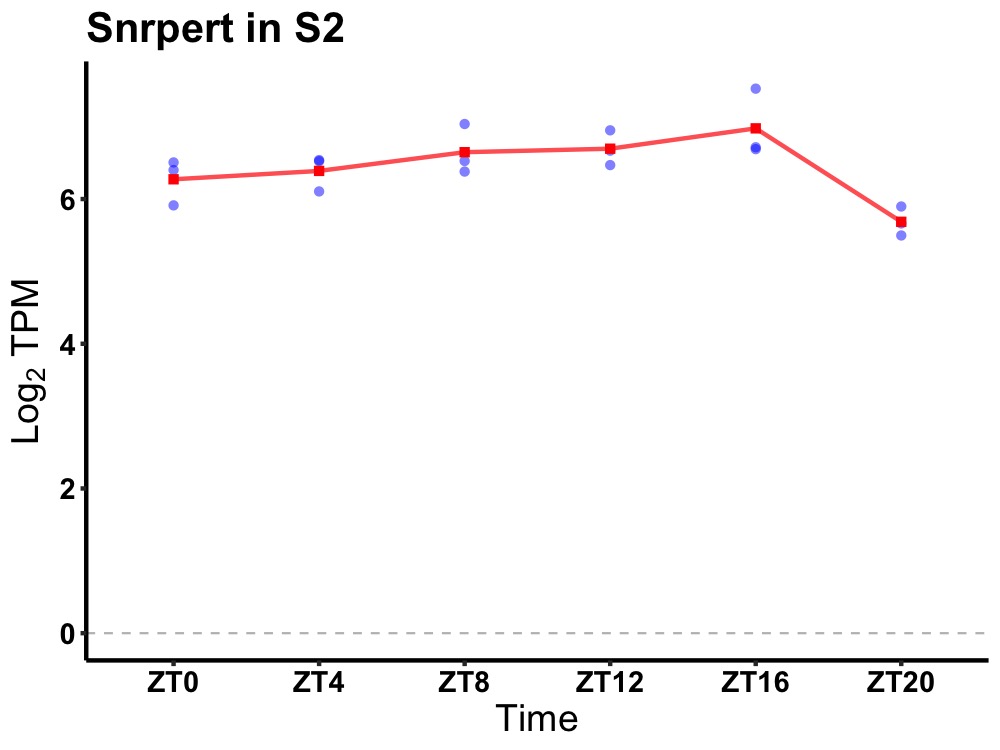 |
small nuclear ribonucleoprotein E, pseudogene | 0.036 | 20 | 12 | 0.25 |
| ENSMUSG00000023020 | Naca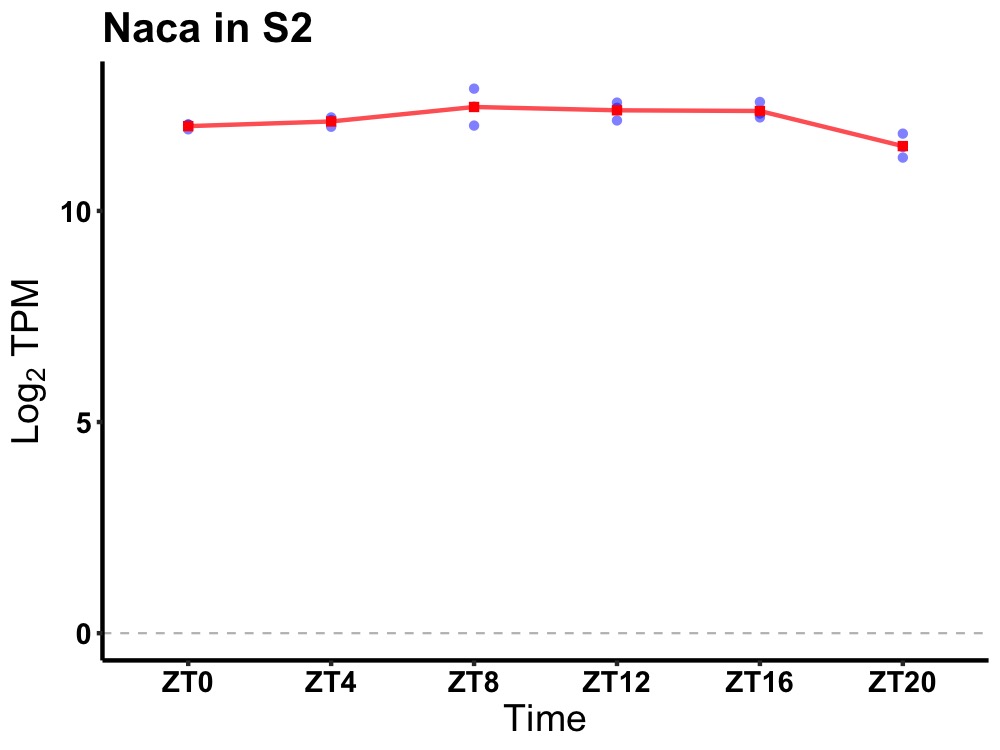 |
nascent polypeptide-associated complex alpha polypeptide | 0.005 | 20 | 12 | 0.25 |
| ENSMUSG00000027180 | Ubxn1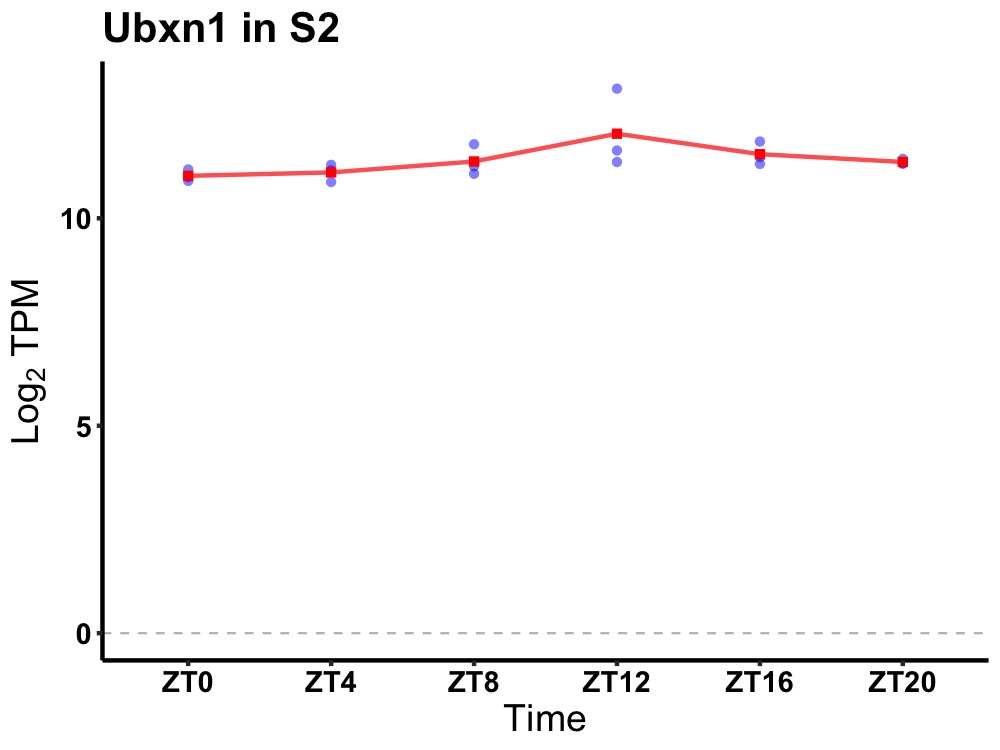 |
UBX domain protein 1 | 0.014 | 24 | 14 | 0.25 |
| ENSMUSG00000028992 | Gm21399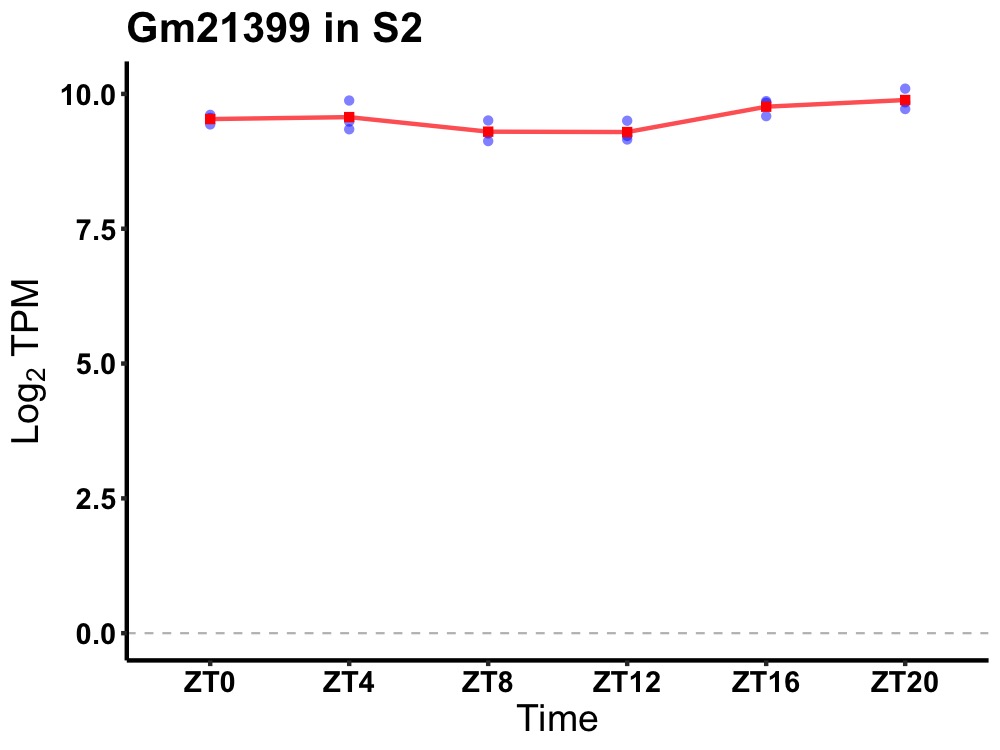 |
predicted gene, 21399 | 0.049 | 24 | 20 | 0.25 |
| ENSMUSG00000031753 | Swi5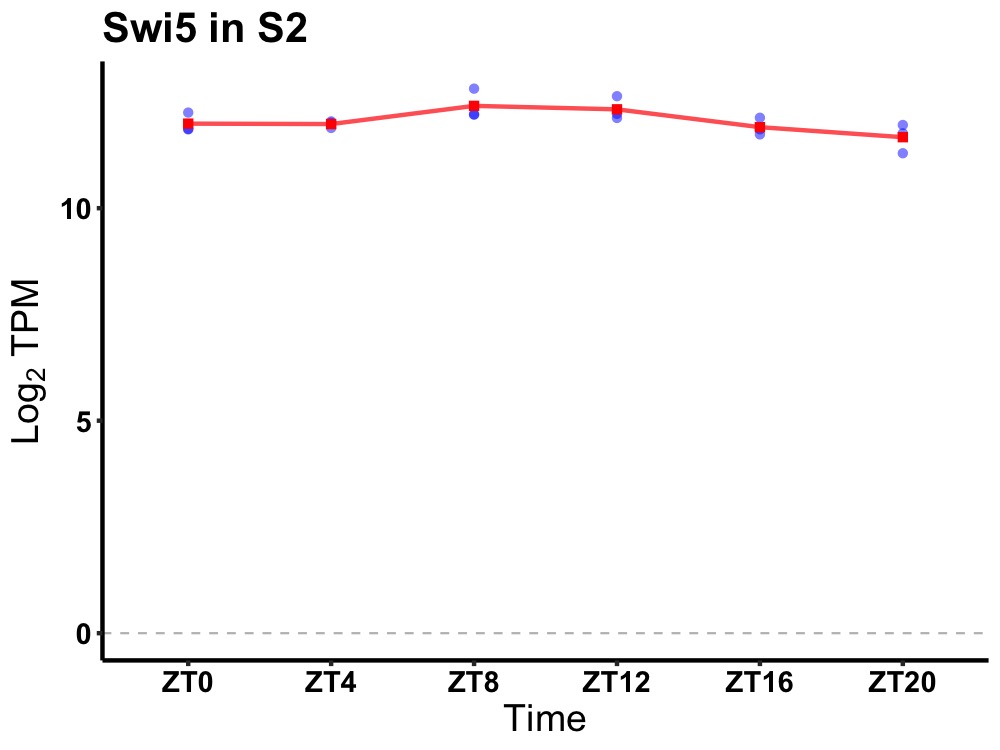 |
SWI5 recombination repair homolog (yeast) | 0.027 | 24 | 10 | 0.24 |
| ENSMUSG00000033793 | Upb1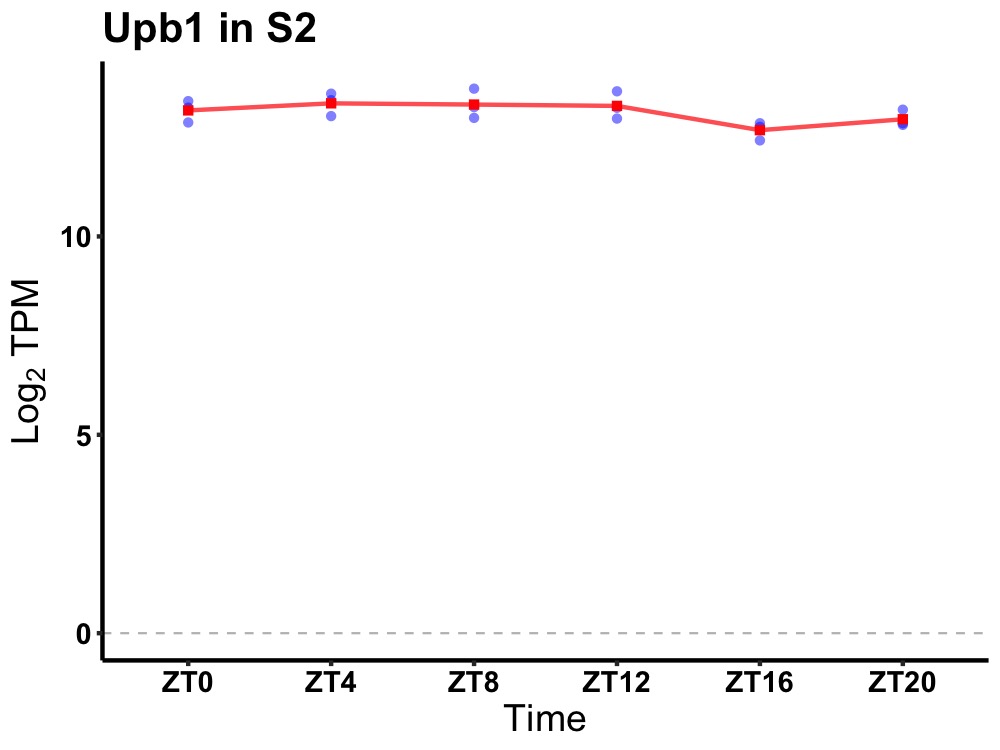 |
ureidopropionase, beta | 0.019 | 24 | 6 | 0.24 |
| ENSMUSG00000032002 | Rpl35a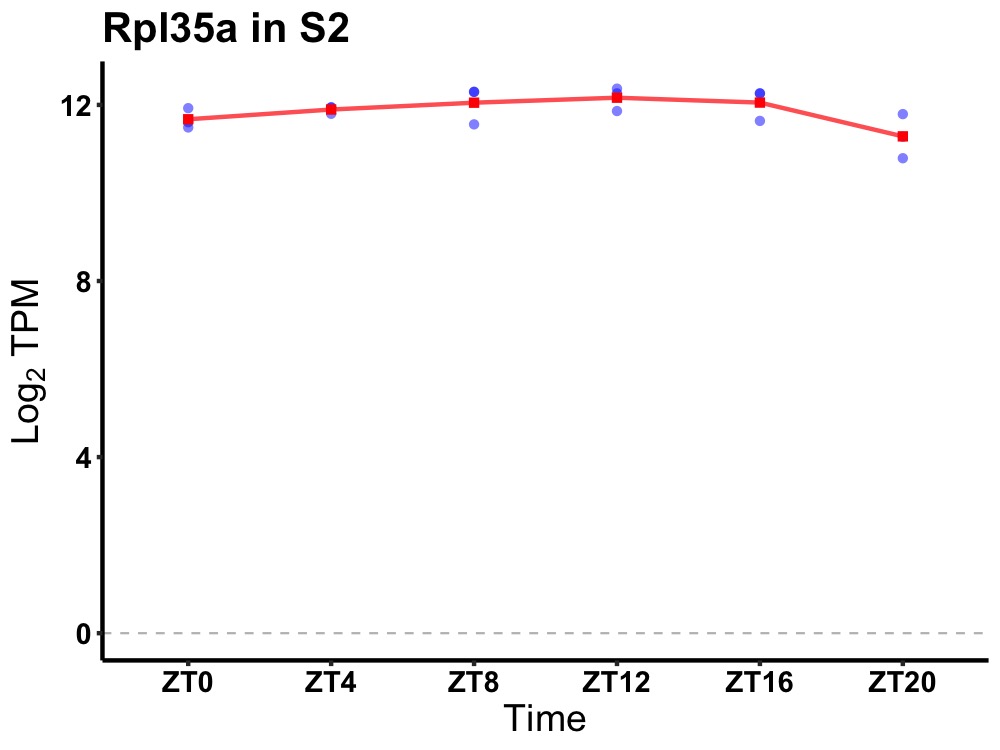 |
ribosomal protein L35A | 0.036 | 20 | 12 | 0.24 |
| ENSMUSG00000020198 | Col4a3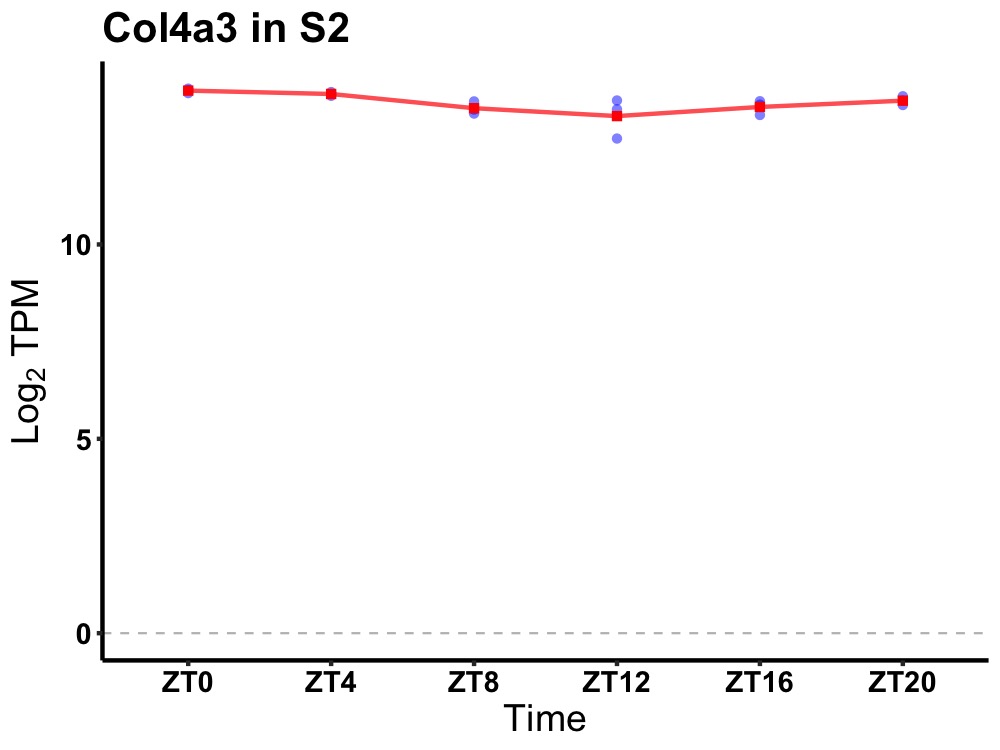 |
collagen, type IV, alpha 3 | 0.002 | 24 | 2 | 0.24 |
| ENSMUSG00000116385 | Mrpl32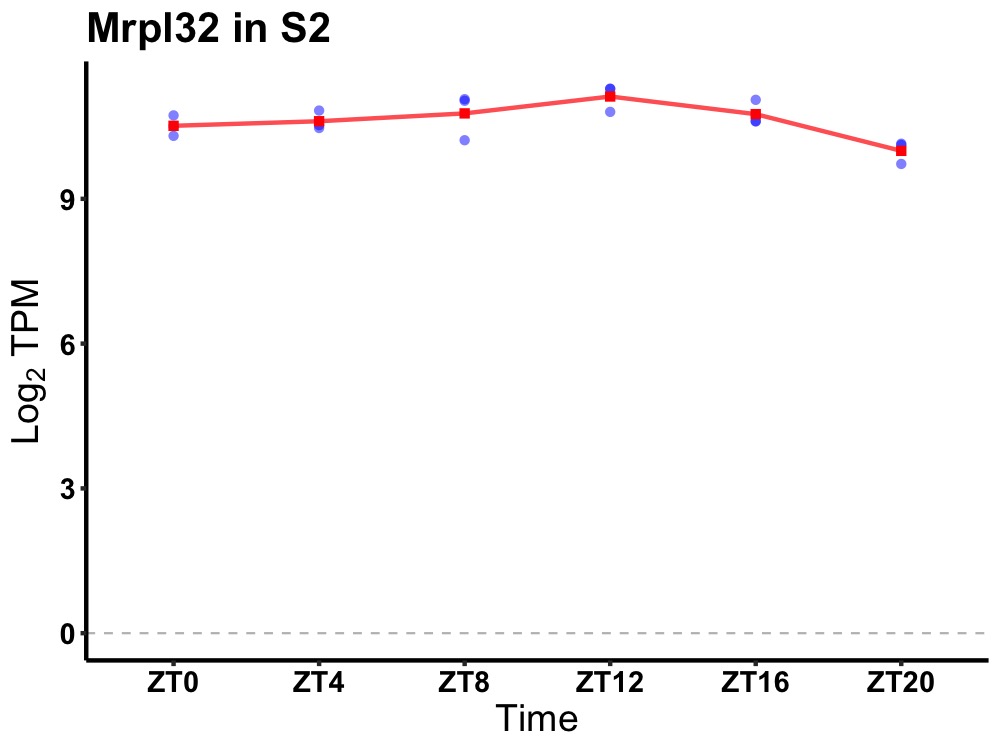 |
mitochondrial ribosomal protein L32 | 0.003 | 20 | 12 | 0.24 |
| ENSMUSG00000037353 | Mrps23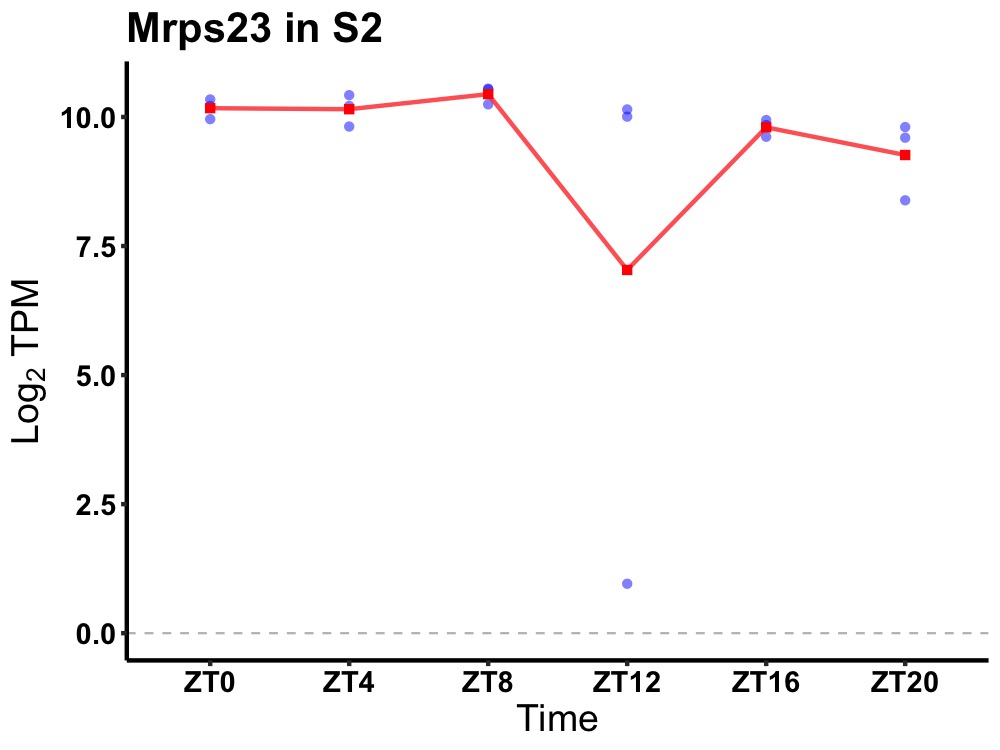 |
28S ribosomal protein S23, mitochondrial | 0.014 | 24 | 8 | 0.24 |
| ENSMUSG00000115505 | Gm9247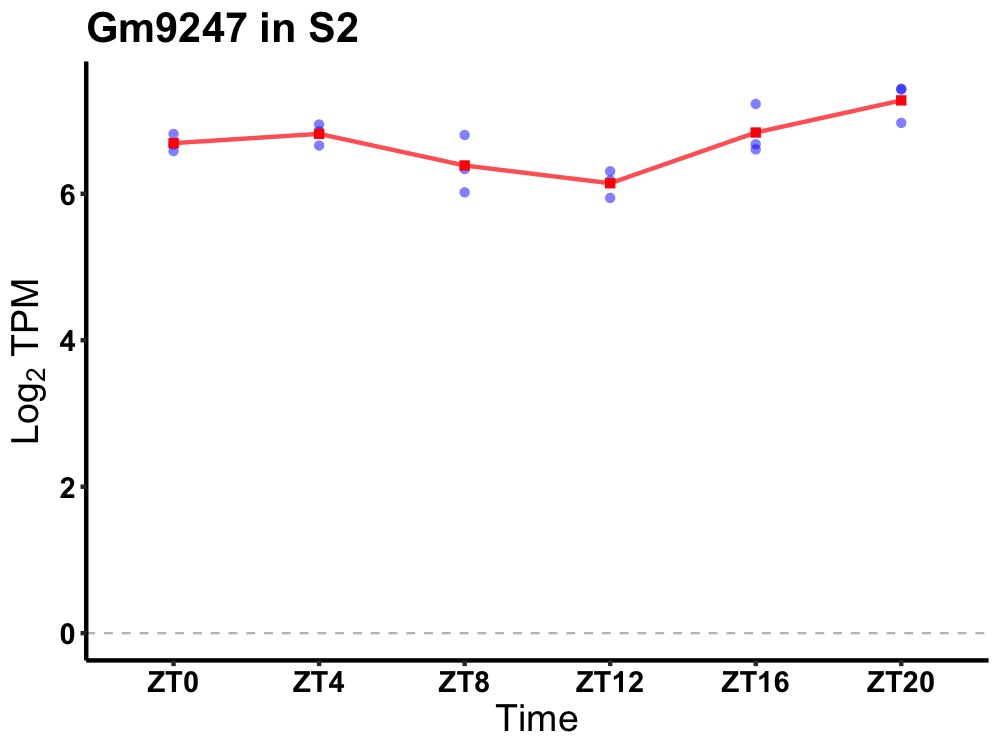 |
predicted gene 9247 | 0.014 | 20 | 2 | 0.24 |
| ENSMUSG00000048118 | H3f3b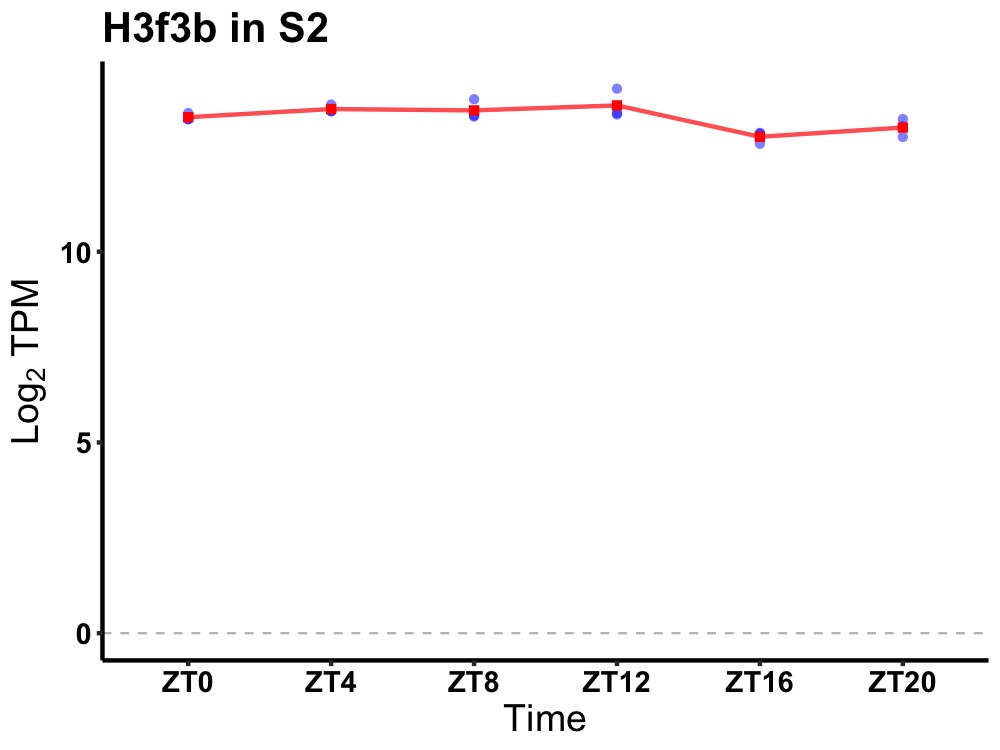 |
H3 histone, family 3B | 0.019 | 24 | 6 | 0.23 |
| ENSMUSG00000063875 | Rps6-ps1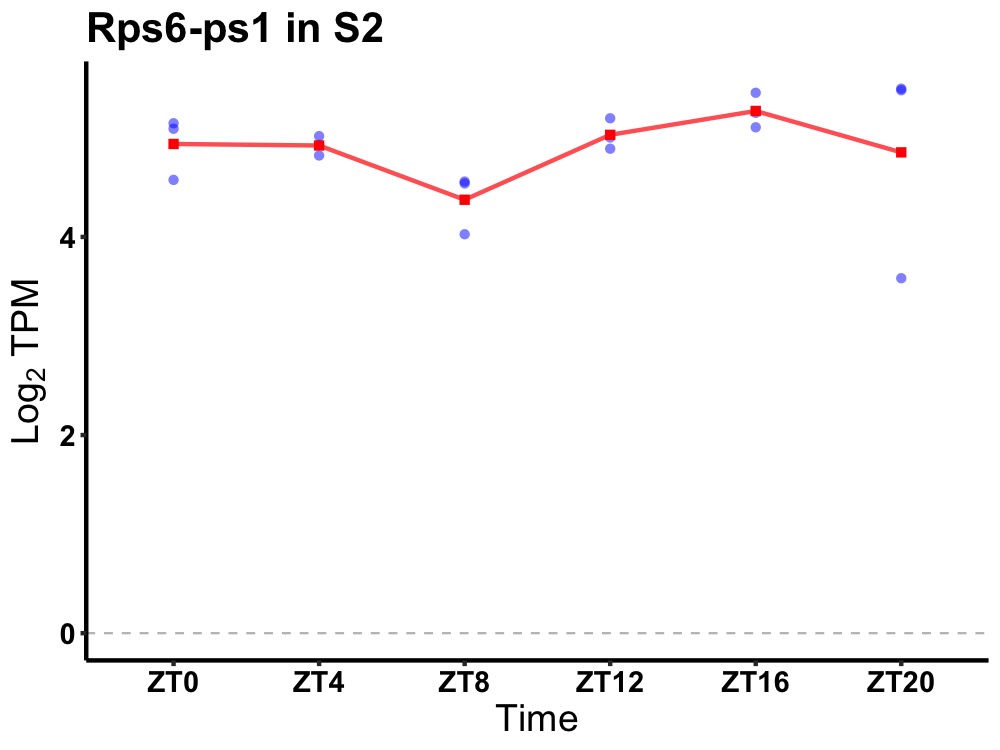 |
ribosomal protein S6, pseudogene 1 | 0.036 | 24 | 20 | 0.23 |
| ENSMUSG00000086174 | Gm6425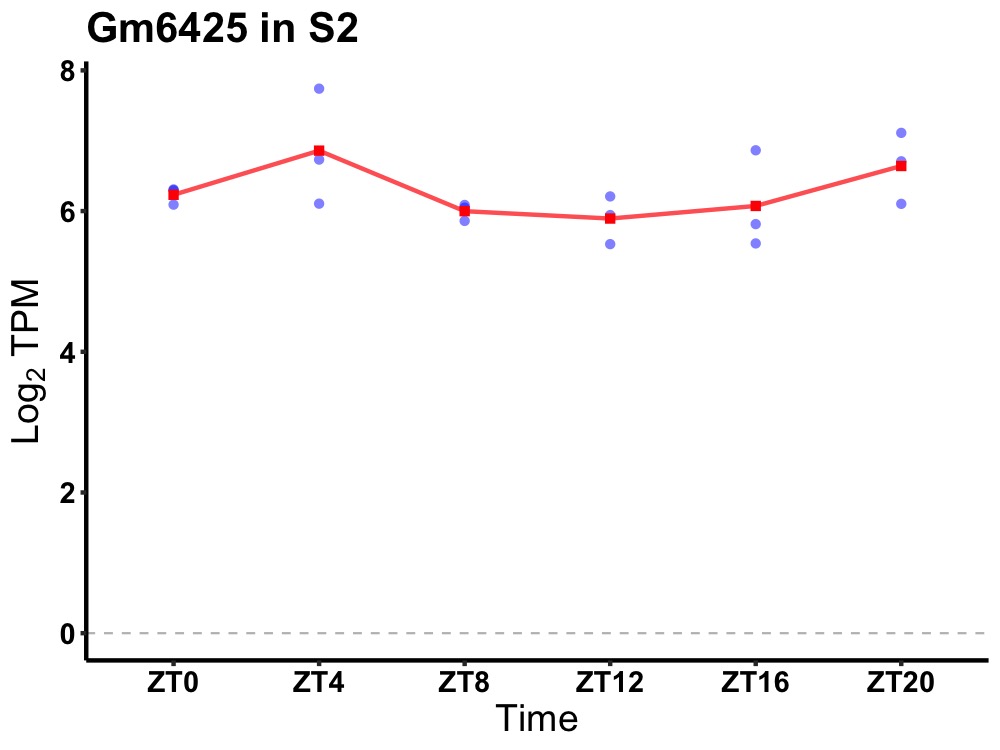 |
predicted pseudogene 6425 | 0.036 | 20 | 4 | 0.23 |
| ENSMUSG00000081729 | Hspa9-ps1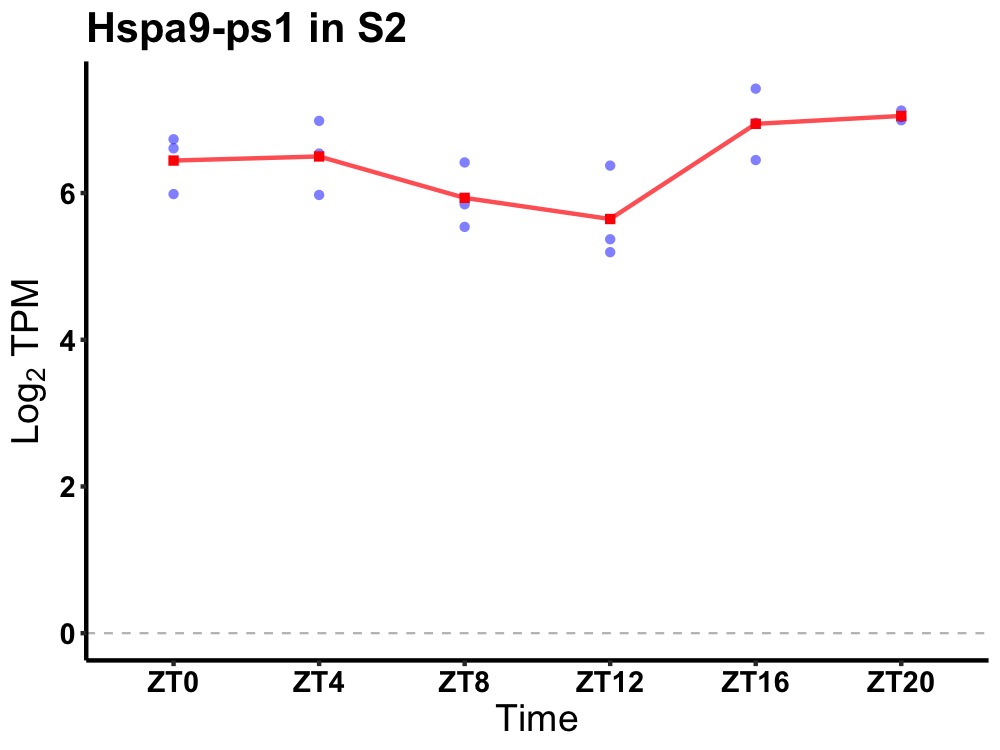 |
heat shock protein 9, pseudogene 1 | 0.019 | 20 | 2 | 0.23 |
| ENSMUSG00000086507 | Mpc2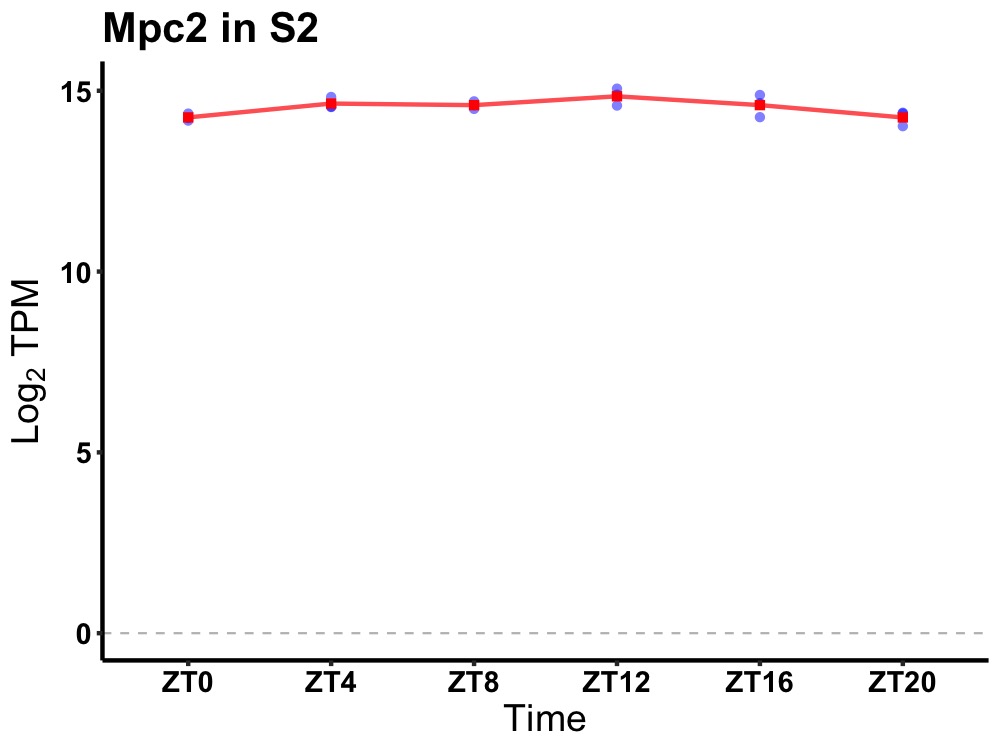 |
mitochondrial pyruvate carrier 2 | 0.014 | 24 | 12 | 0.22 |
| ENSMUSG00000040370 | Rpl26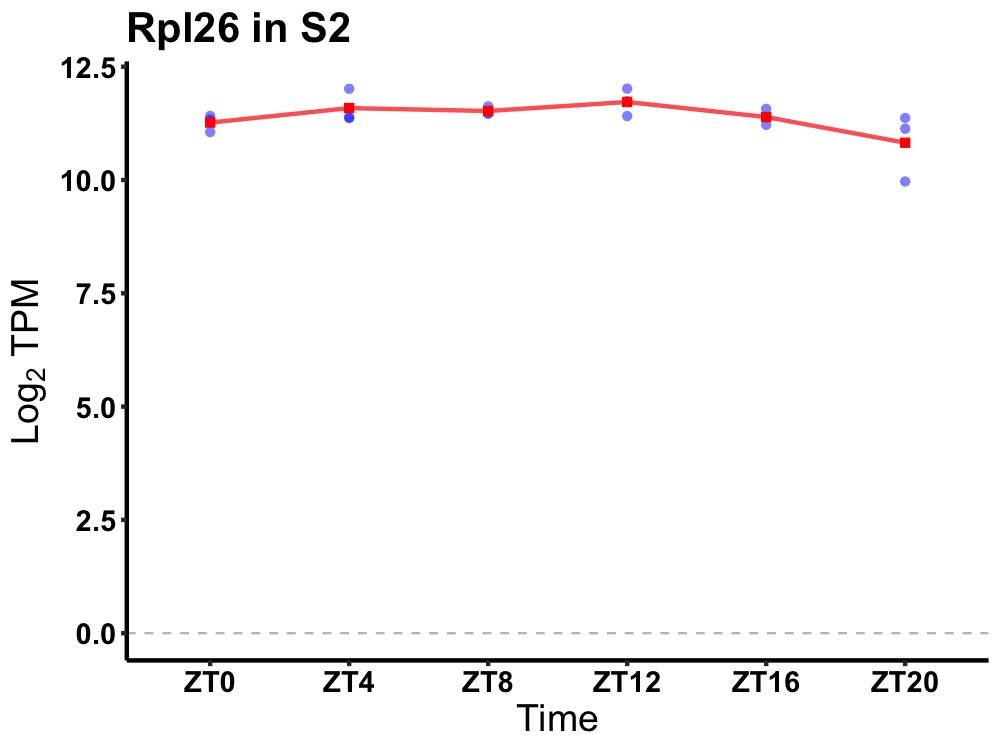 |
ribosomal protein L26 | 0.036 | 24 | 10 | 0.22 |
| ENSMUSG00000028167 | Rpl13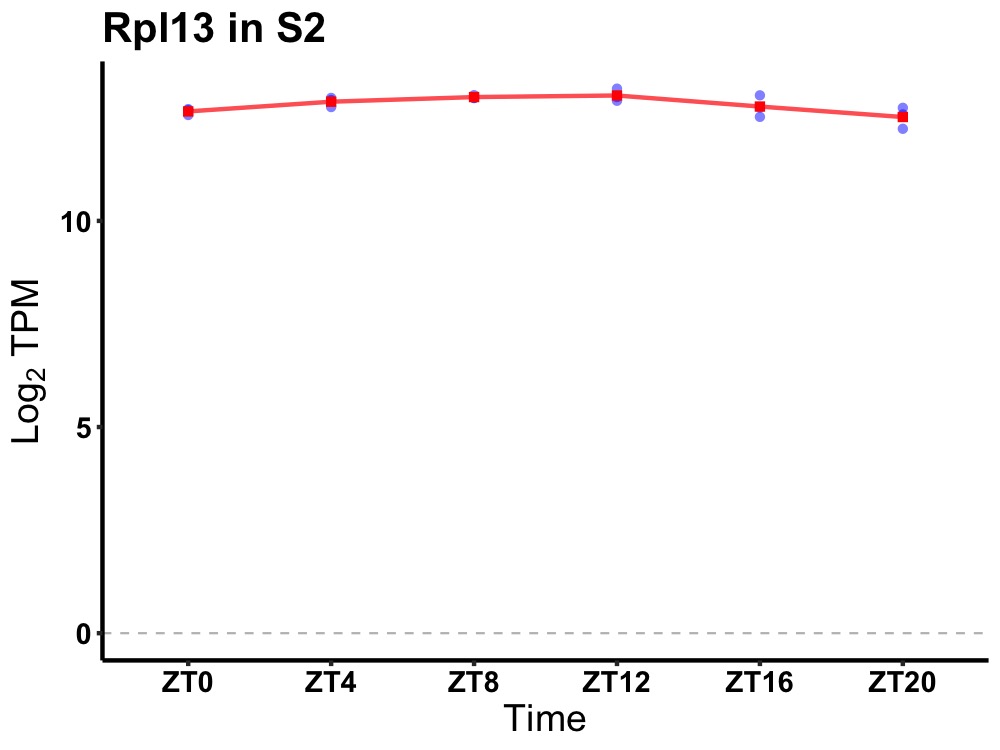 |
ribosomal protein L13 | 0.014 | 24 | 10 | 0.22 |
| ENSMUSG00000039519 | Gng5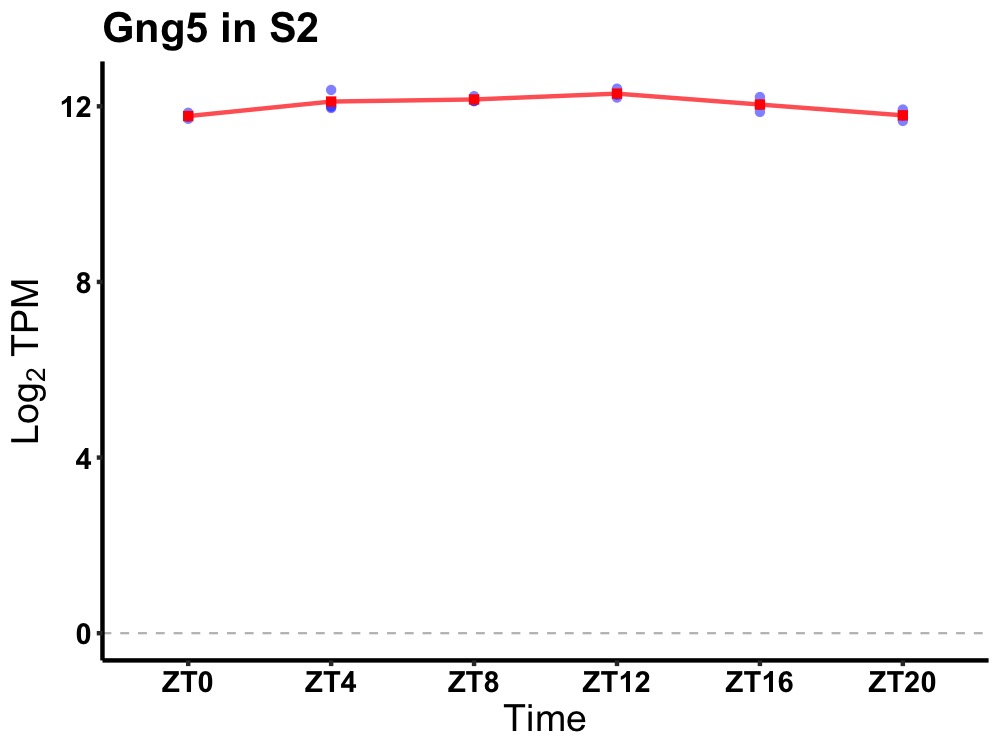 |
guanine nucleotide binding protein (G protein), gamma 5 | 0.001 | 24 | 12 | 0.22 |
| ENSMUSG00000042558 | Selenbp1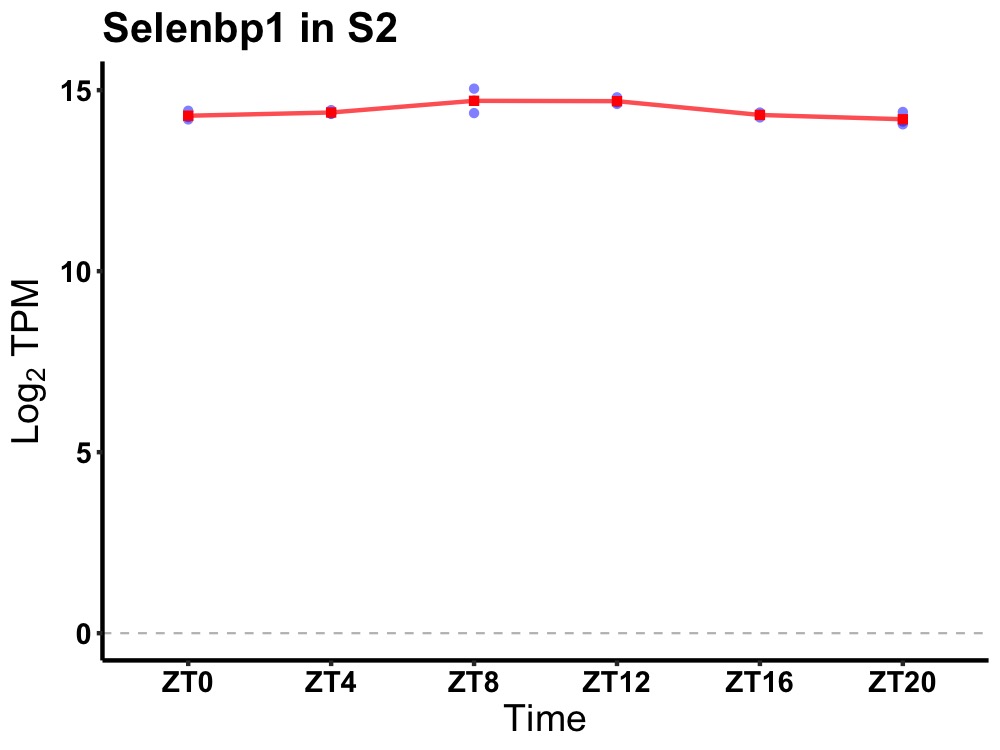 |
selenium binding protein 1 | 0.007 | 24 | 10 | 0.21 |
| ENSMUSG00000013160 | Abhd14b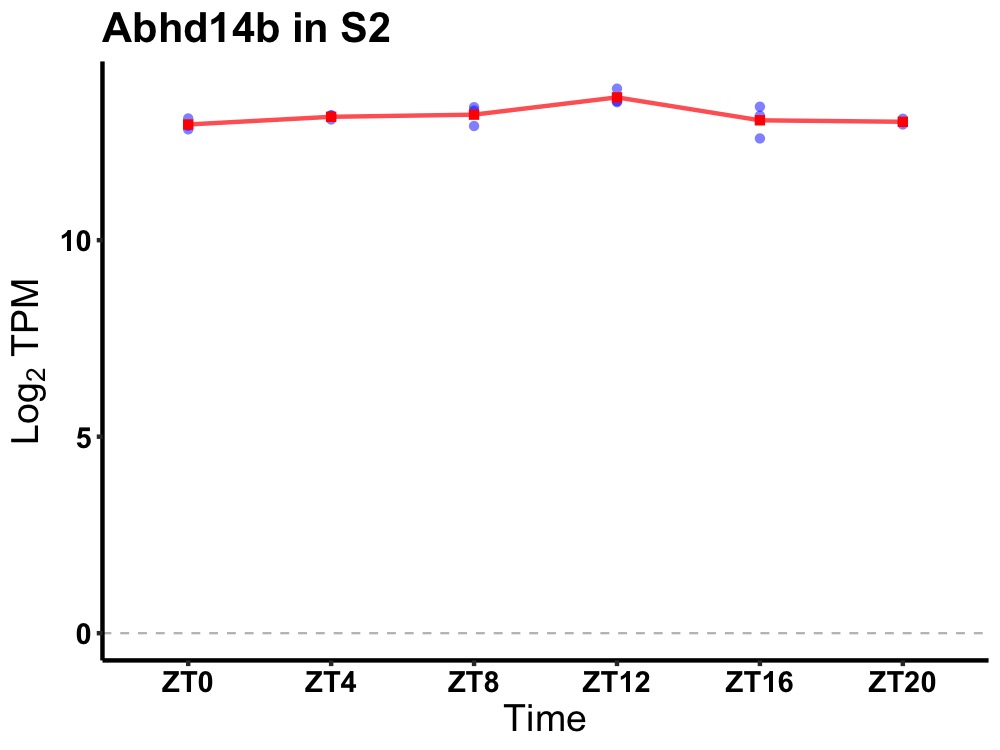 |
abhydrolase domain containing 14b | 0.036 | 24 | 12 | 0.21 |
| ENSMUSG00000114179 | Mrps21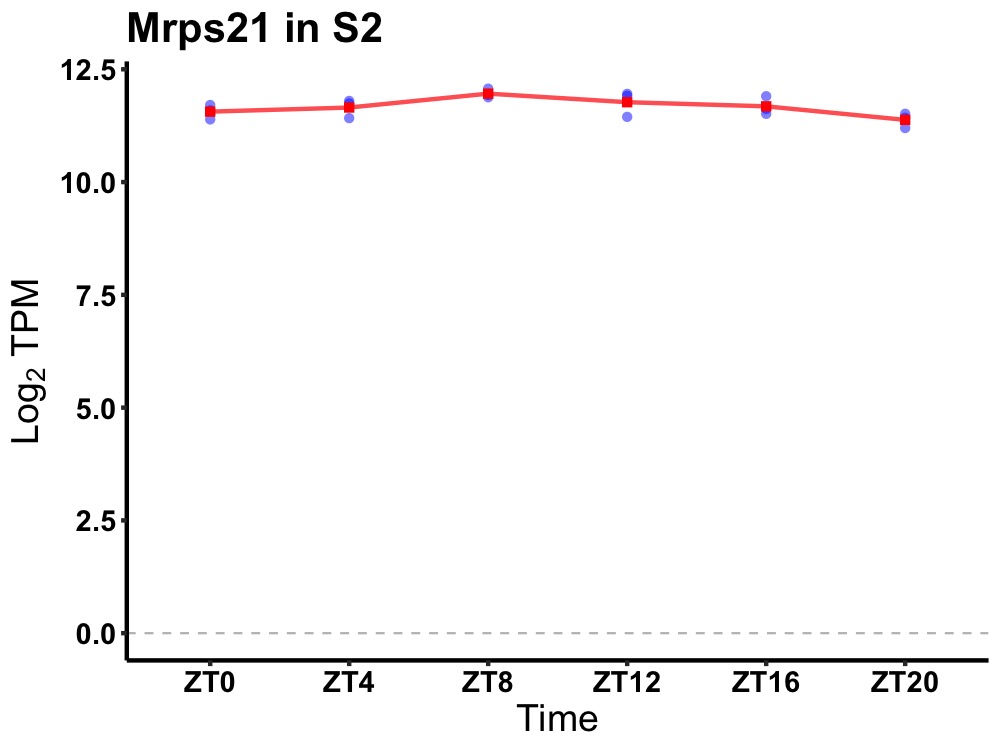 |
mitochondrial ribosomal protein S21 | 0.027 | 24 | 10 | 0.21 |
| ENSMUSG00000033335 | Fkbp2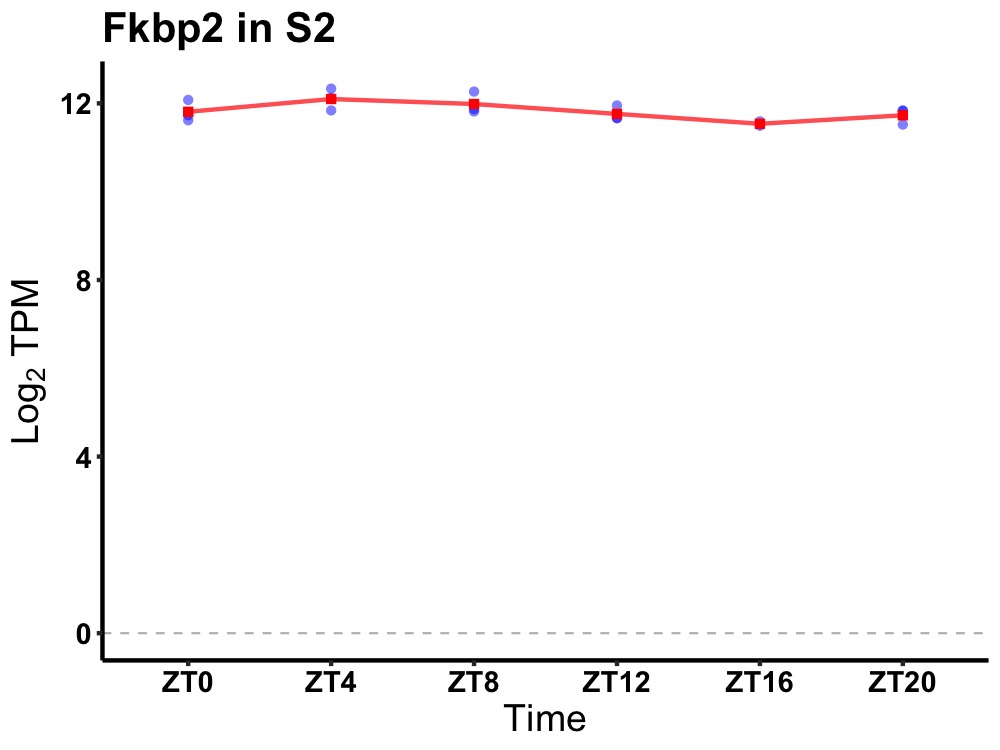 |
FK506 binding protein 2 | 0.014 | 24 | 6 | 0.20 |
| ENSMUSG00000042210 | Fxyd2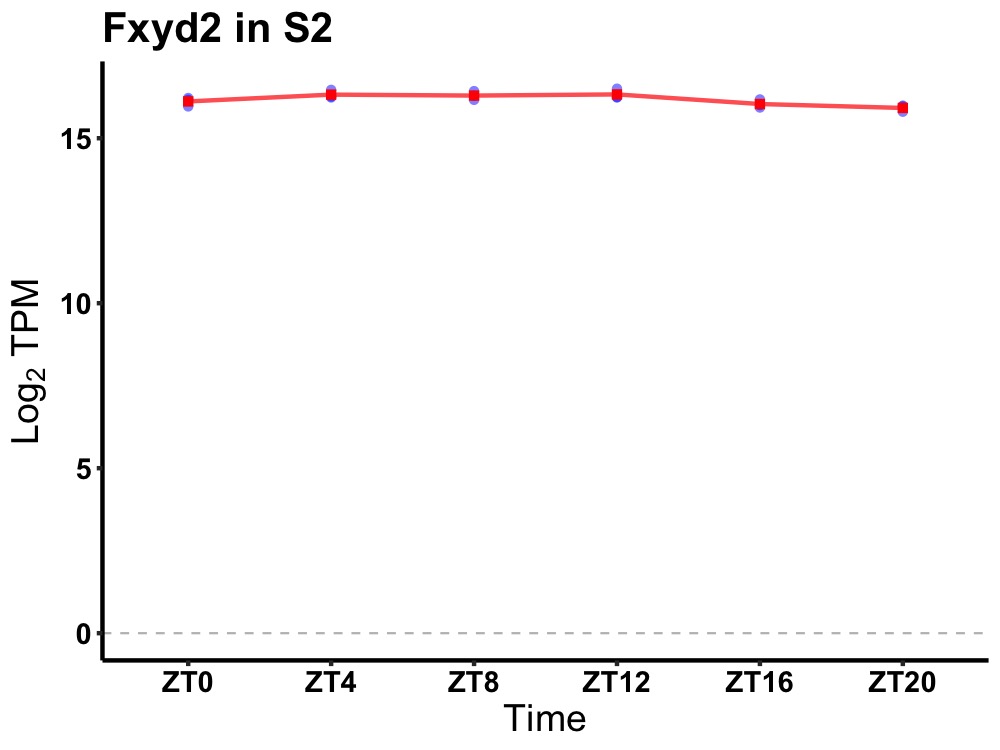 |
FXYD domain-containing ion transport regulator 2 | 0.007 | 24 | 8 | 0.20 |
| ENSMUSG00000082368 | Gm11225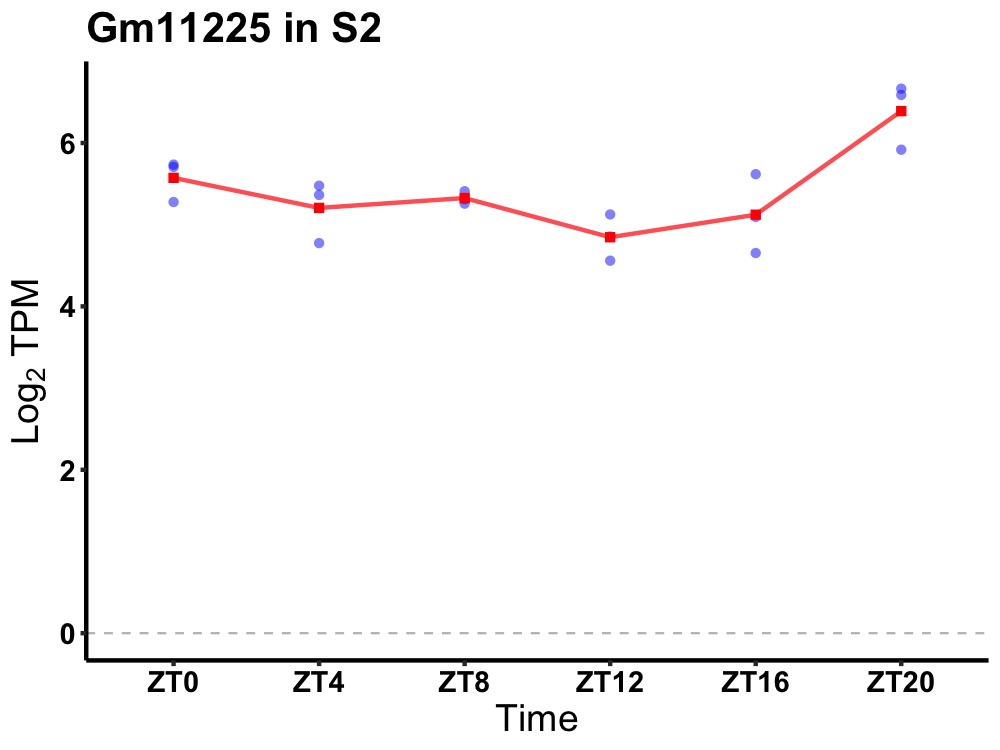 |
predicted gene 11225 | 0.010 | 20 | 2 | 0.20 |
| ENSMUSG00000038762 | mt-Nd4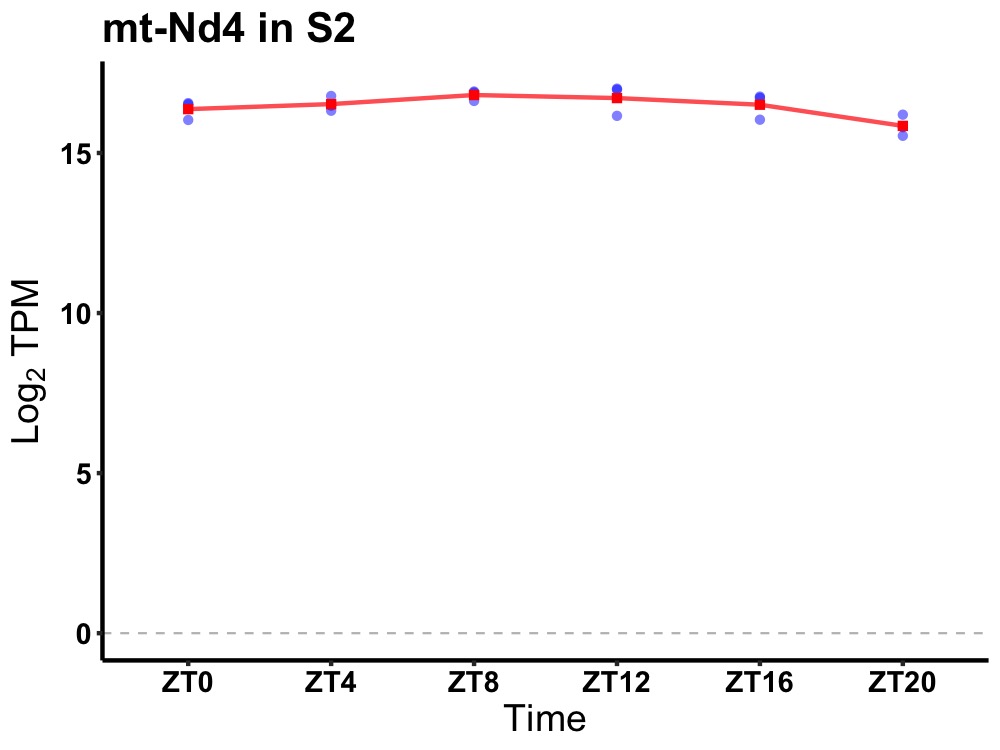 |
mitochondrially encoded NADH dehydrogenase 4 | 0.027 | 20 | 12 | 0.20 |
| ENSMUSG00000083914 | Rps18-ps1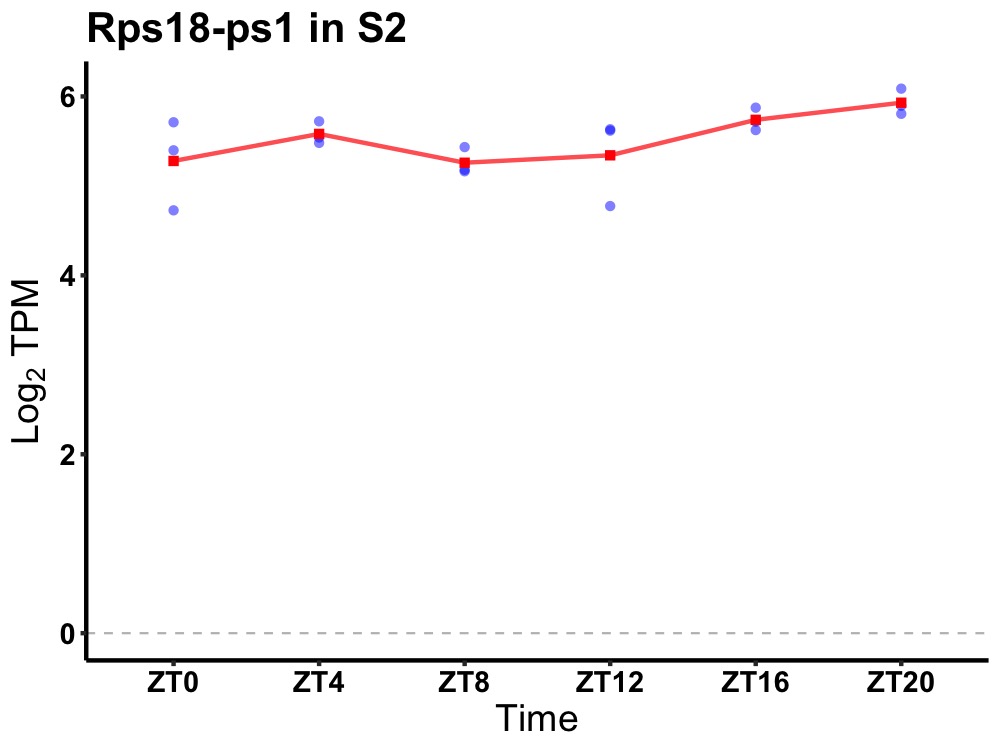 |
ribosomal protein S18, pseudogene 1 | 0.049 | 24 | 20 | 0.20 |
| ENSMUSG00000004843 | Psmb3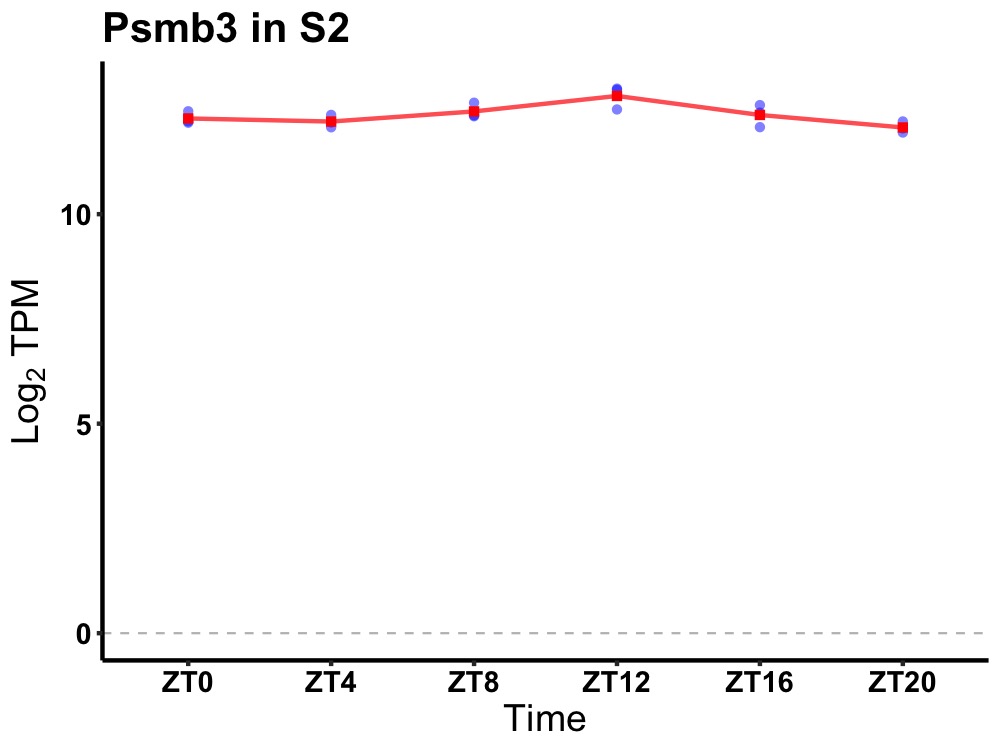 |
proteasome (prosome, macropain) subunit, beta type 3 | 0.019 | 20 | 12 | 0.19 |
| ENSMUSG00000013622 | Taldo1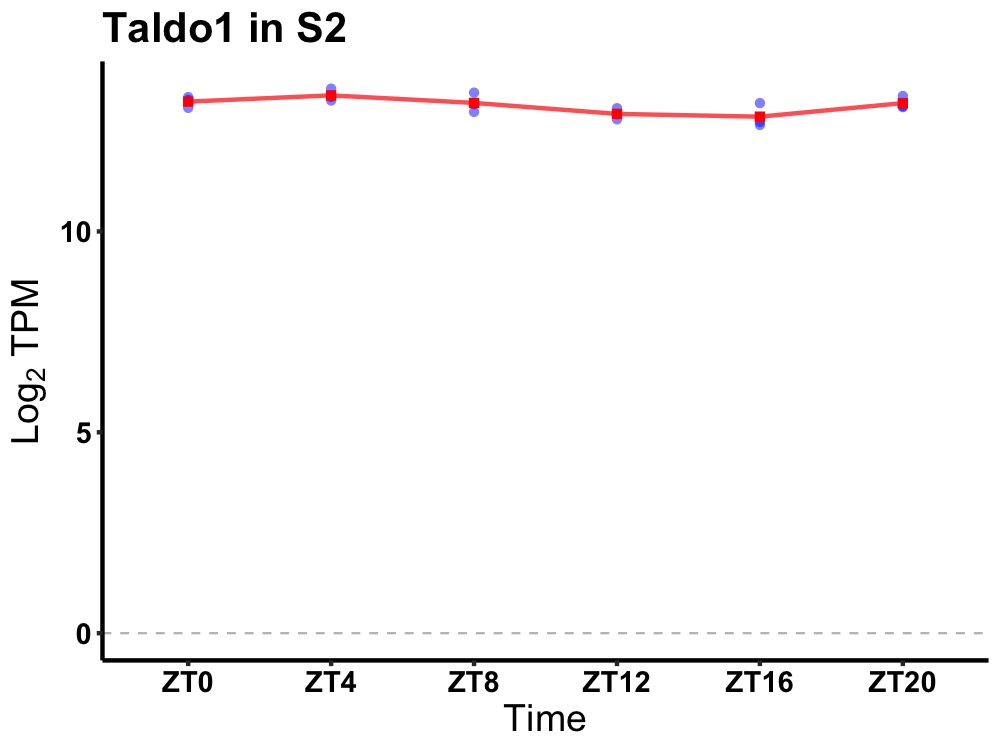 |
transaldolase 1 | 0.036 | 20 | 6 | 0.19 |
| ENSMUSG00000028745 | Rps19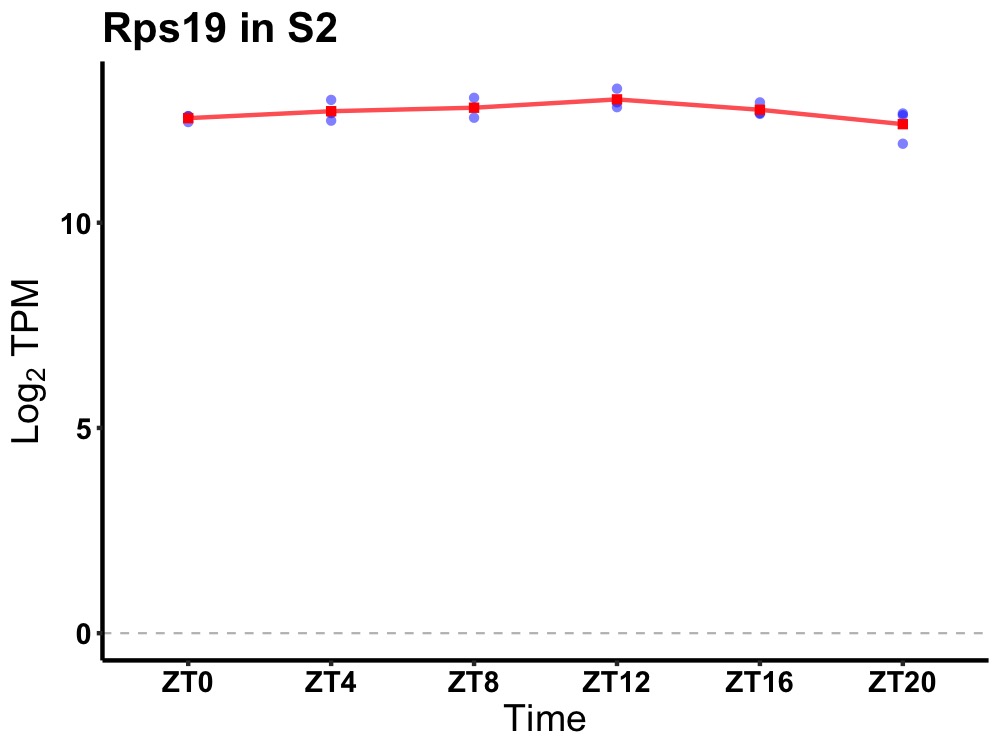 |
ribosomal protein S19 | 0.036 | 24 | 12 | 0.18 |
| ENSMUSG00000063953 | Cisd1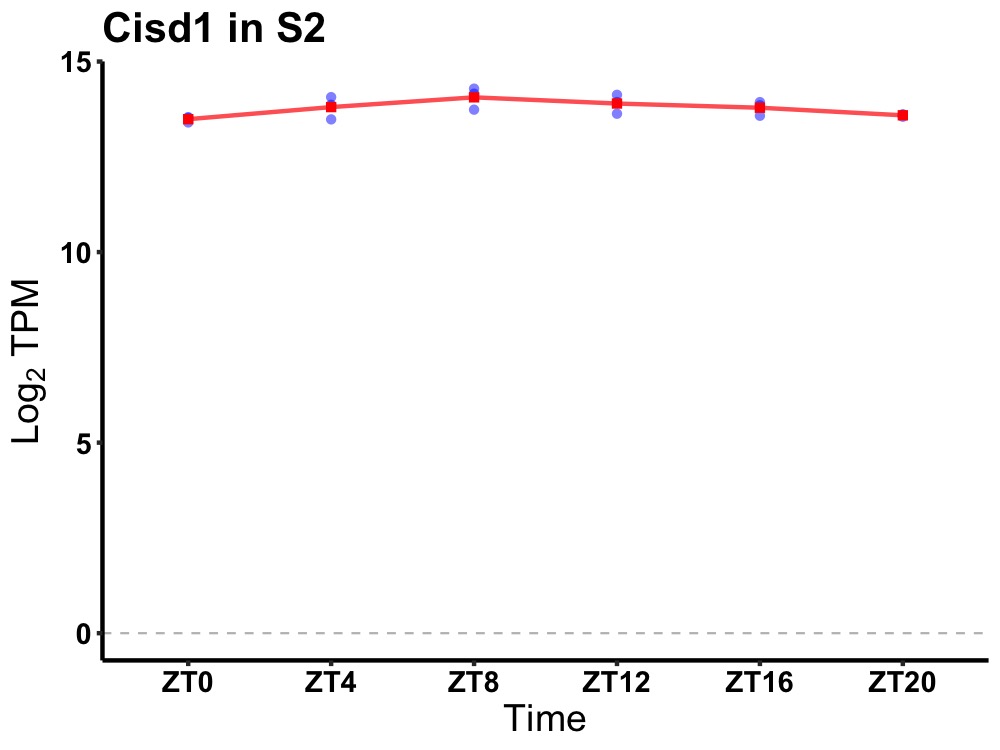 |
CDGSH iron sulfur domain 1 | 0.005 | 20 | 10 | 0.18 |
| ENSMUSG00000028333 | Selenof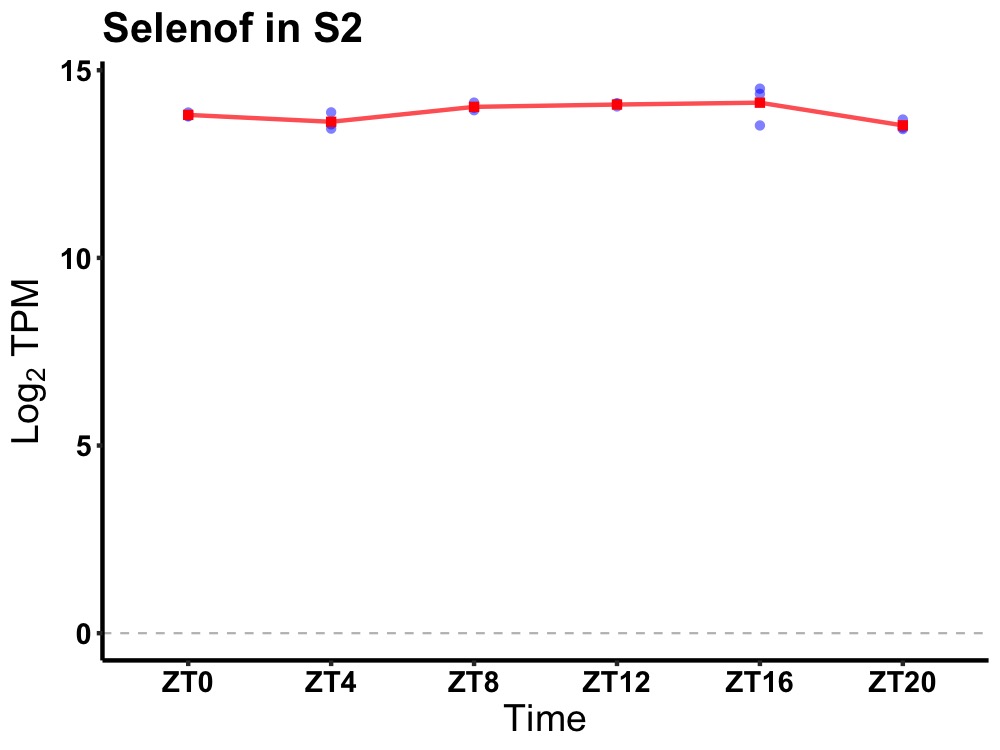 |
selenoprotein F | 0.027 | 20 | 14 | 0.18 |
| ENSMUSG00000083885 | Mrpl54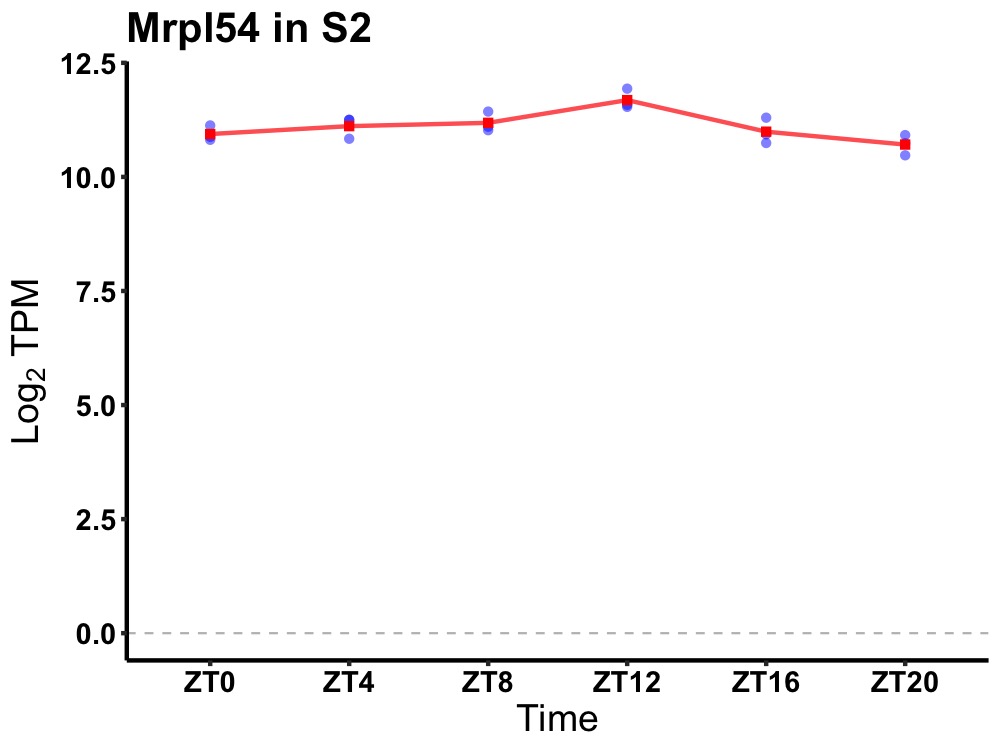 |
mitochondrial ribosomal protein L54 | 0.007 | 20 | 12 | 0.18 |
| ENSMUSG00000079418 | Pebp1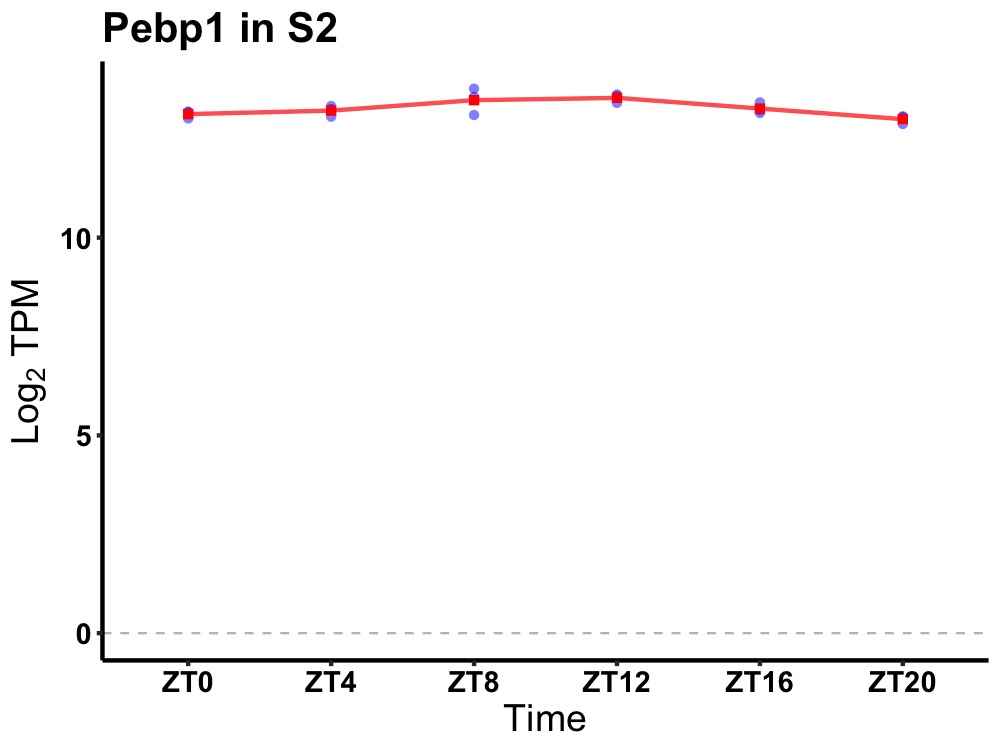 |
phosphatidylethanolamine binding protein 1 | 0.007 | 20 | 12 | 0.18 |
| ENSMUSG00000036138 | Tmem176b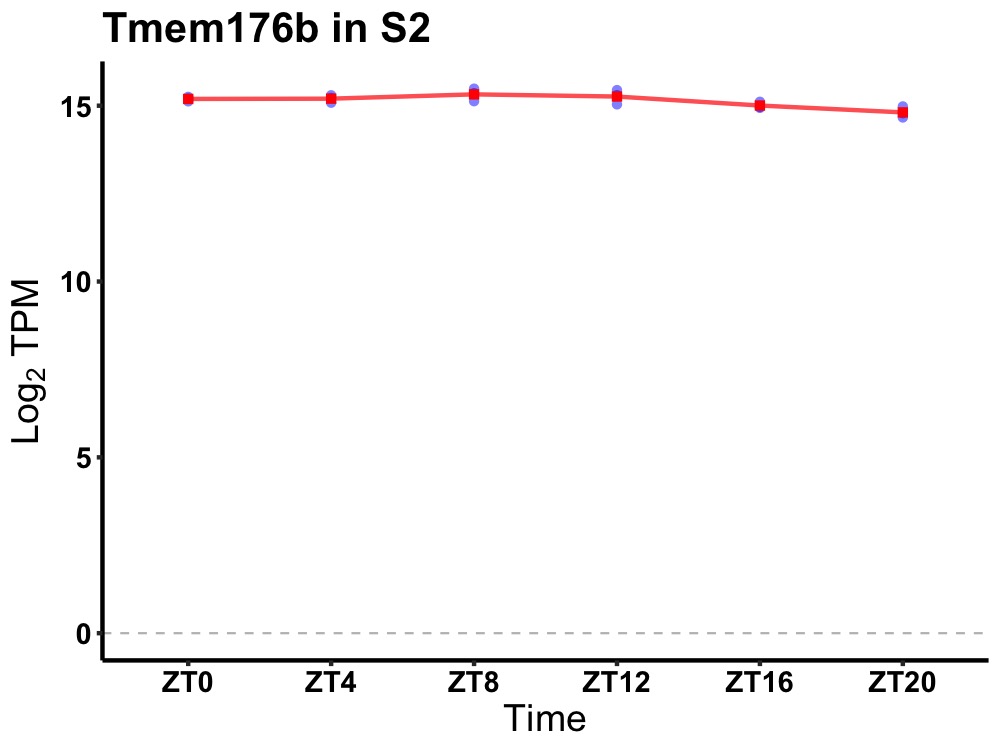 |
transmembrane protein 176B | 0.010 | 24 | 8 | 0.18 |
| ENSMUSG00000027349 | Glud1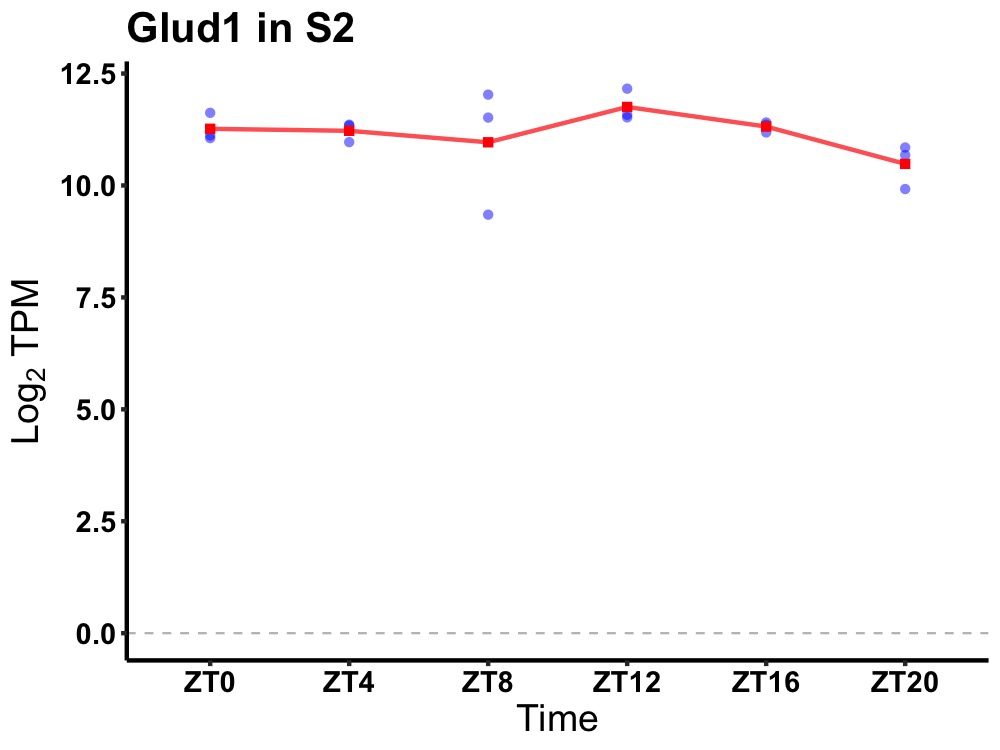 |
glutamate dehydrogenase 1 | 0.036 | 20 | 12 | 0.18 |
| ENSMUSG00000067212 | Erh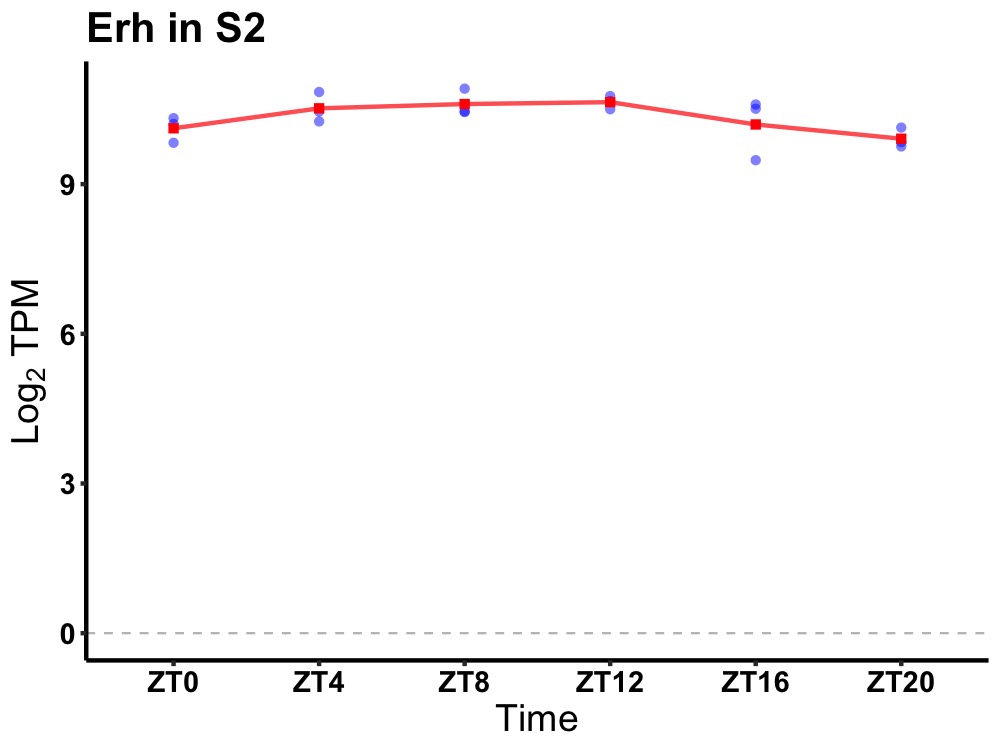 |
ERH mRNA splicing and mitosis factor | 0.049 | 20 | 12 | 0.17 |
| ENSMUSG00000117788 | Prdx5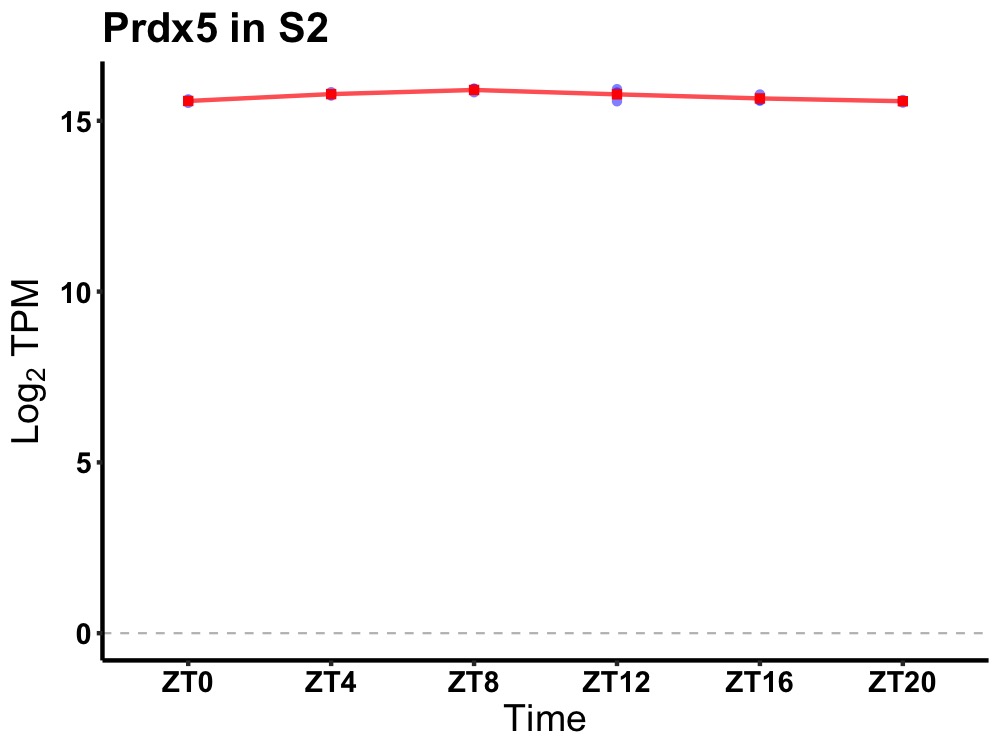 |
peroxiredoxin 5 | 0.007 | 24 | 10 | 0.17 |
| ENSMUSG00000022641 | Uqcrq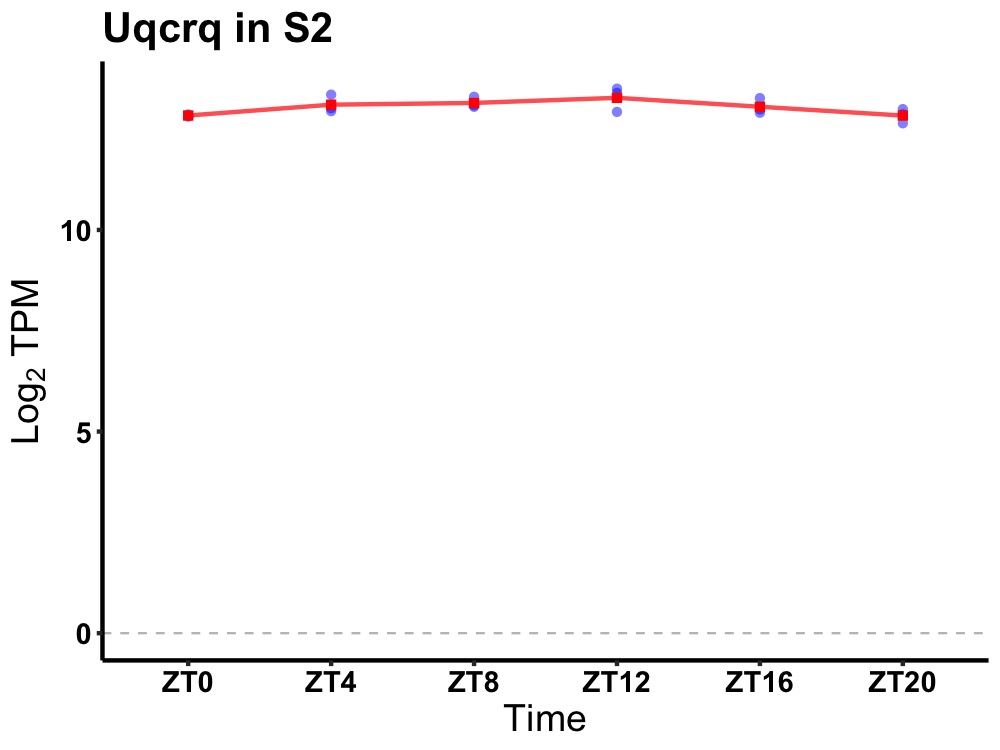 |
ubiquinol-cytochrome c reductase, complex III subunit VII | 0.019 | 24 | 12 | 0.17 |
| ENSMUSG00000094683 | Atp5g3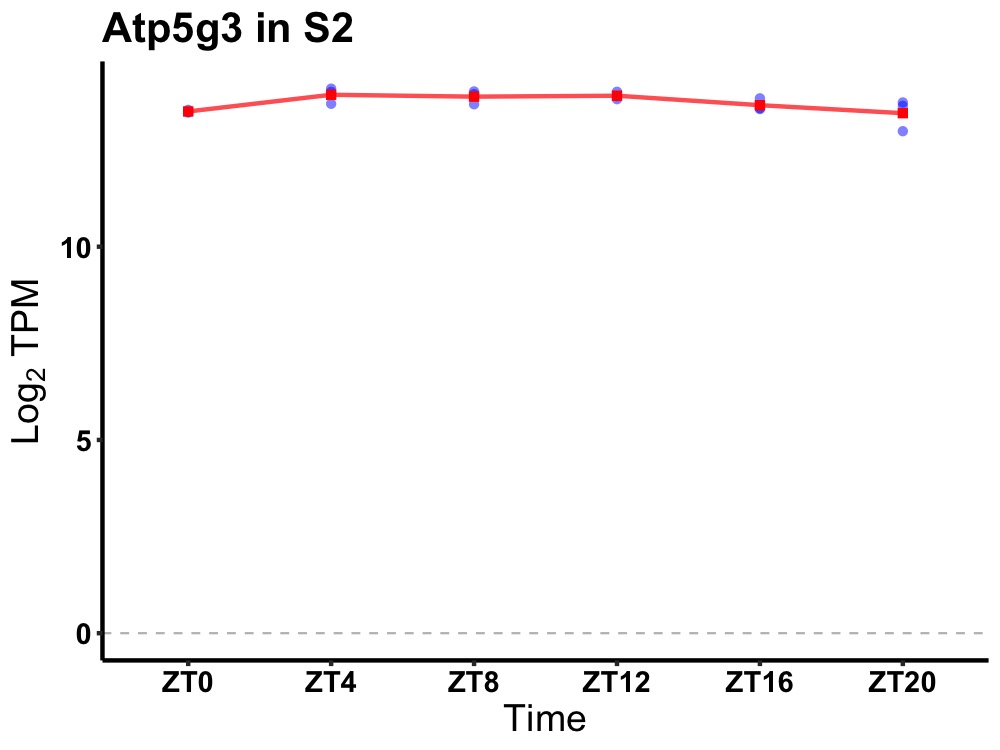 |
ATP synthase, H+ transporting, mitochondrial F0 complex, subunit C3 (subunit 9) | 0.027 | 20 | 10 | 0.17 |
| ENSMUSG00000039105 | Uqcrb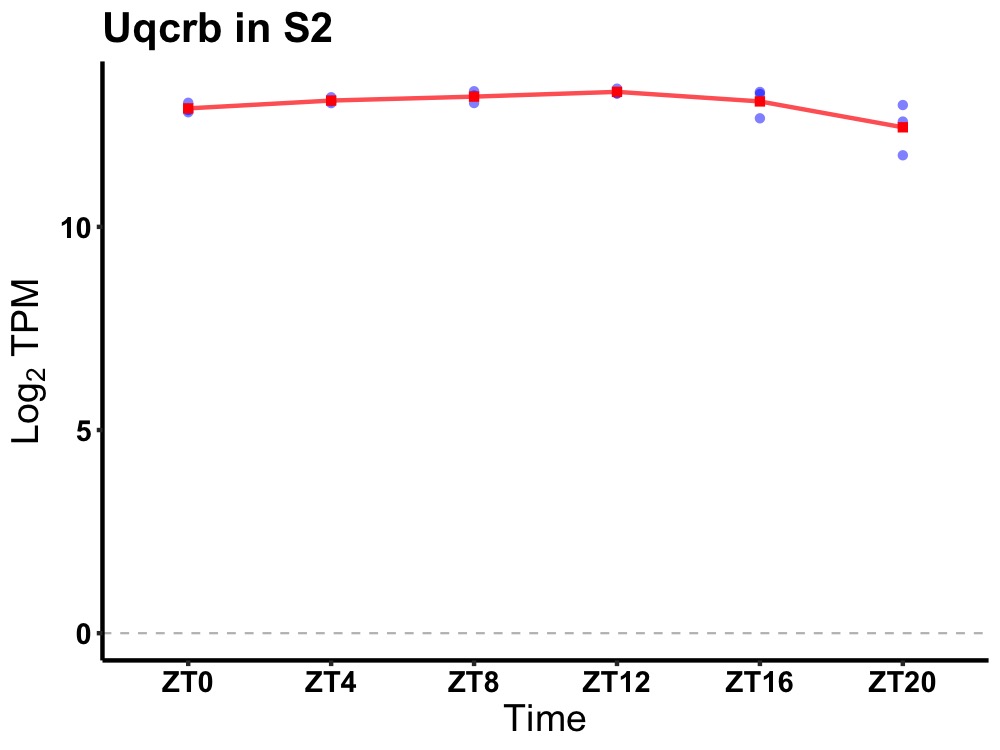 |
ubiquinol-cytochrome c reductase binding protein | 0.003 | 20 | 12 | 0.16 |
| ENSMUSG00000083327 | Vcp-rs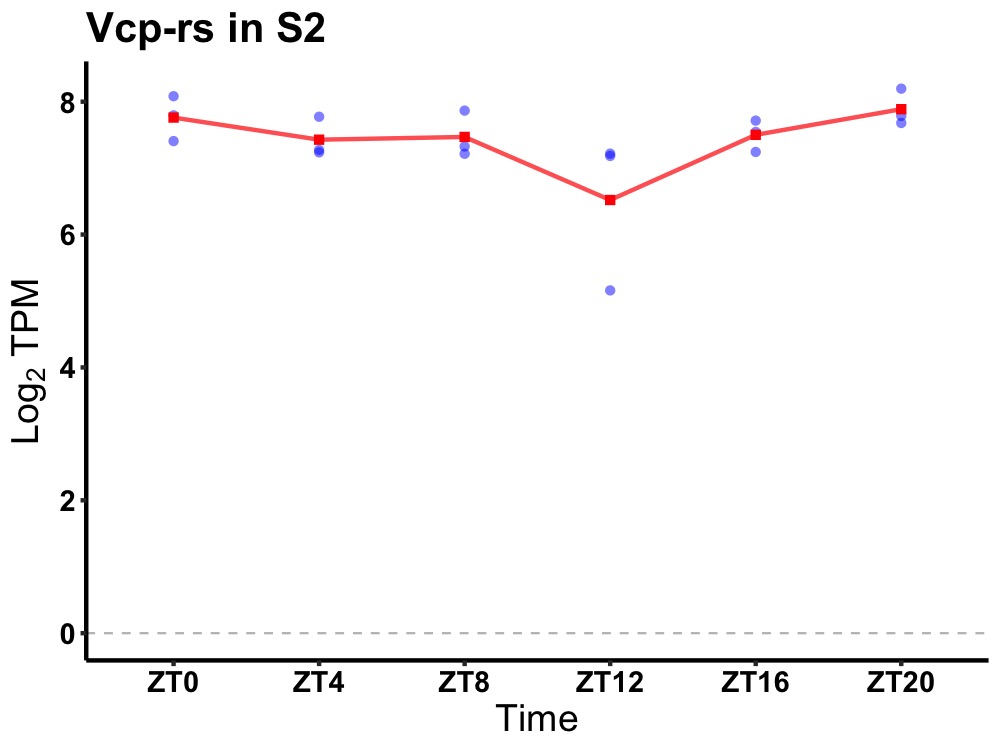 |
valosin containing protein, related sequence | 0.036 | 20 | 2 | 0.16 |
| ENSMUSG00000022244 | Hykk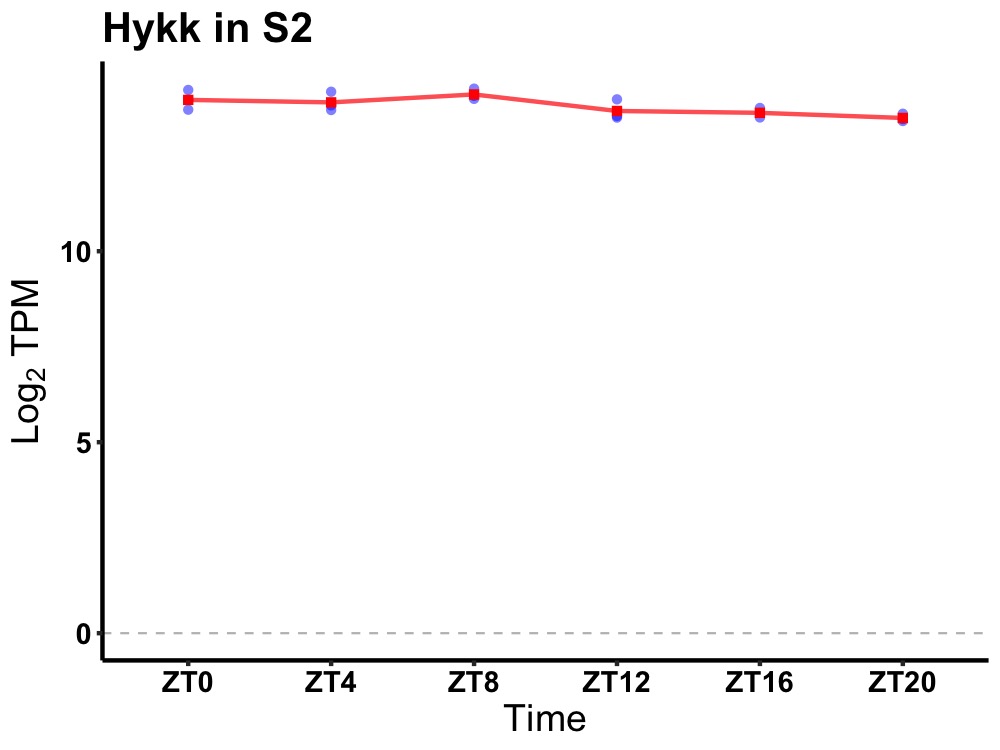 |
hydroxylysine kinase 1 | 0.049 | 24 | 8 | 0.16 |
| ENSMUSG00000026156 | Acot13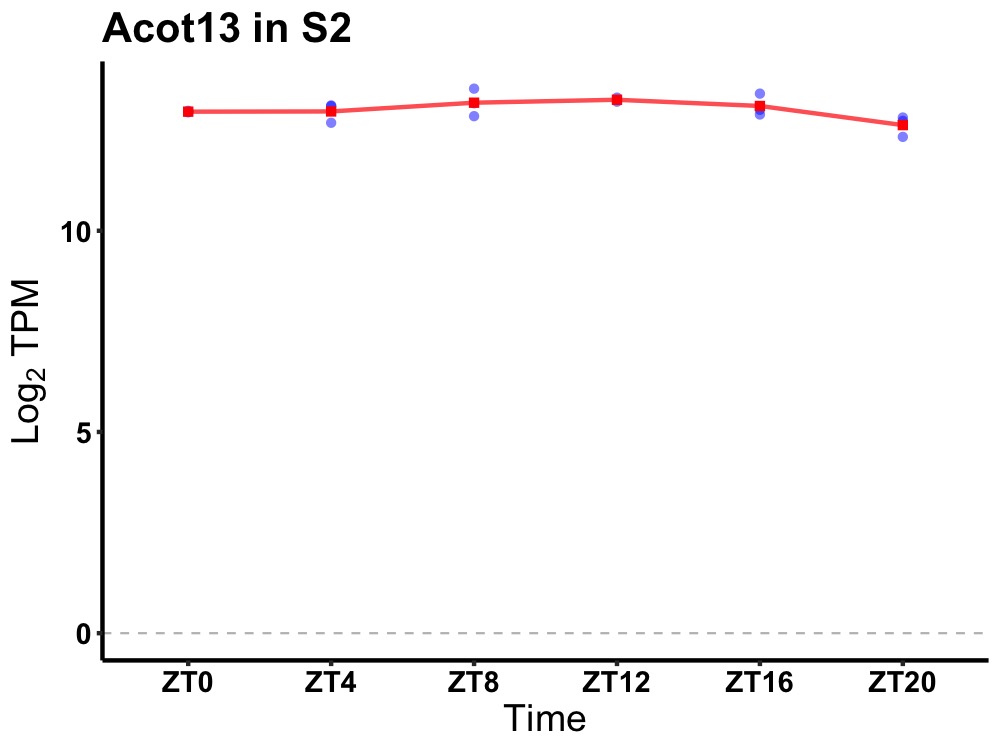 |
acyl-CoA thioesterase 13 | 0.027 | 20 | 12 | 0.16 |
| ENSMUSG00000018474 | Mrpl28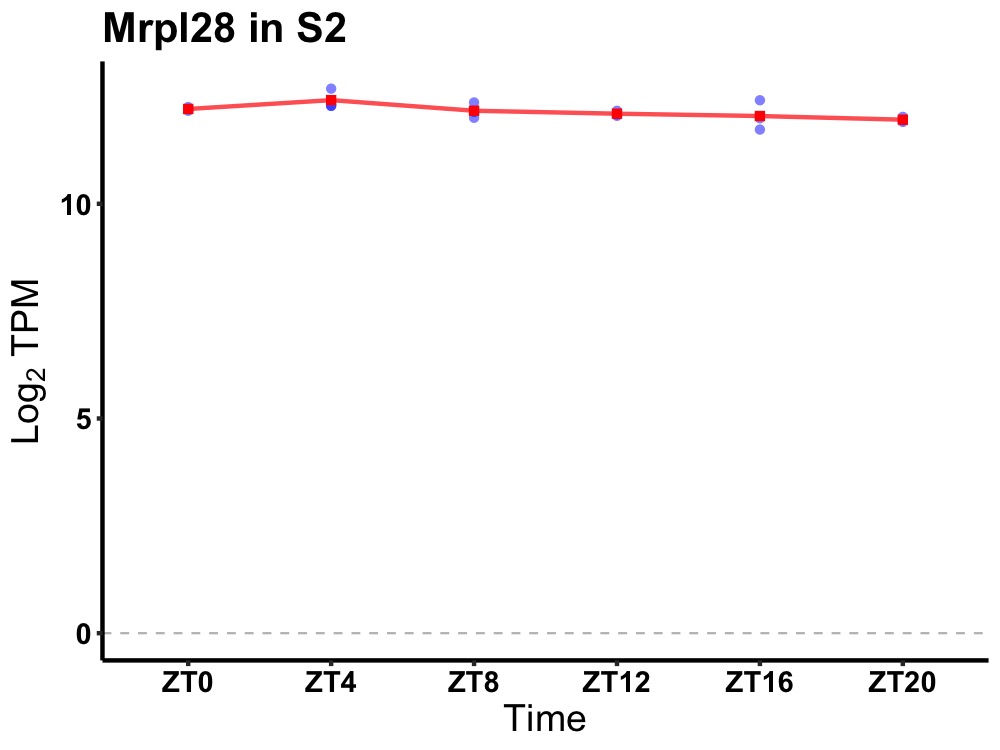 |
mitochondrial ribosomal protein L28 | 0.049 | 24 | 6 | 0.16 |
| ENSMUSG00000117939 | AC110212.1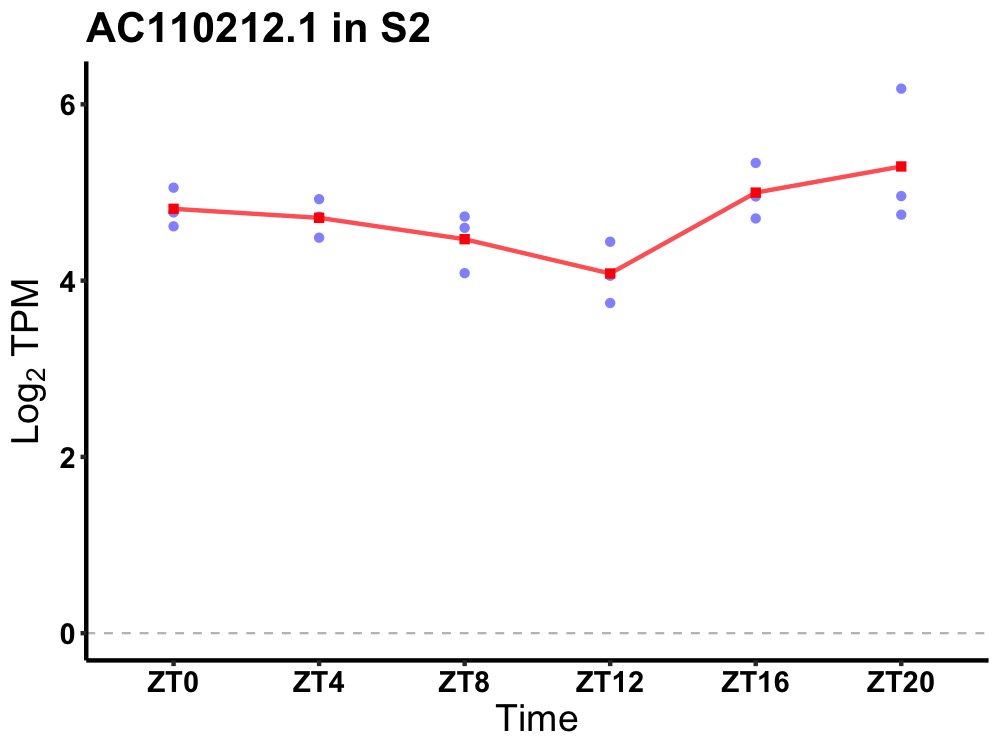 |
none | 0.049 | 20 | 2 | 0.15 |
| ENSMUSG00000034254 | Atp5f1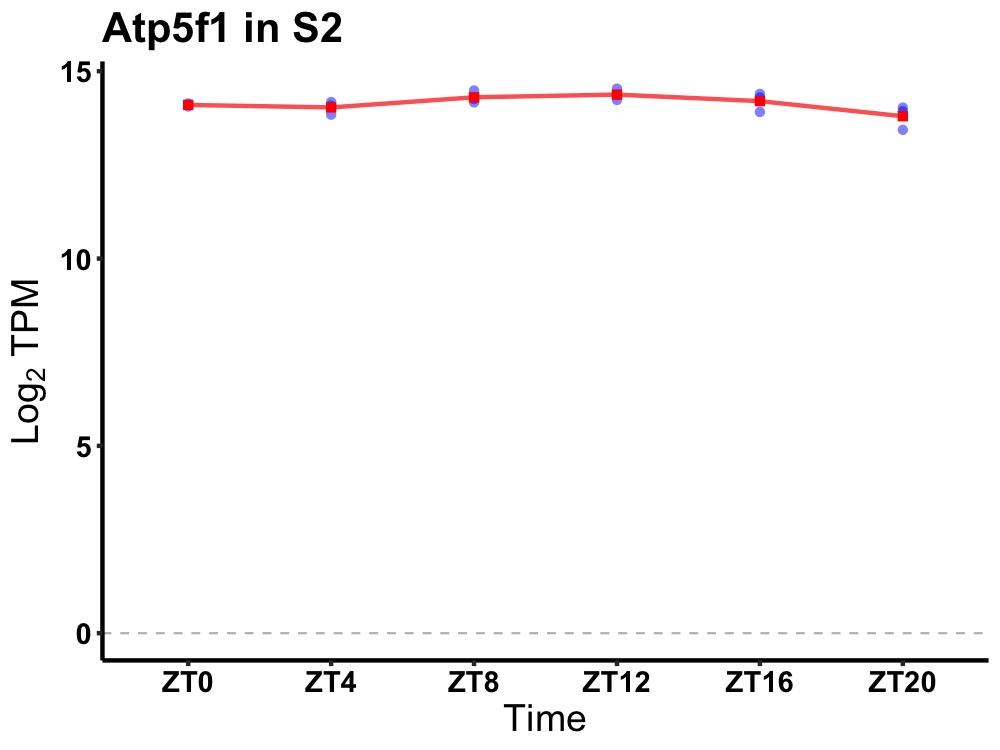 |
ATP synthase, H+ transporting, mitochondrial F0 complex, subunit B1 | 0.014 | 20 | 12 | 0.15 |
| ENSMUSG00000022016 | Ndufs2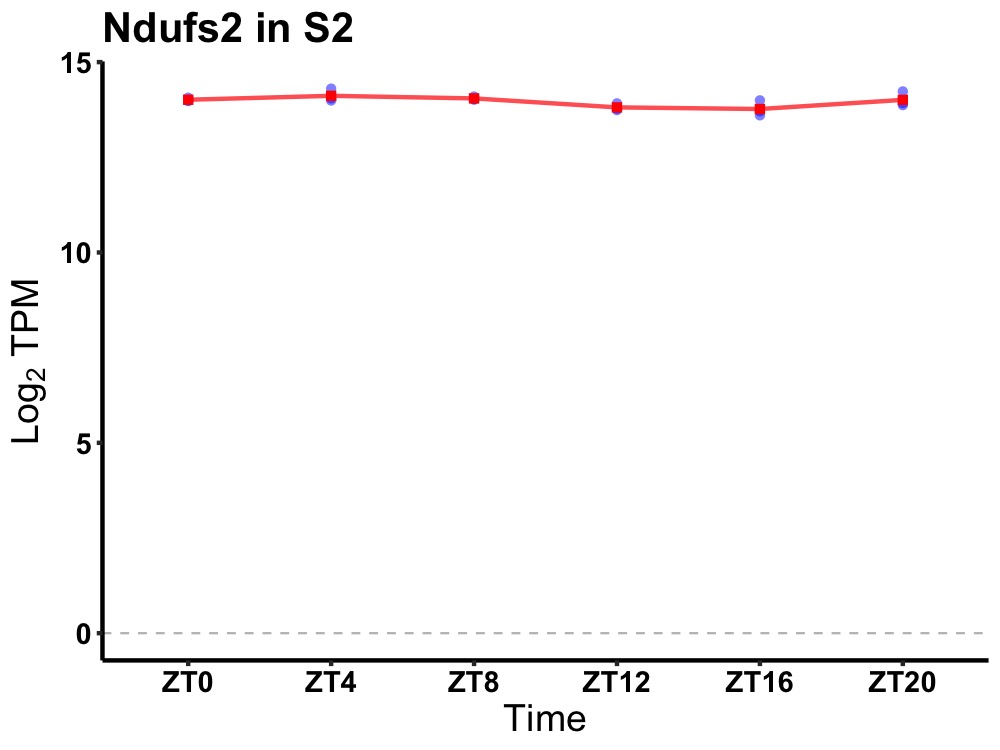 |
NADH:ubiquinone oxidoreductase core subunit S2 | 0.049 | 20 | 6 | 0.15 |
| ENSMUSG00000022370 | Gm1821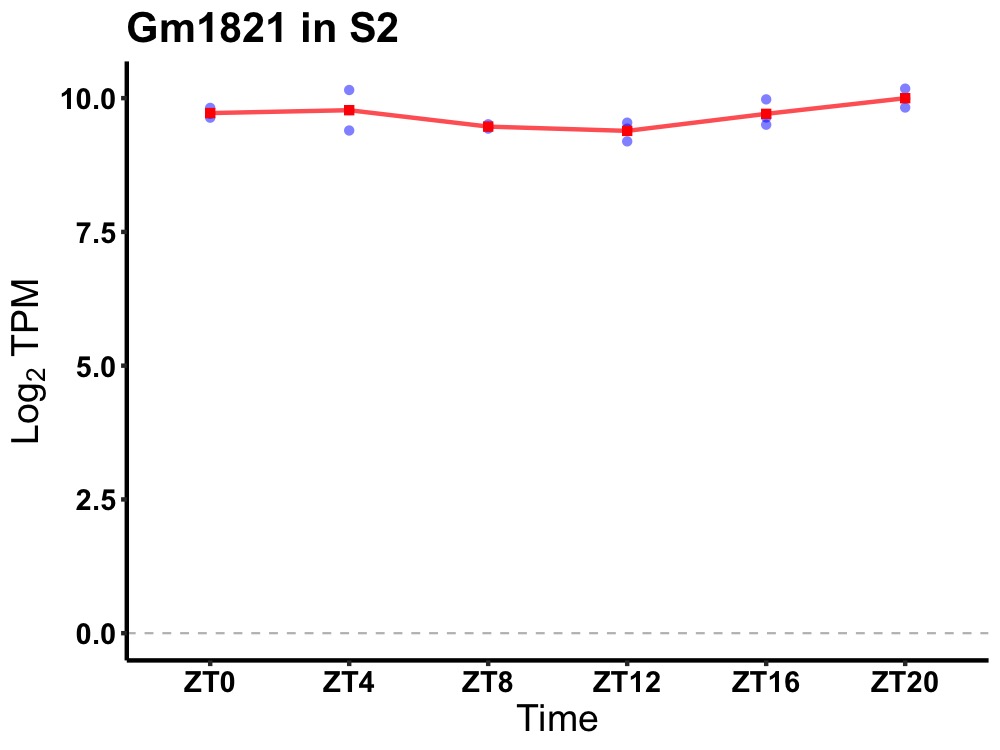 |
predicted gene 1821 | 0.019 | 20 | 2 | 0.15 |
| ENSMUSG00000037542 | Pank1 |
pantothenate kinase 1 | 0.049 | 20 | 12 | 0.14 |
| ENSMUSG00000090098 | Chchd2 |
coiled-coil-helix-coiled-coil-helix domain containing 2 | 0.036 | 24 | 10 | 0.14 |
| ENSMUSG00000025428 | Cox7a2 |
cytochrome c oxidase subunit 7A2 | 0.049 | 20 | 12 | 0.13 |
| ENSMUSG00000117815 | Cd63 |
CD63 antigen | 0.027 | 20 | 12 | 0.12 |
| ENSMUSG00000052456 | Cox7c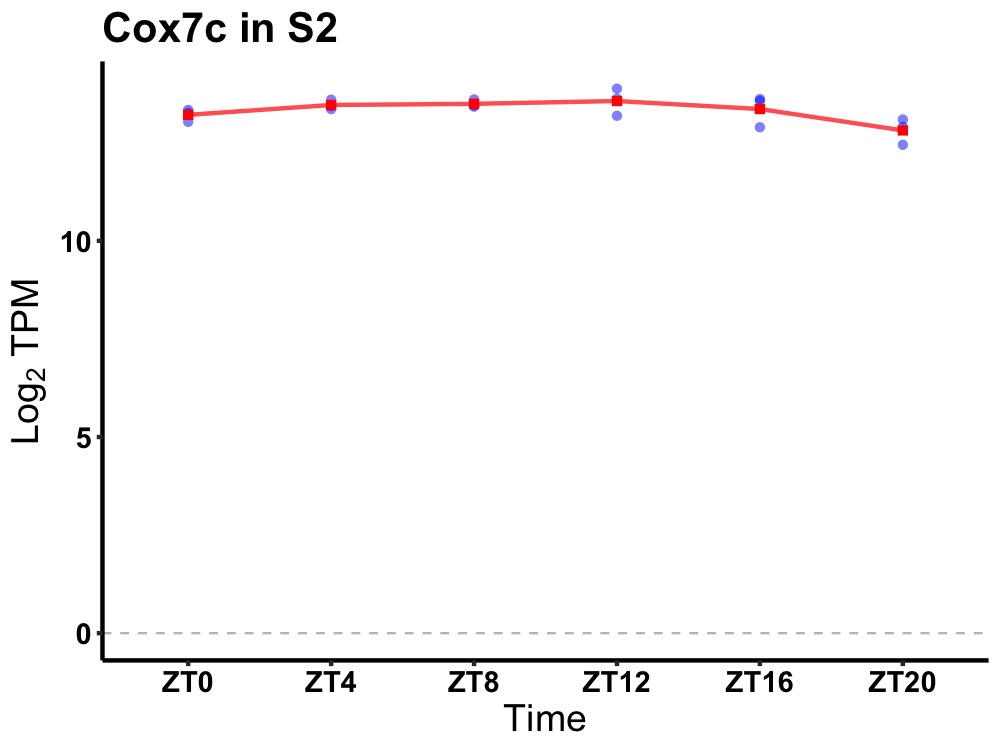 |
cytochrome c oxidase subunit 7C | 0.036 | 20 | 12 | 0.12 |
| ENSMUSG00000080772 | Rpl17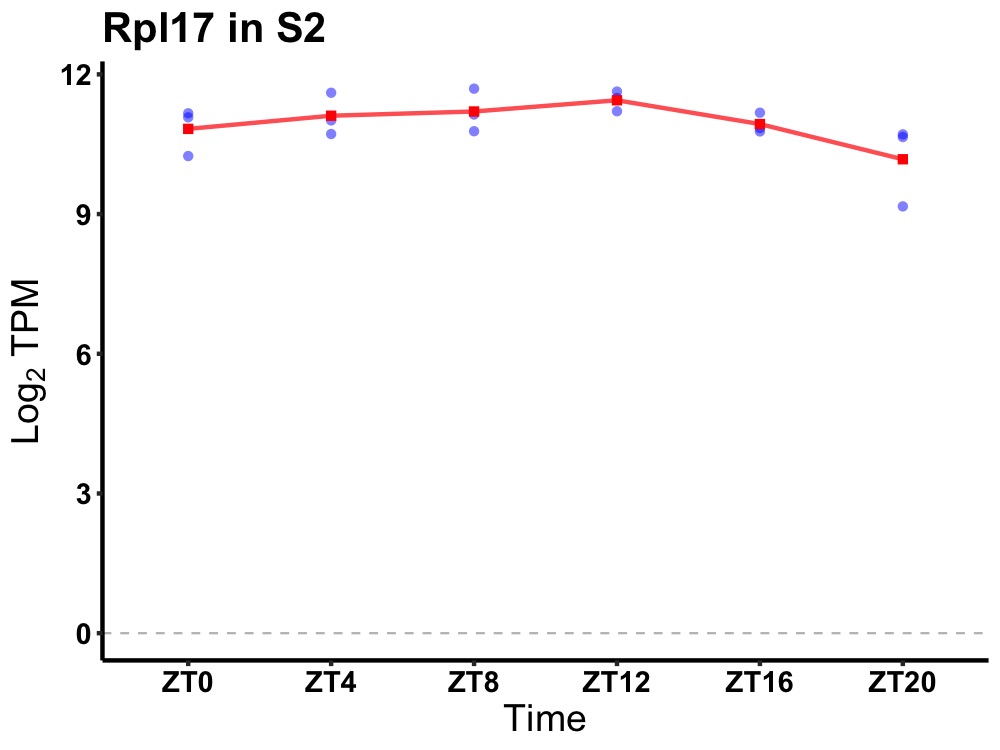 |
ribosomal protein L17 | 0.049 | 20 | 12 | 0.10 |
| ENSMUSG00000063362 | Cox6b1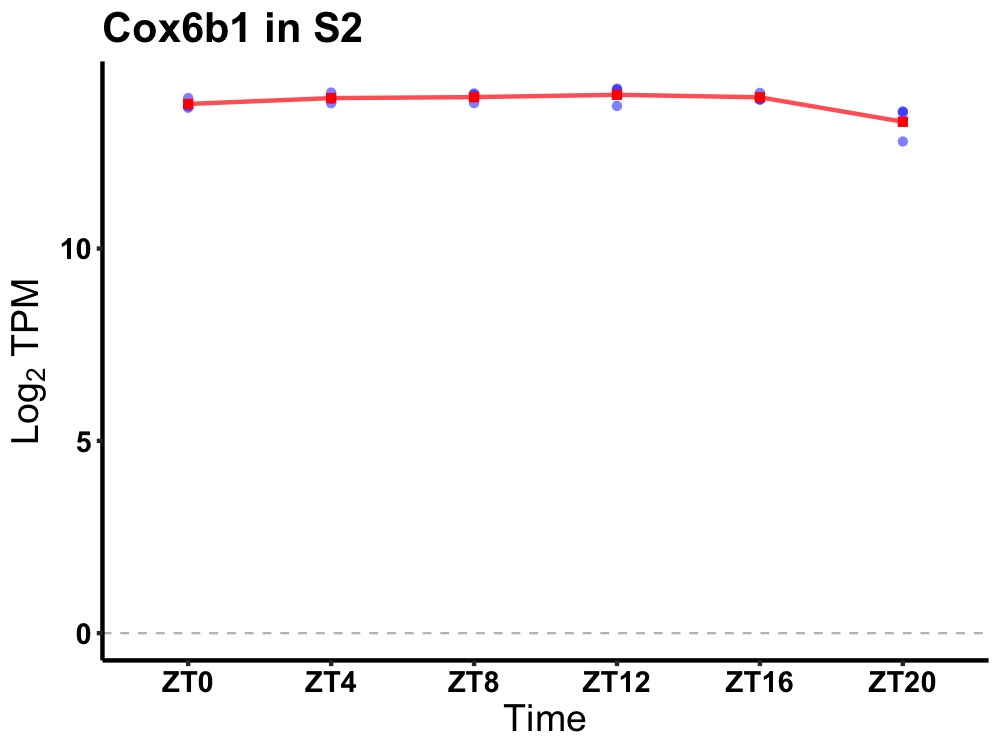 |
cytochrome c oxidase, subunit 6B1 | 0.049 | 20 | 12 | 0.08 |
| Ensembl ID | Gene Symbol | Annotation | PST | |||
| P-value | Period, hr | Phase, hr | Amplitude (Log2[TPM]) | |||
