KinasePredictor
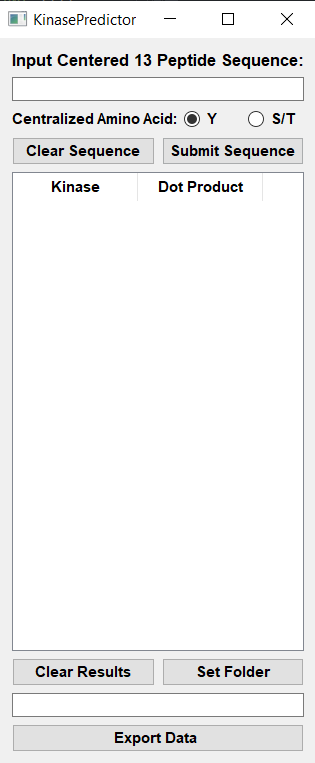
KinasePredictor takes a centralized phosphorylated amino acid sequence and generates a list of predictive scores for each protein kinase. KinasePredictor was created by Brian Poll, Kirby Leo, and Mark A. Knepper in the Epithelial Systems Biology Laboratory at the National Heart, Lung and Blood Institute as part of its Kidney Systems Biology Project. Please contact Mark Knepper with any comments or questions.
Current Version: 0.8
Click here to download KinasePredictor. See Readme file for instructions.
The full database of all kinases included in KinasePredictor can be viewed at this webpage: (Kinase Logos Database)
KinasePredictor data curated from Sugiyama et al. (PMID: 31324866)
The source code for KinasePredictor can be downloaded here: Download
This software is implemented using Python (Python 3.8.5)