Protein Kinases Phylogenetic Tree + Substrate Preference Logos
Below is a phylogenetic tree of human protein kinases and their associated substrate preference logos. Hover over each end node or label of the phylogenetic tree as shown to display the amino acid preferences flanking the post-translational modified site as identified by PTMLogo. See bottom of page for additional information.
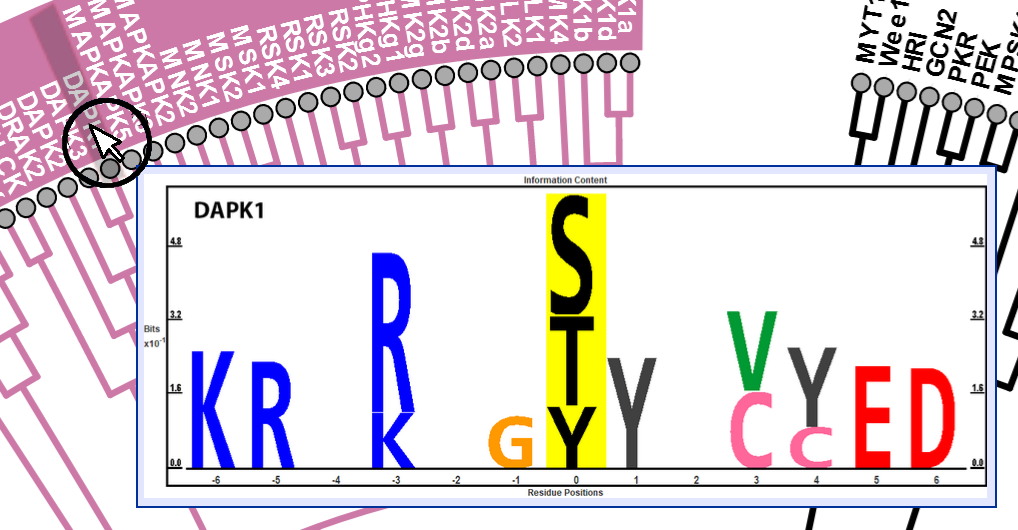
Kinases were organized based on sequence comparison of their catalytic domains as published in Manning et al. PMID: 31324866. Amino acid preferences flanking the post-translational modified site were identified by PTMLogo. All logos were generated from a minimum of 30 amino acid sequences, using Chi-squared filtering alpha = 0.0001. PTMLogo published by Saethang et al. 2019, PMID: 31318409. This phylogenetic tree was created by Brian Poll, Kirby Leo, and Mark A. Knepper in the Epithelial Systems Biology Laboratory at the National Heart, Lung and Blood Institute as part of its Kidney Systems Biology Project. Please contact Mark Knepper with any comments or questions.
This phylogenetic tree includes 351 protein kinases. A complete table of kinases, substrate motifs, and associated metadata can be found on our other webpage here: Kinase Sequence Motifs.
Click to download a complete pdf of all kinase motifs, including mutant and atypical kinases not shown here.
Kinase data used to create the substrate preference motifs was curated from the original publication (PMID: 31324866). These sequences and can be downloaded in an excel file here. Phylogenetic tree generated using Interactive Tree of Life (iTOL) doi: 10.1093/nar/gkab301.